Back to Journals » OncoTargets and Therapy » Volume 13
Long Non-Coding RNA DLEU1 Up-Regulates BIRC6 Expression by Competitively Sponging miR-381-3p to Promote Cisplatin Resistance in Nasopharyngeal Carcinoma
Authors Li H, Huang J, Yu S, Lou Z
Received 6 November 2019
Accepted for publication 15 February 2020
Published 9 March 2020 Volume 2020:13 Pages 2037—2045
DOI https://doi.org/10.2147/OTT.S237456
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Leo Jen-Liang Su
Hangbo Li, Jia Huang, Sa Yu, Zhiping Lou
Department of Otolaryngology, Zhuji People’s Hospital, Zhuji 311800, People’s Republic of China
Correspondence: Zhiping Lou
Department of Otolaryngology, Zhuji People’s Hospital, #9 Jianmin Road, Zhuji 311800, People’s Republic of China
Email [email protected]
Background: Cisplatin (DDP) resistance has become an obstacle to chemotherapy for nasopharyngeal carcinoma (NPC) patients. Recent evidences indicate that long noncoding RNAs (lncRNAs) are involved in tumorigenesis and chemoresistance. However, the potential role of lncRNAs in NPC progression remains largely unknown.
Methods: First, lncRNA expression profiling in NPC was performed via microarray analysis. To explore the involvement of DLEU1 in DDP resistance, loss-of-function experiments were employed in vitro and in vivo. Bioinformatics analysis, luciferase reporter assay, qRT-PCR, and Western blot assays were used to investigate the underlying mechanisms.
Results: Here, we identified 153 differentially expressed lncRNAs. Among them, DLEU1 was remarkably up-regulated in NPC tissues and associated with worse outcome. Knock-down of DLEU1 could sensitize NPC cells to DDP in vitro and in vivo. Further investigations revealed that DLEU1 positively regulated BIRC6 expression via its competing endogenous RNA (ceRNA) activity on miR-381-3p. We also observed that BIRC6 overexpression or miR-381-3p silence could significantly reverse DLEU1-dependent DDP resistance.
Conclusion: Our data suggest that DLEU1 acts as an oncogene to promote DDP resistance and BIRC6 expression in NPC through interacting with miR-381-3p, which may help to develop new strategy against NPC chemoresistance.
Keywords: DLEU1, cisplatin resistance, nasopharyngeal carcinoma, BIRC6
Introduction
Nasopharyngeal carcinoma (NPC) is a head and neck carcinoma with a very unique pattern of geographical distribution. In 2018, there were about 129, 000 newly diagnosed NPC cases, and 70% were in east and southeast Asia.1 Cisplatin (DDP) is often the first choice in chemotherapy regimens for NPC patients.2 However, long-term treatment with DDP will lead to drug resistance which is a main reason for chemotherapy failure.3 Hence, it is of great importance to investigate the molecular mechanisms underlying the DDP resistance and develop effective therapeutic strategies for NPC treatment.
Long noncoding RNAs (lncRNAs) are RNA molecules longer than 200 nucleotides. lncRNAs could function as competing endogenous RNA (ceRNA) to competitively bind with miRNA response elements and reduce their effect on mRNAs.4,5 For instance, up-regulated lncRNA LOC100129148 was associated with unfavorable outcome in patients with NPC and silence of LOC100129148 suppressed cell proliferation and KLF12 expression through sponging miR-539-5p.6 Wang et al reported that lncRNA CCAT1 enhanced paclitaxel resistance of NPC cells via miR-181a/CPEB2 axis.7 These findings indicate that dysregulated lncRNAs play crucial roles in NPC pathogenesis and chemoresistance.
lncRNA DLEU1 (deleted in lymphocytic leukemia 1) is located on chromosome 13q14.3 which is frequently deleted in B-cell chronic lymphocytic leukemia.8 Recent studies demonstrated that DLEU1 was highly expressed and contributed to tumorigenesis and development in a variety of cancers, such as ovarian, colorectal and endometrial cancer.9–11 In Burkitt lymphoma, however, overexpression of DLEU1 decreased cell proliferation, increased programmed cell death and improved survival, indicating DLEU1 may function as a tumor suppressor.12 Taken together, the functions and molecular basis of DLEU1 are complex, yet its function in NPC progression is still unclear.
