Back to Journals » Cancer Management and Research » Volume 12
Long Non-Coding RNA KCNQ1OT1 Promotes Multidrug Resistance in Chordoma by Functioning as a Molecular Sponge of miR-27b-3p and Subsequently Increasing ATF2 Expression
Authors Li L, Lv G, Wang B, Ma H
Received 20 February 2020
Accepted for publication 13 April 2020
Published 25 August 2020 Volume 2020:12 Pages 7847—7853
DOI https://doi.org/10.2147/CMAR.S250611
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Harikrishna Nakshatri
Lei Li, Guohua Lv, Bing Wang, Hong Ma
Department of Spine Surgery, The Second Xiangya Hospital, Central South University, Changsha, Hunan 410011, People’s Republic of China
Correspondence: Hong Ma Tel +86 731-85295124
Email [email protected]
Background: Chordoma, a rare bone tumor, occurs most commonly at the sacrococcygeal and skull base region. To date, chemotherapy is used to treat patients with advanced-stage chordoma. However, multidrug resistance (MDR) greatly hinders the effect of chemotherapy in chordoma. Here, we studied the correlation between KCNQ1OT1 and chemotherapy resistance.
Methods: RT-PCR assay was used to examine KCNQ1OT1, miR-27b-3p, and ATF2 mRNA expression. CCK8 assay was exercised to detect IC50 values of cisplatin in chordoma cells. ATF2 protein expression was detected by Western blot.
Results: KCNQ1OT1 was increased in chemotherapy-resistant patients and cisplatin-resistant cells, and downregulation of KCNQ1OT1 expression weakened MDR in chordoma. In addition, KCNQ1OT1 promoted MDR in chordoma by sponging miR-27b-3p and subsequently increasing ATF2 expression.
Conclusion: KCNQ1OT1 is proved to be strikingly raised in the chemotherapy-resistant group and to promote MDR in chordoma. Our findings demonstrated the role of the KCNQ1OT1/miR-27b-3p/ATF2 axis in MDR of chordoma, which provides new insight into the molecular mechanism of chordoma MDR, and may determine the effect of therapy after receiving chemotherapy by detecting the expression of KCNQ1OT1 in serum.
Keywords: KCNQ1OT1, MDR, miR-27b-3p, ATF2, chordoma
Introduction
Chordoma, a rare bone tumor, occurs most commonly at the sacrococcygeal and skull base region.1,2 The characteristics of chordoma are highly recurrent, aggressive, and locally invasive. To date, surgery is the most effective therapy approach in early-stage chordoma, but not including the advanced-stage disease at the first diagnosis. In contrast, chemotherapy is used as therapy in patients with advanced-stage chordoma. A recent study showed that chemotherapy has made new progress in treating chordoma.3,4 However, multidrug resistance (MDR) greatly hinders the effect of chemotherapy in chordoma.5 Therefore, it is very important to explore the molecular mechanisms of MDR to establish new treatment strategies.
Long non-coding RNA (lncRNA), transcripts with a length exceeding 200 nucleotides, are unable to encode protein.6 Recently, many studies pointed out lncRNA is associated with tumorigenesis and metastasis. For example, lncRNA LINC-PINT inhibits tumor cell invasion through a highly conserved sequence element.7 LncRNA XIST regulates the miR-137-EZH2 axis to promote tumor metastasis in colorectal cancer.8 Upregulation of lncRNA plncRNA-1 promotes metastasis and induces epithelial–mesenchymal transition in hepatocellular carcinoma.9 Additionally, lncRNA promotes or inhibits tumorigenesis and metastasis by functioning as a molecular sponge. For example, lncRNA-AC130710 targeting by miR-129-5p is upregulated in gastric cancer and associates with poor prognosis.10 LncRNA TUG1 sponges miR-145 to promote cancer progression and regulate glutamine metabolism via the Sirt3/GDH axis.11 LncRNA GAS5 inhibits the proliferation and progression of prostate cancer by targeting miR-103 through the AKT/mTOR signaling pathway.12 Notably, lncRNAs are rarely studied in chordoma, especially with regard to MDR. Therefore, it is of great significance to study the function of lncRNA in MDR of chordoma.
LncRNA RNA KCNQ1OT1 has been identified as a key modulator in lung adenocarcinoma,13 hepatocellular carcinoma,14 glioma,15 and colorectal cancer.16 Nevertheless, the expression and functions of KCNQ1OT1 in MDR of chordoma remain poorly known. Thus, we researched the expression and regulation of KCNQ1OT1 in MDR of chordoma. Moreover, we firstly find that KCNQ1OT1 promotes multidrug resistance in chordoma by functioning as a molecular sponge of miR-27b-3p and consequently increasing ATF2 expression.
