Back to Journals » OncoTargets and Therapy » Volume 13
Long Non-Coding RNA GATA6-AS1 Sponges miR-324-5p to Inhibit Lung Cancer Cell Proliferation and Invasion
Authors Wang Z, Pan L, Yang L, Lv P, Mai S, Wang Y
Received 2 April 2020
Accepted for publication 4 July 2020
Published 30 September 2020 Volume 2020:13 Pages 9741—9751
DOI https://doi.org/10.2147/OTT.S256336
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Prof. Dr. Takuya Aoki
Zhenxing Wang,1 Liming Pan,2 Liangliang Yang,1 Peiyun Lv,1 Shixiong Mai,1 Yue Wang1
1Department of Thoracic Surgery, China-Japan Union Hospital of Jilin University, Changchun, Jilin, People’s Republic of China; 2Department of Hand Surgery, The First Bethune Hospital of Jilin University, Changchun, Jilin, People’s Republic of China
Correspondence: Yue Wang
Department of Thoracic Surgery, China-Japan Union Hospital of Jilin University, Changchun, Jilin, People’s Republic of China
Email [email protected]
Background: Long non-coding RNAs (lncRNAs), which are key regulators of gene expression, are involved in lung cancer progression. Although numerous differentially expressed lncRNAs have been reported, merely a limited number of studies have been performed to verify their functions in lung cancer.
Methods: RNA sequencing data were re-analyzed to investigate the GATA6-AS1 expression in lung cancer. RT-qPCR was performed to verify the expression of GATA6-AS1 in collected tissue samples and cell lines. CCK-8 and transwell assays were carried out to evaluate the role of GATA6-AS1 in lung cancer cells. Dual-luciferase reporter assay and bioinformatic analysis were used to explore the miRNA which can be sponged by GATA6-AS1 in lung cancer cells.
Results: Currently, we focused on exploring the role and mechanisms of GATA6-AS1 in lung cancer. Expression of GATA6-AS1 was decreased in lung cancer based on the analysis of RNA sequencing dataset, TCGA data and RT-qPCR of clinical tissue samples. Via overexpression of GATA6-AS1, it was revealed that GATA6-AS1 inhibited lung cancer cell proliferation and invasion. Oncogene miR-324-5p was predicted to interact with GATA6-AS1. RT-qPCR and dual-luciferase reporter assay verified the regulation of miR-324-5p by GATG6-AS1 in lung cancer cells. Overexpression of GATA6-AS1 increased the expression of FBXO11 and SP1, two target genes of miR-324-5p. We further showed that miR-324-5p mimic reversed the effect of GATA6-AS1 overexpression in lung cancer cells.
Conclusion: Overall, our findings demonstrated GATA6-AS1 as a novel tumor suppressor in lung cancer.
Keywords: GATA6-AS1, miR-324-5p, lung cancer
Background
Lung cancer is the most common cancer type globally, representing approximately 11.6% cancer cases in 2018.1 Although there are numerous studies on the biological mechanism of lung cancer, the mortality of patients with lung cancer has remained high in recent years, and the five-year overall survival (OS) of patients with lung cancer is around 10%.2 The major obstacle to treat lung cancer is the metastasis of lung cancer cells, which is closely related to patient deaths.3 Therefore, the mechanism of lung cancer needs to be thoroughly investigated.
Long non-coding RNAs (lncRNAs) are molecules with more than 200 nucleotides.4 Previously known as “genomic junk”,5 recent data suggest that lncRNAs are involved in regulation of gene expression via multiple ways.6,7 According to competing endogenous RNA (ceRNA) theory, lncRNA can interact with microRNA (miRNA) and releases mRNA from the binding of miRNA.8,9 Via modulating key genes in signaling network, lncRNAs are implicated in almost all biological processes such as cell differentiation, metastasis and proliferation.10,11 Thus, dysregulation of lncRNAs contributes to sustained cell proliferation and strong invasive capability of cells during carcinogenesis.12–14 Currently, many lncRNAs have been experimentally verified as oncogenes or tumor suppressors in lung cancer.15,16 LncRNA GATA6-AS1 is a recently discovered cancer-related lncRNA.17,18 However, its function in lung cancer remains unknown.