In this study, we investigated the expression and role of DLEU1 in NPC. We reported the finding that DLEU1 expression was increased in NPC tissues and cell lines, and associated with poor overall survival. Functionally, DLEU1 promoted DDP resistance and up-regulated BIRC6 through reducing miR-381-3p expression. These findings indicate that DLEU1 could be a new therapeutic target and prognostic marker in NPC.
Materials and Methods
Cell Culture and Tissue Samples
Human NPC cell lines (5-8F, 6-10B, C666-1, CNE1, CNE2, HNE-1, HONE-1, and SUNE-1) were purchased from American Type Culture Collection (ATCC) and maintained in RPMI-1640 (Invitrogen, Grand Island, NY, USA). Human nasopharyngeal epithelial cell line NP69 was obtained from the Cell Bank of Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences (Shanghai, China) and cultured in keratinocyte-SFM (Invitrogen). HEK-293T cells were purchased from ATCC and grown in DMEM (Invitrogen). A total of 67 NPC tissues were obtained from patients who received primary surgery at Zhuji People’s Hospital. Twelve normal nasopharyngeal epithelial tissues were collected from patients who had nasal operations. Our study protocol was approved by the research ethics committee of Zhuji People’s Hospital (ZJSRMYY-2017H-052). Written informed consent was signed by all participants.
Oligonucleotide and Plasmid Transfection
The siRNA for DLEU1 (si-DLEU1), negative control siRNA (si-NC), miR-381-3p mimics, miR-381-3p inhibitor (miR-381-3p-in), miRNA negative control (miRNA-NC) and pcDNA-BIRC6 were commercially synthesized by GenePharma (Shanghai, China). Transient transfection was performed with 50 nM oligonucleotides using Lipofectamine 2000. Subsequent experiments were performed at 48 hrs post-transfection. Short hairpin RNA (shRNA) targeting DLEU1 (sh-DLEU1) and non-specific control (sh-NC) were synthesized by GenePharma and cloned into pSuper-retro-puromycin vectors. For a stable cell transfection, SUNE-1 cells were transfected with sh-DLEU1 or sh-NC and then selected by 0.5 μg/mL puromycin.
MTT Assay
Cells were plated into 96-well plates (5000 cells/well) and treated with different concentrations of DDP (0, 5, 10, 15 and 20 μg/mL). Forty-eight hours after incubation, cell viability was assessed by MTT assay. The spectrophotometric absorbance was measured at 490 nm.
qRT-PCR
Total RNA was extracted from tissues and cell lines using TRIzol reagent (Ambion, Life Technologies, USA). cDNA of miRNA and mRNA or lncRNA was synthesized using TaqMan miR Reverse Transcriptase Kit (Applied Biosystems-Life Technologies) and ImProm-II Reverse Transcription System (Promega), respectively. qPCR assays were performed using the SYBR Premix EX Taq™ (Takara). Their relative expression levels were calculated using 2−ΔΔCT and normalized to U6 and β-actin.
Western Blot
Cell lysate was prepared using RIPA lysis buffer with protease inhibitors. Equal amounts of protein were separated by 10% SDS-PAGE, transferred to PVDF membranes, and then probed with anti-BIRC6 (Cell Signaling Technology) at 4°C overnight followed by incubation with HRP-conjugated secondary antibody for 1 hr at room temperature. Protein was normalized with β-actin (Cell Signaling Technology).
Microarray Analysis
Total RNA was extracted from three NPC tissues and three normal nasopharyngeal epithelial tissues using TRIzol Reagent (Invitrogen) and purified by an RNeasy Mini Kit (Qiagen, Valencia, CA). cDNA was synthesized, labeled, and hybridized to the LncRNA Expression Microarray (Arraystar, Rockville, MD). After washing, slides were analyzed on the lncRNAs microarray. Differentially expressed lncRNAs were identified with thresholds of P < 0.05 and fold change ≥2.0 using GeneSpring GX v12.1 software package (Agilent Technologies, Santa Clara, CA, USA).