Materials and Methods
Serum Sample of Chordoma Patients
Serum samples were collected from chordoma patients between August 2016 and October 2019. The study was approved by the Institutional Research Ethics Committee of the Second Xiangya Hospital with Central South University. Consent was gained from all subjects in the written form and all methods were performed in accordance with the HIPAA guidelines and Declaration of Helsinki.
Cell Culture
The chordoma cell lines JHC7 and U-CH1 were provided by the Institute of Biochemistry and Cell Biology, Shanghai Institute for Biological Sciences, Chinese Academy of Sciences (Shanghai, China). JHC7 cells were cultured in DMEM medium containing 100 U/mL of streptomycin/penicillin and 10% fetal bovine serum (FBS, Gibco, USA). All cells were subjected to incubation in humidified 5% CO2 at 37°C.
Drug Sensitivity Assay
The half maximal inhibitory concentration (IC50) was detected by performing a CCK8 assay following the manufacturers’ instructions. Briefly, after 48 h, CCK8 (10 μL/well, Dojindo, Japan) was added into each well and incubated for 2 h. Then, the solution was detected spectrophotometrically at 450 nm.
Quantitative Real-Time PCR
Trizol reagent (Invitrogen Life Technologies, Carlsbad, CA, USA) was used to harvest the total RNA. Then, the total RNA was subjected to synthesis by using Transcriptor First Strand cDNA Synthesis Kit (Roche Life Science, Shanghai, China). The qPCR was used with SYBR Premix Ex Taq kit (TaKaRa, China). The primers are shown below: KCNQ1OT1, forward 5ʹ-TGCAGAAGACAGGACACTGG-3ʹ, reverse 5ʹ-CTTTGGTGGGAAAGGACAGA-3ʹ; GAPDH, forward 5ʹ-TGCACCACCAACTGCTTAGC-3ʹ, reverse 5ʹ-GGCATGCACTGTGGTCATGAG-3ʹ.
Western Blot Analysis
Western blot analysis was used according to standard protocols. Briefly, total protein in cells was collected by using RIPA lysis buffer. The SDS-PAGE was used to separate the proteins and was transferred to the nitrocellulose membrane. The primary antibody ATF2 and GAPDH were from Santa Cruze, USA. The second antibody was obtained from Cell Signaling, Danvers, MA, USA.
Statistical Analysis
All data are shown as mean ± standard deviation. The Student’s t-test was implemented for statistical analysis by Graphpad Prism 7.0. P<0.05 were considered to be statistically significant.
Results
KCNQ1OT1 Was Increased in Chemotherapy Resistant Patients and Cisplatin-Resistant Cell
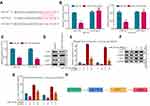
To explore the relationship between KCNQ1OT1 and MDR in chordoma, we detected the expression of KCNQ1OT1 in chordoma patients’ serum after four cycles of cisplatin combined with methotrexate chemotherapy. The result showed the expression of KCNQ1OT1 in the chemotherapy-resistant group (SD+PD, n=43) was increased compared to the chemotherapy-sensitive group (CR+PR, n=40) according to the Response Evaluation Criteria in Solid Tumors (RECIST) (Figure 1A). Notably, there was no difference between the chemotherapy-resistant group and the chemotherapy-resistant group before treatment. In addition, Kaplan–Meier survival analysis showed the survival of the chemotherapy-resistant group was lower than the chemotherapy-sensitive group (Figure 1B). The RT-PCR also showed that KCNQ1OT1 expression in cisplatin-resistant chordoma cell JHC7 and U-CH1 cell (JHC7/R and U-CH1/R cell) was increased compared to JHC7 and U-CH1 cell (Figure 1C). Thus, these results suggest KCNQ1OT1 is related to chordoma MDR.