Many miRNAs are pivotal for lung cancer development.19,20 For example, miR-324-5p, which is a recently defined oncogenic miRNA in lung cancer,21,22 is frequently upregulated in lung cancer and facilitates lung cancer cell proliferation and invasion.21 In addition, miR-324-5p also targets FBXO11, a regulator of epithelial–mesenchymal transition (EMT), to promote development of drug resistance for lung cancer,22 whereasthe mechanism underlying the aberrant expression of miR-324-5p in lung cancer remains elusive.
The current study aimed to study the function and mechanisms of GATA6-AS1 in lung cancer. We found that GATA6-AS1 was a significantly downregulated lncRNA in lung cancer. GATA6-AS1 sponged oncogene miR-324-5p and inhibited cell proliferation and invasion in lung cancer cells.
Patients and Methods
Patient Samples
Lung cancer tumor tissues and the adjacent normal tissues were collected during surgery from 100 patients with lung cancer. Written informed consents were obtained from all patients. The protocol of the experiments was approved by the Medical Ethics Committee of the Third Hospital of Jilin University (Approval number: JLU201508-04). Samples were stored at −80°C before RNA extraction and RT-qPCR.
Cell Culture
Normal human bronchial epithelial cell line (16HBE) was obtained from the Institute of Biochemistry and Cell Biology of the Chinese Academy of Sciences (Shanghai, China). Lung cancer cell lines NCI-H1975, A549, NCI-H460 and NCI-H1299 were purchased from American Type Culture Collection (Manassas, VA). These cells were cultured in DMEM (Thermo Fisher Scientific) supplemented with 10% FBS (Hyclone; Thermo Fisher Scientific) in a humid incubator with 5% CO2 at 37°C.
Bioinformatic Analysis
The expression pattern of GATA6-AS1 in multiple cancer types was analyzed by the GEPIA software (http://gepia.cancer-pku.cn/) with datasets from TCGA project (normal and tumor tissues) and GTEx project (normal tissues). The correlation between GATA6-AS1 expression and OS of patients with lung cancer (n=1144, including 672 lung adenocarcinoma (LUAD) and 271 lung squamous cell carcinoma (LUSC)) was analyzed by Kaplan–Meier Plotter software (https://kmplot.com/analysis/index.php?p=serviceandcancer=lung). The patients were split by median expression of GATA6-AS1 (572 cases in high expression group and 572 cases in low expression group). The potential miRNAs that may interact with GATA6-AS1 were predicted by miRDB software (http://mirdb.org/).
RT-qPCR
Trizol (Invitrogen) was used to extract RNA from cells and tissues. The RNA was reverse-transcribed into cDNA with PrimeScriptTM RT Kit (TaKaRa, Tokyo, Japan). RT-qPCR reaction was carried out with 2× EsayTaq PCR SuperMix (TransGen Biotech, Beijing, China). The products were run on 2% agarose gels and stained with ethidium bromide. LncRNA/mRNA was normalized to GAPDH and miRNA was normalized to U6 with the 2−ΔΔCt method.23
Overexpression of GATA6-AS1
Human GATA6-AS1 sequence was obtained by PCR method from A549 cDNA and inserted into pcDNA3 plasmid. The constructs were transfected into cells with Lipofectamine 3000 reagent (Invitrogen). The cells were cultured for another 48 hours and then subjected to RNA extraction and RT-qPCR to confirm the transfection efficiency.
Overexpression and Downregulation of miR-324-5p
miR-324-5p mimic, miR-324-5p inhibitor, and miR-NC were obtained from GenePharma (Suzhou, China), and transfected into cells with Lipofectamine 3000 reagent (Invitrogen). The cells were cultured for another 48 hours and then subjected to RNA extraction and RT-qPCR to confirm the transfection efficiency.