Xenografts
Twenty 5-week-old female BALB/c nude mice were randomly divided into sh-DLEU1 group and sh-NC group. About 1×107 SUNE-1 cells were inoculated subcutaneously into each mouse. When the tumors reached a volume of approximately 100 mm3, each group of mice were intraperitoneally injected with DDP (5 mg/kg) every 2 days. After 4 weeks of treatment, all mice were sacrificed and the tumors were surgically removed. The animal assay was approved by the animal management committee of Zhuji People’s Hospital, and all experimental procedures and animal care were in accordance with the institutional ethics guidelines for animal experiments (ZJSRMYY-2017D-068).
Luciferase Reporter Assay
A fragment of the wild type (wt) or mutant (mut) DLEU1 and BIRC6 3’UTR that contained the putative miR-381-3p binding sites was cloned into pGL3-REPORT luciferase vector. HEK293T cells (1×104) were seeded into 24-well plates and transiently co-transfected wt or mut reporter plasmid with miR-381 mimics or miRNA-NC. Dual luciferase reporter assays (Promega) were analyzed 48 hrs after transfection according to the manufacturer’s instructions.
Statistical Analysis
Data are expressed as mean ± SD of triplicate experiments. Statistical analysis was performed using Student’s t-test on Graphpad Prism 6 software. Survival curves were assayed using the Kaplan-Meier method. P value<0.05 was considered statistically significant.
Results
DLEU1 Is Overexpressed in NPC and Predicts Poor Overall Survival
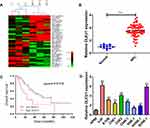
To explore the potential involvement of lncRNAs in NPC progression, lncRNA microarray analysis was performed between NPC tissues and normal nasopharyngeal epithelial tissues. The result showed that 153 lncRNAs were differentially expressed (82 up-regulated and 71 down-regulated) with fold change >2.0 and P < 0.05 (Figure 1A). Of interest, DLEU1 expression was up-regulated in NPC tissues. The expression level of DLEU1 was validated to be up-regulated 3.53-fold in 67 NPC tissues compared with that of 12 normal nasopharyngeal epithelial tissues (Figure 1B). Clinicopathological features showed that up-expression of DLEU1 was strongly associated with clinical stage (Table 1; P=0.019), T stage (P=0.032) and chemotherapy response (P=0.012). Furthermore, Kaplan–Meier survival analysis revealed that high level of DLEU1 predicted poor outcome (P=0.016; Figure 1C). We also determined the expression profiles of DLEU1 in normal epithelium cells and NPC cell lines. As shown in Figure 1D, DLEU1 was up-regulated in all 8 NPC cell lines, especially in 5-8F and SUNE-1 cells. These findings suggest a crucial role of DLEU1 in NPC development.
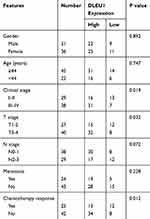 |
Table 1 Correlation Analysis Between DLEU1 Expression and Clinicopathologic Features of NPC Patients |
Knock-Down of DLEU1 Sensitizes NPC Cells to DDP in vitro and vivo
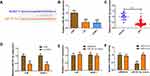
DDP is an important chemotherapy regimen for NPC treatment. To investigate whether DLEU1 could affect DDP resistance in NPC development, loss-of-function experiment was performed via transfecting specific DLEU1 siRNA into 5-8F and SUNE-1 cells (Figure 2A). When exposing to increasing concentrations of DDP, cells transfected with si-DLEU1 were more sensitive to DDP than cells transfected with si-NC (Figure 2B and C). To further evaluate the effect of DLEU1 on DDP resistance in vivo, SUNE-1 cells stably expressing sh-DLEU1 or sh-NC were subcutaneously injected into nude mice following by treatment with DDP. Four weeks after treatment, both tumor volume and weight in sh-DLEU1 group were decreased than control group (Figure 2D–F). Collectively, these data indicate up-regulated DLEU1 may contribute to NPC development through increasing DDP resistance.