Downregulation of KCNQ1OT1 Expression Weakens MDR in Chordoma
To ascertain the function of KCNQ1OT1 in chordoma MDR, chordoma cells were treated with si-KCNQ1OT1 (JHC7/si-KCNQ1OT1, JHC7/R/si-KCNQ1OT1, U-CH1/si-KCNQ1OT1, and U-CH1/R/si-KCNQ1OT1) or si-NC (JHC7/si-NC, JHC7/R/si-NC, U-CH1/si-NC, and U-CH1/R/si-NC), and RT-PCR was performed to detect KCNQ1OT1 expression (Figure 2A). Afterward, the CCK8 assay was used to examine the IC50 values of cisplatin in chordoma cells. The results showed IC50 values of cisplatin in JHC7/si-KCNQ1OT1, JHC7/R/si-KCNQ1OT1, U-CH1/si-KCNQ1OT1, and U-CH1/R/si-KCNQ1OT1 cells were less than JHC7/si-NC, JHC7/R/si-NC, U-CH1/si-NC, and U-CH1/R/si-NC, respectively (Figure 2B). In addition, JHC7/R and U-CH1/R cells with stable KCNQ1OT1 downregulation were established (Figure 2C). The result showed that the KCNQ1OT1 downregulation group with cisplatin treatment remarkably inhibited the growth curve of the tumor compared to the control group (Figure 2D) and tumor weight in the KCNQ1OT1 downregulation group with cisplatin treatment was conspicuously lighter compared to in the control group (Figure 2E). It can be suggested that downregulation of KCNQ1OT1 expression weakens MDR of chordoma in vitro and vivo.
KCNQ1OT1 Sponged miR-27b-3p in a Competitive Manner to Promote MDR
To detect whether lncRNA directly interacts with miRNA to modulate the expression of miRNA targets in MDR, the expression distribution of KCNQ1OT1 in JCH7/R and U-CH1/R cell was firstly detected. The results showed KCNQ1OT1 was mostly located in the cytoplasm of JCH7/R and U-CH1/R cell (Figure 3A). Potential binding sites between miR-27b-3p and KCNQ1OT1 were predicted by the DIANA tool, and luciferase reporter analysis was performed to verify their complementary combinations (Figure 3B and C). The RT-PCR showed the expression of miR-27b-3p in JCH7/R and U-CH1/R cell was decreased compared to JCH7 and U-CH1 cell (Figure 3D) and the expression of miR-27b-3p in JHC7/R/si-KCNQ1OT and U-CH1/R/si-KCNQ1OT was increased compared to JCH7/R and U-CH1/R cell (Figure 3E). RIP assay pointed out the direct binding between miR-27b-3p and KCNQ1OT1 (Figure 3F). In addition, the expression of miR-27b-3p in the chemotherapy-resistant group was decreased compared to the chemotherapy-sensitive group (Figure 3G) and the KCNQ1OT1 expression was inversely correlated with miR-27b-3p in the chemotherapy-resistant group by Pearson correlation analysis (Figure 3H). In addition, JCH7/R and U-CH1/R cell were transfected by the miR-27b-3p mimics, and RT-PCR demonstrated the miR-219a-5p expression was increased in miR-27b-3p mimics compared to miR-NC (Figure 3I). IC50 values of cisplatin in JHC7/R/miR-27b-3p mimics and U-CH1/R/miR-27b-3p mimic cells were less than JHC7/R/miR-NC and U-CH1/R/miR-NC, respectively (Figure 3J). Thus, KCNQ1OT1 sponged miR-27b-3p in a competitive manner to promote MDR.
KCNQ1OT1 Promotes MDR in Chordoma by Sponging miR-27b-3p and Subsequently Increasing ATF2 Expression
To identify potential targets for miR-27b-3p, we used TargetScan, miRanda, and PicTar to predict target protein and ATF2 was considered to be a target gene of miR-27b-3p, based on a putative target sequence of ATF2 3ʹ UTR (Figure 4A). In addition, fluorescence analysis indicated that increased expression of miR-27b-3p significantly enhanced the luciferase activity of wild type ATF2 3ʹ UTR (Figure 4B), but not in mutant type ATF2 3ʹ UTR, suggesting that ATF2 is a direct target of miR-27b-3p in chordoma cells. We further used RT-PCR and Western blot assays to detect mRNA and protein of ATF2 after increasing miR-27b-3p expression. The results showed that increased expression of miR-27b-3p obviously decreased ATF2 expression on mRNA levels (Figure 4C) and protein levels (Figure 4D) in JHC7/R and U-CH1/R cell. In addition, JHC7/R/si-KCNQ1OT1 and U-CH1/R/si-KCNQ1OT1 cell were transfected with the miR-27b-3p inhibitor and si-ATF2 or si-NC, and RT-PCR and Western blot demonstrated the ATF2 expression with transfected miR-27b-3p inhibitor was increased compared to transfected miR-NC and ATF2 expression with transfected si-ATF2 was decreased compared to transfected si-NC in mRNA level (Figure 4E) and protein level (Figure 4F). Transfection of si-ATF2 can reduce IC50 values of cisplatin, and transfected miR-27b-3p inhibitor can increase IC50 values of cisplatin in transfected si-ATF2 (Figure 4G). Therefore, KCNQ1OT1 promotes MDR in chordoma by sponging miR-27b-3p and consequently increasing ATF2 expression (Figure 4H).