Western Blotting
FBXO11 (ab181801, 1:1000), SNAIL (ab229701, 1:2000), and GAPDH (ab8245, 1:10,000) antibodies were purchased from Abcam (Cambridge, UK). HRP-conjugated antibodies to rabbit (ABL3012-2, 1:10,000) and mouse (ABL3031-2, 1:10,000) were products of AbSci (College Park, MD). Lysates were acquired from cells via RIPA lysis buffer (Thermo Fisher Scientific). Proteins were separated via an SDS-PAGE gel and transferred to the PVDF membrane. Membrane was blocked in 5% milk, primary antibody and secondary antibody sequentially. The blot was finally developed with the ECL Western blotting substrate (Pierce; Thermo Fisher Scientific). The quantification of bands was performed with Image J V.1.6.0 software (NIH).
Cell Proliferation and Cell Invasion Assays
For cell proliferation assay, cells were seeded in 96-well plates, transfected with indicated agent and maintained for indicated periods. The medium was mixed with CCK-8 solution (DoJinDo, Tokyo, Japan) and incubated for 2 hours. Absorbance at 450 nM was measured to reflect cell proliferation ability.
For cell invasion assay, treated cells were seeded in 12-well plates. The upper chamber was pre-coated with Matrigel (BD Corning) and filled with serum-free medium. In the lower chamber, the well was filled with medium containing 10% FBS. After 48 hours, the invaded cells were fixed with 4% PFA and stained with 0.1% (w/v) crystal violet. Three random fields of each group were observed and the cell number was counted to reflect cell invasive capacity.
Dual-Luciferase Reporter Assay
GATA6-AS1 was subcloned from pcDNA3 to pmiRGLO luciferase plasmid. FBXO11 3ʹUTR was obtained by PCR method from A549 cDNA and inserted into pmiRGLO. Site mutations were introduced into pmiRGLO-GATA6-AS1 and pmiRGLO-FBXO11 3ʹUTR with QuikChange Lightning Site-Directed Mutagenesis Kit (Agilent Technologies, Santa Clara, CA). Cells were seeded in 24-well plates, co-transfected with luciferase constructs and pcDNA3 with or without miR-324-5p mimic. After additional incubation for 48 hours, the luciferase activity of each well was determined with a Dual-Luciferase Reporter System (Promega, Madison, WI).
Statistical Analysis
The data were presented as mean ± SD by the calculation of Graphpad Prism 6. Student’s t test was applied to compare differences between two groups. Differences among three groups were analyzed with one-way ANOVA followed by Tukey’s test. Pearson correlation analysis was used to study the expression correlation. P less than 0.05 was statistically significant.
Results
LncRNA GATA6-AS1 Was Decreased in Lung Cancer
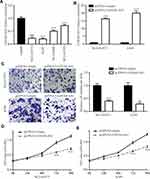
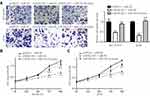
To investigate the role of GATA6-AS1 in lung cancer, we firstly used online tools to analyze the level of GATA6-AS1 in a panel of 33 cancer types from TCGA datasets and GTEx data. It was found that there was decreased expression of GATA6-AS1 in 11 out of 33 cancer types including LUAD and LUSC (Figure 1A). Moreover, upregulation of GATA6-AS1 was not seen in any types of cancer. We collected 100 pairs of lung cancer tumor tissues and the matched normal tumor samples. The RT-qPCR result suggested that GATA6-AS1 was decreased in most cases (80%) which was consistent with that of TCGA data (Figure 1B). Low expression of GATA6-AS1 was associated with metastasis of lung cancer, while GATA6-AS1 level was not associated with age, gender, smoking history, histologic type, T status or N status (Table 1). GATA6-AS1 was significantly lower in tumors of metastasis compared with those without metastasis (Figure 1C). Furthermore, to study the association between GATA6-AS1 expression and the prognosis of patients with lung cancer, we performed Kaplan–Meier plot analysis in patients with lung cancer. Interestingly, high expression of GATA5-AS1 was associated with prolonged OS in patients with lung cancer (Figure 1D). The data implied a pivotal role of GATA6-AS1 in lung cancer.