DLEU1 Binds to miR-381-3p and Suppresses Its Expression
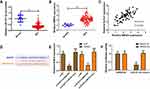
lncRNAs mainly exert their biological functions through ceRNA mechanism. We speculated whether DLEU1 could interact with miRNAs to regulate DDP-resistance. Bioinformatics analysis revealed that DLEU1 contains binding sites for miR-381-3p (Figure 3A). miR-381-3p expression in 5-8F and SUNE-1 cells was lower than that of NP69 cells (Figure 3B). Besides, miR-381-3p level was down-regulated 2.55-fold in NPC tissues compared with normal tissues (Figure 3C). Then we explored the regulatory relationship between DLEU1 and miR-381-3p in 5-8F and SUNE-1 cells. Knock-down of DLEU1 negatively regulated miR-381-3p expression (Figure 3D). Meanwhile, miR-381-3p mimics have no significant influence on DLEU1 level (Figure 3E). Luciferase reporter assay showed that co-transfection of miR-381-3p mimics with DLEU1-wt, rather than the DLEU1-mut, significantly reduced luciferase activity (Figure 3F). These results indicate that miR-381-3p is a key downstream target of DLEU1 to promote DDP resistance in NPC.
DLEU1 Acts as a ceRNA to Up-Regulate BIRC6 Expression via Sponging miR-381-3p
Recently, BIRC6 was identified as an oncogene involved in chemoradiotherapy resistance. Thus, we aimed to verify whether DLEU1 could regulate BIRC6 expression by interacting with miR-381-3p in NPC cells. Compared with NP69 cells, BIRC6 level was higher in 5-8F and SUNE-1 cells (Figure 4A). We also observed an increased level of BIRC6 in NPC tissues (Figure 4B). Our results showed that there was a strong correlation between DLEU1 and BIRC6 in NPC tissues (R=0.7015, P<0.0001; Figure 4C). Bioinformatics analysis showed that the 3’UTR sequence of BIRC6 was complementary to the seed sequence of miR-381-3p (Figure 4D). In addition, BIRC6 protein level was down-regulated when transfected with si-DLEU1, and miR-381-3p inhibitor could abolish BIRC6 inhibition induced by DLEU1 knock-down (Figure 4E). Furthermore, luciferase reporter assay showed BIRC6-wt and miR-381-3p mimics co-transfection significantly decreased luciferase activity, but BIRC6-mut failed to change luciferase activity (Figure 4F).
DLEU1 Promotes DDP Resistance via miR-381-3p and BIRC6-Dependent Manner
To further confirm the functional role of miR-381-3p and BIRC6 in DLEU1-modulated DDP resistance, miR-381-3p inhibitor or pcDNA-BIRC6 was co-transfected with si-DLEU1 into 5-8F and SUNE-1 cells. As shown in Figure 5A and B, cells co-transfected si-DLEU1 with miR-381-3p inhibitor exhibited a less sensitive ability to DDP compared with cells transfected with si-DLEU1 alone as determined using MTT assay. Consistently, BIRC6 overexpression significantly reversed the inhibition of DDP resistance induced by DLEU1 knock-down in both into 5-8F and SUNE-1 cells (Figure 5C and D). These findings indicate that DLEU1 promotes DDP resistance at least partly through interacting with miR-381-3p and BIRC6.
Discussion
Abnormally expressed DLEU1 has been identified in several types of human cancer and could function as oncogene or tumor suppressor to affect cancer progression.9–12 In our microarray analysis, we found that DLEU1 expression increased in NPC tissues. We also observed that high level of DLEU1 predicted an unfavorable prognosis of patients with NPC. In addition, knock-down of DLEU1 reduced DDP resistance in vivo and in vitro. These findings suggest that DLEU1 acts as an oncogene in NPC. Further mechanistic analysis demonstrated that DLEU1 exerted its role by sponging miR-381-3p and up-regulating BIRC6 expression in NPC cells. Recently, DLEU1 was reported to enhance resistance to DDP in bladder cancer cells through regulating miR-99b/HS3ST3B1 axis, which is in line with our findings.13 Furthermore, DLEU1 overexpression was observed to be associated with rituximab and/or cyclophosphamide resistance in Burkitt lymphoma cells.12 As a kind of lncRNA, DLEU1 can interact with many molecules, like miRNAs and RNA-binding proteins. There would be various underlying mechanisms of DLEU1-induced DDP resistance, which are worthy of further study.