Discussion
Recently, several studies have reported that lncRNA plays a significant role in progression, metastasis, and MDR in tumor. For example, overexpression of lncRNA DANCR is associated with advanced tumor progression and poor prognosis in patients with colorectal cancer.17 Knockdown of lncRNA ANRIL suppresses cell proliferation, metastasis, and invasion via regulating miR-122-5p expression in hepatocellular carcinoma.18 LncRNA UCA1 enhances cell proliferation and 5-fluorouracil resistance in colorectal cancer by inhibiting miR-204-5p.19 To date, the relationship between lncRNA and tumors has rarely been studied. To our knowledge, there are only two papers on the role of lncRNA in chordoma.20,21 In this study, we firstly reported KCNQ1OT1 expression was increased in the chemotherapy-resistant group and cisplatin-resistant chordoma cell and downregulation of KCNQ1OT1 weakened MDR of chordoma in vitro and vivo. In addition, KCNQ1OT1 sponged miR-27b-3p in a competitive manner to promote MRD in chordoma.
MDR still greatly hinders the effect of chemotherapy in chordoma and the regulatory mechanism of MDR has been unclear. Therefore, in this study, we researched the molecular mechanisms of MDR. Early studies pointed out that overexpression of miR-27b-3p weakens MDR in multiple types of tumors. For example, the downregulation of miR-27b-3p enhances tamoxifen resistance in breast cancer by increasing NR5A2 and CREB1 expression.22 Notably, research indicated suppressing ATF2 expression decreases the cisplatin-resistance in tumor.23 In this study, we found upregulation of miR-27b-3p weakens MDR in chordoma and ATF2 is a direct target of miR-27b-3p.
In conclusion, KCNQ1OT1 is proved to be strikingly raised in the chemotherapy-resistant group and promote MDR in chordoma. In addition, KCNQ1OT1 promotes multidrug resistance in chordoma by functioning as a molecular sponge of miR-27b-3p and subsequently increasing ATF2 expression. Therefore, our findings demonstrated the function of the KCNQ1OT1/miR-27b-3p/ATF2 axis in MDR of chordoma, which provides new insight into the molecular mechanism of chordoma MDR, and may determine the effect of therapy after receiving chemotherapy by detecting the expression of KCNQ1OT1 in serum.
Funding
Supported by the Natural Science Foundation of Hunan Province, China (Grant No. 2019JJ50888).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Gulluoglu S, Turksoy O, Kuskucu A, Ture U, Bayrak OF. The molecular aspects of chordoma. Neurosurg Rev. 2016;39(2):
2. Patel P, Brooks C, Seneviratne A, Hess DA, Seguin CA. Investigating microenvironmental regulation of human chordoma cell behaviour. PLoS One. 2014;9(12):e115909. doi:10.1371/journal.pone.0115909
3. Chen H, Garbutt CC, Spentzos D, Choy E, Hornicek FJ, Duan Z. Expression and therapeutic potential of SOX9 in chordoma. Clin Cancer Res. 2017;23(17):5176–5186. doi:10.1158/1078-0432.CCR-17-0177
4. Cao X, Lu Y, Liu Y, et al. Combination of PARP inhibitor and temozolomide to suppress chordoma progression. J Mol Med (Berl). 2019;97(8):1183–1193. doi:10.1007/s00109-019-01802-z
5. Gagliardi F, Boari N, Riva P, Mortini P. Current therapeutic options and novel molecular markers in skull base chordomas. Neurosurg Rev. 2012;35(1):
6. Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10(3):155–159. doi:10.1038/nrg2521
7. Marin-Bejar O, Mas AM, Gonzalez J, et al. The human lncRNA LINC-PINT inhibits tumor cell invasion through a highly conserved sequence element. Genome Biol. 2017;18(1):202. doi:10.1186/s13059-017-1331-y