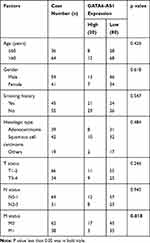 |
Table 1 Correlation Between GATA6-AS1 Expression and Clinical Pathological Factors of Patients |
Overexpression of GATA6-AS1 Affected Lung Cancer Cell Proliferation and Invasion
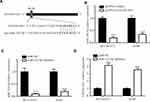
We next detected GATA6-AS1 level in lung cancer cells by the RT-qPCR method. The results showed that GATA6-AS1 was decreased in lung cancer cell lines NCI-H1975, A549, NCI-H460, and NCI-H1299 compared with normal human bronchial epithelial cell line 16-HBE (Figure 2A). Due to the low expression of GATA6-AS1, we selected NCI-H1975 and A549 for further studies. GATA6-AS1 sequence was inserted into pcDNA3 plasmid. Transfection of GATA6-AS1 significantly elevated GATA6-AS1 expression in NCI-H1975 and A549 cells (Figure 2B). With transwell assay, we found that GATA6-AS1 overexpression decreased invaded cell number of NCI-H1975 and A549 cells (Figure 2C), indicating the hampered invasion ability of lung cancer cells. Moreover, the CCK-8 assay showed that GATA6-AS1 overexpression also inhibited cell proliferation ability of NCI-H1975 and A549 cells (Figure 2DE).
GATA6-AS1 Regulated Oncogene miR-324-5p in Lung Cancer Cells
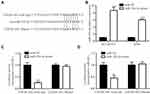
We used miRDB software to predict potential binding sites for miRNAs in GATA6-AS1 sequence. Among 41 binding sites, we noticed that there was a site for oncogene miR-324-5p in GATA6-AS1 (Figure 3A). In NCI-H1975 and A549 cells, GATA6-AS1 overexpression decreased miR-324-5p levels (Figure 3B). Furthermore, transfection of miR-324-5p inhibitor decreased miR-324-5p expression in NCI-H1975 and A549 cells (Figure 3C), and the downregulation of miR-324-5p elevated GATA6-AS1 levels in these cells (Figure 3D).
GATA6-AS1 Sponged miR-324-5p
We constructed luciferase vector containing GATA6-AS1-wild type or GATA6-AS1-Mutant (with point-mutation in the predicted binding site) (Figure 4A). miR-324-5p was increased in NCI-H1975 and A549 cells upon transfection of miR-324-5p mimic (Figure 4B). In the dual-luciferase reporter assay, it was observed that miR-324-5p mimic repressed luciferase value of GATA6-AS1-wild type without affecting the luciferase activity of GATA6-AS1-Mutant in NCI-H1975 cells (Figure 4C). Similar results were observed in A549 cells (Figure 4D).
GATA6-AS1 Promoted FBXO11 Expression
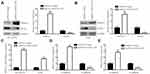
FBXO11, a key regulator of EMT, was a verified target of miR-324-5p in lung cancer cells.22 Consistent with the regulatory association between GATA6-AS1 and miR-324-5p, GATA6-AS1 overexpression elevated FBXO11 protein expression in NCI-H1975 cells (Figure 5A). As acknowledged, FBXO11 facilitated ubiquitin-mediated degradation of SNAIL protein.24 Consistently, at the protein level, GATA6-AS1 decreased SNAIL expression in NCI-H1975 cells (Figure 5A), and similar results were observed in A549 cells (Figure 5B). RT-qPCR manifested that GATA6-AS1 upregulated FBXO11 mRNA levels in NCI-H1975 and A549 cells (Figure 5C). Moreover, GATA6-AS1 increased E-cadherin mRNA levels and decreased N-cadherin mRNA levels in NCI-H1975 and A549 cells (Figure 5DE). These data suggested that GATA6-AS1 might regulate cell invasion and proliferation through EMT.