A growing body of evidence has demonstrated that miRNAs play an important role in NPC progression.14 Recent studies have suggested that miR-381-3p level was decreased in several cancers and overexpression of miR-381-3p inhibited cell proliferation and metastasis, and induced cell cycle arrest and apoptosis.15–19 However, the functions and mechanisms of miR-381-3p in NPC are still unclear. In our study, miR-381-3p was observed to be down-regulated in NPC. In addition, qRT-PCR and luciferase reporter assay showed that DLEU1 served as a ceRNA for miR-381-3p. Furthermore, silence of miR-381-3p reversed DDP sensitization-induced by DLEU1 knock-down. Taken together, these results suggest that miR-381-3p was a direct target of DLEU1.
BIRC6 gene encodes a 528 kDa protein that consists of N-terminal BIR domain and C-terminal ubiquitin-conjugating domain in mammals.20 BIRC6 is a key regulator for multiple biological processes, such as anti-apoptosis, cytoprotection and regulation of cytokinesis.21 Increasing evidences indicated that BIRC6 is frequently overexpressed in various tumors and associated with poor outcome.22–25 Several studies also demonstrated that BIRC6 participated in tumor cell chemoresistance, including imatinib, enzalutamide and cisplatin.26–28 Here, we observed that BIRC6 level was higher in NPC tissues compared with normal controls and strongly correlated with DLEU1 expression. DLEU1 knock-down reduced BIRC6 expression while BIRC6 knock-down reversed DLEU1-modulated DDP resistance in NPC cells. These data indicate that DLEU1 promotes DDP resistance at least partly through regulating BIRC6 expression.
In summary, we demonstrated that DLEU1 was significantly up-regulated and predicted poor overall survival of NPC patients. Our data support that DLEU1 acts as oncogene to promote DDP resistance at least partly through miR-381-3p/BIRC6 axis. More importantly, DLEU1 might serve as a potential therapeutic sensitizer in NPC treatment.
Disclosure
The authors declare that they have no competing interests in this work.
References
1. Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.21492
2. Chen YP, Chan ATC, Le QT, et al. Nasopharyngeal carcinoma. Lancet. 2019;394(10192):64–80. doi:10.1016/S0140-6736(19)30956-0
3. Huncharek M, Kupelnick B. Combined chemoradiation versus radiation therapy alone in locally advanced nasopharyngeal carcinoma: results of a meta-analysis of 1528 patients from six randomized trials. Am J Clin Oncol. 2002;25(3):219–223. doi:10.1097/00000421-200206000-00002
4. Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43(6):904–914. doi:10.1016/j.molcel.2011.08.018
5. Salmena L, Poliseno L, Tay Y, et al. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146(3):353–358. doi:10.1016/j.cell.2011.07.014
6. Sun KY, Peng T, Chen Z, et al. Long non-coding RNA LOC100129148 functions as an oncogene in human nasopharyngeal carcinoma by targeting miR-539-5p. Aging (Albany NY). 2017;9(3):999–1011. doi:10.18632/aging.101205
7. Wang Q, Zhang W, Hao S. LncRNA CCAT1 modulates the sensitivity of paclitaxel in nasopharynx cancers cells via miR-181a/CPEB2 axis. Cell Cycle. 2017;16(8):795–801. doi:10.1080/15384101.2017.1301334
8. Liu Y, Corcoran M, Rasool O, et al. Cloning of two candidate tumor suppressor genes within a 10 kb region on chromosome 13q14, frequently deleted in chronic lymphocytic leukemia. Oncogene. 1997;15(20):2463–2473. doi:10.1038/sj.onc.1201643
9. Wang LL, Sun KX, Wu DD, et al. DLEU1 contributes to ovarian carcinoma tumourigenesis and development by interacting with miR-490-3p and altering CDK1 expression. J Cell Mol Med. 2017;21(11):3055–3065. doi:10.1111/jcmm.13217
10. Liu T, Han Z, Li H, et al. LncRNA DLEU1 contributes to colorectal cancer progression via activation of KPNA3. Mol Cancer. 2018;17(1):118. doi:10.1186/s12943-018-0873-2
11. Shao W, Li Y, Chen F, et al. Long non-coding RNA DLEU1 contributes to the development of endometrial cancer by sponging miR-490 to regulate SP1 expression. Pharmazie. 2018;73(7):379–385. doi:10.1691/ph.2018.8352
12. Lee S, Luo W, Shah T, et al. The effects of DLEU1 gene expression in Burkitt lymphoma (BL): potential mechanism of chemoimmunotherapy resistance in BL. Oncotarget. 2017;8(17):27839–27853. doi:10.18632/oncotarget.15711
13. Li Y, Shi B, Dong F, et al. Long non-coding RNA DLEU1 promotes cell proliferation, invasion, and confers cisplatin resistance in bladder cancer by regulating the miR-99b/HS3ST3B1 axis. Front Genet. 2019;10:280. doi:10.3389/fgene.2019.00280
14. Spence T, Bruce J, Yip KW, et al. MicroRNAs in nasopharyngeal carcinoma. Chin Clin Oncol. 2016;5(2):17. doi:10.21037/cco
15. Yang X, Ruan H, Hu X, et al. miR-381-3p suppresses the proliferation of oral squamous cell carcinoma cells by directly targeting FGFR2. Am J Cancer Res. 2017;7(4):913–922.