8. Liu X, Cui L, Hua D. Long noncoding RNA XIST regulates miR-137-EZH2 axis to promote tumor metastasis in colorectal cancer. Oncol Res. 2018;27(1):99–106. doi:10.3727/096504018X15195193936573
9. Dong L, Ni J, Hu W, Yu C, Li H. Upregulation of long non-coding RNA PlncRNA-1 promotes metastasis and induces epithelial-mesenchymal transition in hepatocellular carcinoma. Cell Physiol Biochem. 2016;38(2):836–846. doi:10.1159/000443038
10. Xu C, Shao Y, Xia T, et al. lncRNA-AC130710 targeting by miR-129-5p is upregulated in gastric cancer and associates with poor prognosis. Tumour Biol. 2014;35(10):9701–9706. doi:10.1007/s13277-014-2274-5
11. Zeng B, Ye H, Chen J, et al. LncRNA TUG1 sponges miR-145 to promote cancer progression and regulate glutamine metabolism via Sirt3/GDH axis. Oncotarget. 2017;8(69):113650–113661. doi:10.18632/oncotarget.21922
12. Xue D, Zhou C, Lu H, Xu R, Xu X, He X. LncRNA GAS5 inhibits proliferation and progression of prostate cancer by targeting miR-103 through AKT/mTOR signaling pathway. Tumour Biol. 2016. doi:10.1007/s13277-016-5429-8
13. Ren K, Xu R, Huang J, Zhao J, Shi W. Knockdown of long non-coding RNA KCNQ1OT1 depressed chemoresistance to paclitaxel in lung adenocarcinoma. Cancer Chemother Pharmacol. 2017;80(2):243–250. doi:10.1007/s00280-017-3356-z
14. Hu H, Yang L, Li L, Zeng C. Long non-coding RNA KCNQ1OT1 modulates oxaliplatin resistance in hepatocellular carcinoma through miR-7-5p/ABCC1 axis. Biochem Biophys Res Commun. 2018;503(4):2400–2406. doi:10.1016/j.bbrc.2018.06.168
15. Gong W, Zheng J, Liu X, et al. Knockdown of long non-coding RNA KCNQ1OT1 restrained glioma cells’ malignancy by activating miR-370/CCNE2 axis. Front Cell Neurosci. 2017;11:84. doi:10.3389/fncel.2017.00084
16. Bian Y, Gao G, Zhang Q, et al. KCNQ1OT1/miR-217/ZEB1 feedback loop facilitates cell migration and epithelial-mesenchymal transition in colorectal cancer. Cancer Biol Ther. 2019;20(6):886–896. doi:10.1080/15384047.2019.1579959
17. Liu Y, Zhang M, Liang L, Li J, Chen YX. Over-expression of lncRNA DANCR is associated with advanced tumor progression and poor prognosis in patients with colorectal cancer. Int J Clin Exp Pathol. 2015;8(9):11480–11484.
18. Ma J, Li T, Han X, Yuan H. Knockdown of LncRNA ANRIL suppresses cell proliferation, metastasis, and invasion via regulating miR-122-5p expression in hepatocellular carcinoma. J Cancer Res Clin Oncol. 2018;144(2):205–214. doi:10.1007/s00432-017-2543-y
19. Bian Z, Jin L, Zhang J, et al. LncRNA-UCA1 enhances cell proliferation and 5-fluorouracil resistance in colorectal cancer by inhibiting miR-204-5p. Sci Rep. 2016;6:23892. doi:10.1038/srep23892
20. Ma X, Qi S, Duan Z, et al. Long non-coding RNA LOC554202 modulates chordoma cell proliferation and invasion by recruiting EZH2 and regulating miR-31 expression. Cell Prolif. 2017;50(6):e12388. doi:10.1111/cpr.12388
21. Chen H, Zhang K, Lu J, Wu G, Yang H, Chen K. Comprehensive analysis of mRNA-lncRNA co-expression profile revealing crucial role of imprinted gene cluster DLK1-MEG3 in chordoma. Oncotarget. 2017;8(68):112623–112635. doi:10.18632/oncotarget.22616
22. Zhu J, Zou Z, Nie P, et al. Downregulation of microRNA-27b-3p enhances tamoxifen resistance in breast cancer by increasing NR5A2 and CREB1 expression. Cell Death Dis. 2016;7(11):e2454. doi:10.1038/cddis.2016.361
23. Tian L, Zhang J, Ren X, et al. Overexpression of miR-26b decreases the cisplatin-resistance in laryngeal cancer by targeting ATF2. Oncotarget. 2017;8(45):79023–79033. doi:10.18632/oncotarget.20784
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.