GATA6-AS1 Regulated FBXO11 via Sponging miR-324-5p
To examine whether miR-324-5p was involved in regulation of FBXO11, we co-transfected GATA6-AS1 and miR-324-5p mimic into lung cancer cells. In NCI-H1975, the Western blotting showed that miR-324-5p mimic reversed upregulation of FBXO11 and downregulation of SNAIL protein levels which were induced by transfection of GATA6-AS1 (Figure 6A). Similarly, miR-324-5p mimic also reversed the effect of GATA6-AS1 overexpression in A549 cells (Figure 6B). At the mRNA level, miR-324-5p mimic also attenuated the upregulation of FBXO11 in NCI-H1975 and A549 cells (Figure 6C). We constructed luciferase vector containing FBXO11 3ʹUTR-wild type or FBXO11 3ʹUTR -Mutant (with point-mutation in the binding site) (Figure 6D). Dual-luciferase reporter assay showed that miR-324-5p mimic repressed luciferase activity of FBXO11 3ʹUTR-wild type which was reversed by GATA6-AS1 in NCI-H1975 and A549 cells, meanwhile, the activity of FBXO11 3ʹUTR-mutant was not affected by miR-324-5p mimic or GATA6-AS1 (Figure 6EF).
GATA6-AS1 Relied on Regulation of miR-324-5p to Control Lung Cell Invasion and Proliferation
In NCI-H1975 and A549 cells, the transwell assay showed that miR-324-5p mimic attenuated the effect of GATA6-AS1 overexpression on cell invasion (Figure 7A). In addition, the CCK-8 assay showed that miR-324-5p mimic attenuated the effect of GATA6-AS1 overexpression on cell proliferation in NCI-H1975 and A549 cells (Figure 7BC). These results manifested a GATA6-AS1/miR-324-5p axis on regulation of lung cancer cell proliferation and invasion.
Discussion
Although the functions of several lncRNAs have been revealed in lung cancer,25,26 the biological roles of many differentially expressed lncRNAs remain unknown in lung cancer. GATA6-AS1 was a cancer-associated lncRNA according to the most recent studies. In gastric cancer, GATA6-AS1 was decreased and inhibited EMT through inactivation of Wnt signaling.17 RNA sequencing data indicated that GATA6-AS1 was one of the most significantly downregulated lncRNAs in LUSC.18 We firstly used online tool to analyze the expression of GATA6-AS1 in TCGA data of various cancer types and GTEx project using GEPIA software. Interestingly, GATA6-AS1 was downregulated in multiple cancer types (11 out of 33) including LUAD and LUSC. This downregulation of GATA6-AS1 was confirmed in our collected tissue samples. Meanwhile, GATA6-AS1 expression was associated with metastasis status of patients. Expression of GATA6-AS1 was also decreased in lung cancer cell lines. In the function assay, it was observed that GATA6-AS1 inhibited cell proliferation and invasion of lung cancer cells, which was similar to the observation in gastric cancer.17 Thus, the data showed that GATA6-AS1 was a tumor suppressor in lung cancer.
The role of miR-324-5p in cancer is controversial. miR-324-5p targeted TGFβ2 and inhibited gallbladder carcinoma cell metastasis.27 miR-324-5p directly repressed ELAVL1 to inhibit cell proliferation and invasion of colorectal cancer.28 On the contrary, in lung cancer, upregulation of miR-324-5p was observed in most patients. Downregulation of miR-324-5p inhibited cell proliferation, colony forming and invasion of lung cancer cells.21 Via bioinformatic analysis, we found that there was a binding site for miR-324-5p in GATA6-AS1 sequence. We further confirmed that there was a mutual regulatory association between GATA6-AS1 and miR-324-5p. One recent study discovered that miR-324-5p directly suppressed FBXO11 expression in lung cancer cells.22 FBXO11 was a member of F-box protein and constituted ubiquitin-protein ligase complex, promoted ubiquitin-mediated degradation of SNAIL, a key regulator of EMT, in cancer cells.24 We experimentally showed that GATA6-AS1 upregulated FBXO11 and downregulated SNAIL protein expression. Moreover, miR-324-5p mimic attenuated the effect of GATA6-AS1 on FBXO11 and SNAIL expression in lung cancer cells. Thus, the data manifested that GATA6/AS1 sponged miR-324-5p to regulate FBXO11 and inhibited cell proliferation and invasion in lung cancer cells.