16. Kong W, Yang L, Li PP, et al. MiR-381-3p inhibits proliferation, migration and invasion by targeting LRP6 in papillary thyroid carcinoma. Eur Rev Med Pharmacol Sci. 2018;22(12):3804–3811. doi:10.26355/eurrev_201806_15264
17. Shang A, Zhou C, Bian G, et al. miR-381-3p restrains cervical cancer progression by downregulating FGF7. J Cell Biochem. 2019;120(1):778–789. doi:10.1002/jcb.27438
18. Wu M, Fan B, Guo Q, et al. Knockdown of SETDB1 inhibits breast cancer progression by miR-381-3p-related regulation. Biol Res. 2018;51(1):39. doi:10.1186/s40659-018-0189-0
19. Li J, Ying Y, Xie H, et al. Dual regulatory role of CCNA2 in modulating CDK6 and MET-mediated cell-cycle pathway and EMT progression is blocked by miR-381-3p in bladder cancer. FASEB J. 2019;33(1):1374–1388. doi:10.1096/fj.201800667R
20. Bartke T, Pohl C, Pyrowolakis G, et al. Dual role of BRUCE as an antiapoptotic IAP and a chimeric E2/E3 ubiquitin ligase. Mol Cell. 2004;14(6):801–811. doi:10.1016/j.molcel.2004.05.018
21. Pohl C, Jentsch S. Final stages of cytokinesis and midbody ring formation are controlled by BRUCE. Cell. 2008;132(5):832–845. doi:10.1016/j.cell.2008.01.012
22. Hu T, Weng S, Tang W, et al. Overexpression of BIRC6 is a predictor of prognosis for colorectal cancer. PLoS One. 2015;10(5):e0125281. doi:10.1371/journal.pone.0125281
23. Gharabaghi MA. Diagnostic investigation of BIRC6 and SIRT1 protein expression level as potential prognostic biomarkers in patients with non-small cell lung cancer. Clin Respir J. 2018;12(2):633–638. doi:10.1111/crj.2018.12.issue-2
24. Zhuang W, Zhang C, Hao F, et al. Baculoviral IAP repeat containing 6 (BIRC6) is a predictor of prognosis in prostate cancer. Med Sci Monit. 2018;24:839–845. doi:10.12659/MSM.904052
25. Low CG, Luk IS, Lin D, et al. BIRC6 protein, an inhibitor of apoptosis: role in survival of human prostate cancer cells. PLoS One. 2013;8(2):e55837. doi:10.1371/journal.pone.0055837
26. Okumu DO, East MP, Levine M, et al. BIRC6 mediates imatinib resistance independently of Mcl-1. PLoS One. 2017;12(5):e0177871. doi:10.1371/journal.pone.0177871
27. Luk IS, Shrestha R, Xue H, et al. BIRC6 targeting as potential therapy for advanced, enzalutamide-resistant prostate cancer. Clin Cancer Res. 2017;23(6):1542–1551. doi:10.1158/1078-0432.CCR-16-0718
28. Dong X, Lin D, Low C, et al. Elevated expression of BIRC6 protein in non-small-cell lung cancers is associated with cancer recurrence and chemoresistance. J Thorac Oncol. 2013;8(2):161–170. doi:10.1097/JTO.0b013e31827d5237
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.