In conclusion, the current data depicted a GATA6-AS1/miR-324-5p/FBXO11 axis in lung cancer and implied that GATA6-AS1 suppressed lung cancer progression. Therefore, GATA6-AS1 was a candidate biomarker for patients with lung cancer.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi:10.3322/caac.21492
2. Thomson CS, Forman D. Cancer survival in England and the influence of early diagnosis: what can we learn from recent EUROCARE results? Br J Cancer. 2009;101(Suppl 2):S102109. doi:10.1038/sj.bjc.6605399
3. Sun M, Liu XH, Wang KM, et al. Downregulation of BRAF activated non-coding RNA is associated with poor prognosis for non-small cell lung cancer and promotes metastasis by affecting epithelial-mesenchymal transition. Mol Cancer. 2014;13:68. doi:10.1186/1476-4598-13-68
4. Vitiello M, Tuccoli A, Poliseno L. Long non-coding RNAs in cancer: implications for personalized therapy. Cell Oncol. 2015;38(1):17–28. doi:10.1007/s13402-014-0180-x
5. Adelman K, Egan E. Non-coding RNA: more uses for genomic junk. Nature. 2017;543(7644):183–185. doi:10.1038/543183a
6. Costa FF. Non-coding RNAs, epigenetics and complexity. Gene. 2008;410(1):9–17. doi:10.1016/j.gene.2007.12.008
7. Pecero ML, Salvador-Bofill J, Molina-Pinelo S. Long non-coding RNAs as monitoring tools and therapeutic targets in breast cancer. Cell Oncol. 2019;42(1):1–12. doi:10.1007/s13402-018-0412-6
8. Abdollahzadeh R, Daraei A, Mansoori Y, Sepahvand M, Amoli MM, Tavakkoly-Bazzaz J. Competing endogenous RNA (ceRNA) cross talk and language in ceRNA regulatory networks: A new look at hallmarks of breast cancer. J Cell Physiol. 2019;234(7):10080–10100. doi:10.1002/jcp.27941
9. Wang LX, Wan C, Dong ZB, Wang BH, Liu HY, Li Y. Integrative analysis of long noncoding RNA (lncRNA), microRNA (miRNA) and mRNA expression and construction of a competing endogenous RNA (ceRNA) network in metastatic melanoma. Med Sci Monit. 2019;25:2896–2907. doi:10.12659/MSM.913881
10. Liang T, Zhou B, Shi L, et al. lncRNA AK017368 promotes proliferation and suppresses differentiation of myoblasts in skeletal muscle development by attenuating the function of miR-30c. FASEB J. 2018;32(1):377–389. doi:10.1096/fj.201700560rr
11. Rossi M, Bucci G, Rizzotto D, et al. LncRNA EPR controls epithelial proliferation by coordinating Cdkn1a transcription and mRNA decay response to TGF-beta. Nat Commun. 2019;10(1):1969. doi:10.1038/s41467-019-09754-1
12. Hu B, Zhang H, Wang Z, Zhang F, Wei H, Li L. LncRNA CCAT1/miR-130a-3p axis increases cisplatin resistance in non-small-cell lung cancer cell line by targeting SOX4. Cancer Biol Ther. 2017;18(12):974–983. doi:10.1080/15384047.2017.1385679
13. Jiao P, Hou J, Yao M, Wu J, Ren G. SNHG14 silencing suppresses the progression and promotes cisplatin sensitivity in non-small cell lung cancer. Biomed Pharmacother. 2019;117:109164. doi:10.1016/j.biopha.2019.109164
14. Li N, Gao WJ, Liu NS. LncRNA BCAR4 promotes proliferation, invasion and metastasis of non-small cell lung cancer cells by affecting epithelial-mesenchymal transition. Eur Rev Med Pharmacol Sci. 2017;21(9):2075–2086.
15. Liu L, Zhou XY, Zhang JQ, et al. LncRNA HULC promotes non-small cell lung cancer cell proliferation and inhibits the apoptosis by up-regulating sphingosine kinase 1 (SPHK1) and its downstream PI3K/Akt pathway. Eur Rev Med Pharmacol Sci. 2018;22(24):8722–8730. doi:10.26355/eurrev_201812_16637
16. Xie X, Liu HT, Mei J, et al. LncRNA HMlincRNA717 is down-regulated in non-small cell lung cancer and associated with poor prognosis. Int J Clin Exp Pathol. 2014;7(12):8881–8886.
17. Li ZT, Zhang X, Wang DW, et al. Overexpressed lncRNA GATA6-AS1 Inhibits LNM and EMT via FZD4 through the Wnt/beta-catenin signaling pathway in GC. Mol Ther Nucleic Acids. 2019;19:827–840. doi:10.1016/j.omtn.2019.09.034
18. Xiong Y, Zhang X, Lin Z, et al. SFTA1P, LINC00968, GATA6-AS1, TBX5-AS1, and FEZF1-AS1 are crucial long non-coding RNAs associated with the prognosis of lung squamous cell carcinoma. Oncol Lett. 2019;18(4):3985–3993. doi:10.3892/ol.2019.10744
19. Alvarez-Garcia I, Miska EA. MicroRNA functions in animal development and human disease. Development. 2005;132(21):4653–4662. doi:10.1242/dev.02073
20. Bian HB, Pan X, Yang JS, Wang ZX, De W. Upregulation of microRNA-451 increases cisplatin sensitivity of non-small cell lung cancer cell line (A549). J Exp Clin Cancer Res. 2011;30:20. doi:10.1186/1756-9966-30-20
21. Lin MH, Chen YZ, Lee MY, et al. Comprehensive identification of microRNA arm selection preference in lung cancer: miR-324-5p and −3p serve oncogenic functions in lung cancer. Oncol Lett. 2018;15(6):9818–9826. doi:10.3892/ol.2018.8557
22. Ba Z, Zhou Y, Yang Z, Xu J, Zhang X. miR-324-5p upregulation potentiates resistance to cisplatin by targeting FBXO11 signalling in non-small cell lung cancer cells. J Biochem. 2019;166(6):517–527. doi:10.1093/jb/mvz066
23. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C (T)) Method. Methods. 2001;25(4):402–408. doi:10.1006/meth.2001.1262
24. Jin Y, Shenoy AK, Doernberg S, et al. FBXO11 promotes ubiquitination of the Snail family of transcription factors in cancer progression and epidermal development. Cancer Lett. 2015;362(1):70–82. doi:10.1016/j.canlet.2015.03.037
25. Castro-Oropeza R, Melendez-Zajgla J, Maldonado V, Vazquez-Santillan K. The emerging role of lncRNAs in the regulation of cancer stem cells. Cell Oncol. 2018;41(6):585–603. doi:10.1007/s13402-018-0406-4
26. Zhang Z, Wang Y, Zhang W, Li J, Liu W, Lu W. Long non-coding RNA SNHG14 exerts oncogenic functions in non-small cell lung cancer through acting as an miR-340 sponge. Biosci Rep. 2019;39:1.
27. Zhang X, Zhang L, Chen M, Liu D. miR-324-5p inhibits gallbladder carcinoma cell metastatic behaviours by downregulation of transforming growth factor beta 2 expression. Artif Cells Nanomed Biotechnol. 2020;48(1):315–324. doi:10.1080/21691401.2019.1703724
28. Gu C, Zhang M, Sun W, Dong C. Upregulation of miR-324-5p inhibits proliferation and invasion of colorectal cancer cells by targeting ELAVL1. Oncol Res. 2019;27(5):515–524. doi:10.3727/096504018X15166183598572
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.