Back to Journals » OncoTargets and Therapy » Volume 13
Long Non-Coding RNA BCAR4 Binds to miR-644a and Targets TLX1 to Promote the Progression of Bladder Cancer
Authors Wang X, He H, Rui W, Xie X, Wang D, Zhu Y
Received 30 September 2019
Accepted for publication 25 February 2020
Published 24 March 2020 Volume 2020:13 Pages 2483—2490
DOI https://doi.org/10.2147/OTT.S232965
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Geoffrey Pietersz
Xiaojing Wang,* Hongchao He,* Wenbin Rui, Xin Xie, Dawei Wang, Yu Zhu
Department of Urology, Ruijin Hospital, School of Medicine, Shanghai Jiaotong University, Shanghai 200025, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Dawei Wang; Yu Zhu
Department of Urology, Ruijin Hospital, School of Medicine, Shanghai Jiaotong University, Shanghai 200025, People’s Republic of China
Email [email protected]; [email protected]
Background: Bladder cancer is a serious threat to human health. It is meaningful to study the pathogenesis of bladder cancer. Long non-coding RNAs (lncRNAs) are reported to promote or inhibit bladder cancer development. However, the role of lncRNA BCAR4 in the regulation of bladder cancer remains unclear.
Purpose: This study was to explore the role of lncRNA BCAR4 in the progression of bladder cancer cell.
Methods: RT-PCR was used to examine the expression of BCAR4 and miR-644a. CCK8 assay, colony formation assay, Transwell assay were used to detect the progression of bladder cancer cells after transfecting of indicated plasmids.
Results: The expression of BCAR4 was higher in bladder cancer cell lines than normal urothelial cell line. Moreover, the expression of BCAR4 was associated with the advanced stage and metastasis of bladder cancer. Through knockdown of BCAR4, we discovered that knockdown of BCAR4 significantly decreased the proliferation, migration and invasive abilities of bladder cancer cells. Mechanically, we showed that BCAR4 can bind to miR-644a directly and targets TLX1. Moreover, we also showed that miR-644a was also highly expressed in bladder cancer cells and inhibition of miR-644a or overexpression of TLX1 can increased the migration abilities of bladder cancer caused by knockdown of BCAR4.
Conclusion: We showed that BCAR4 sponged miR-644a to modulate the expression of TLX1 and promote bladder cancer development.
Keywords: BCAR4, miR-644a, TLX1, bladder cancer, proliferation and migration
Introduction
Bladder cancer is a malignant tumor that occurs in the bladder mucosa.1 It is the most common malignant tumor in the urinary system and one of the ten most common tumors in the whole body.2 Bladder cancer can occur at any age, even in children.3 The incidence increased with age, with a high incidence of 50 to 70 years old. The incidence of bladder cancer in males is 3 to 4 times that in females.4 Bladder cancer seriously affects people’s normal life, not only destroys the level of metabolism, but also does great harm to the urinary system.5,6 Therefore, it is very important to study the pathogenesis of bladder cancer.7
Long non-coding RNAs (lncRNAs) play important roles in cancer.8 Genome association studies have found that a large number of lncRNAs are associated with various types of cancer.9,10 LncRNA may inhibit or promote the proliferation and metastasis of tumors. Because of the diversity and specificity of genomic expression of lncRNAs, lncRNAs have been identified as new tumor molecular markers and therapeutic targets.10 There are many reports that discovered lncRNAs participate in the regulation of bladder cancer.11 BCAR4 was a long non-coding RNA that was identified to regulate many cancers progression.12–14 Wei et al reported that BCAR4 can promote the proliferation of glioma cells through EGFR/PI3K/AKT signaling.15 Moreover, Siddique H found that BCAR4 can act as a prognostic marker for colorectal cancer.16 However, the role of BCAR4 in the regulation of bladder cancer remains unclear.
MicroRNAs, referred to as miRNAs, are a class of non-coding single stranded RNA molecules with about 22 nucleotides (nt), which are cut by Dicerase from a 70nt pre-microRNAs (pre-microRNAs).17,18 MicroRNAs are involved in a series of important processes in life, including development, hematopoiesis, organogenesis, apoptosis and cell proliferation, and even cancer.19,20 For example, miR-644a was proved to regulate the progression of gastric cancer, hepatocellular carcinoma and breast cancer.21,22 Studies have found some microRNAs closely related to bladder cancer.23,24 However, the role of miR-644a in the regulation of bladder cancer remain unknown and it is not clear what specific mechanisms that these microRNAs promote or inhibit the occurrence of bladder cancer.
TLX1, also known as HOX11 was proved to be an important transcription factor involved in the regulation of embryonic spleen organogenesis.25 Moreover, TLX1 was also proved to regulate the progression of many cancer and Vanden Bempt et al reported that TLX1 was participated in the regulation of T Cell Acute Lymphoblastic Leukemia.26,27 However, the role of TLX1 in the regulation of bladder remains elusive.
Materials and Methods
Samples
Human bladder cancer samples and adjacent healthy bladder tissues were obtained from 38 bladder cancer patients under surgery at Ruijin Hospital. All samples were kept in liquid nitrogen before use. This study was approved by the Ethics Committee of Taiyuan Second People’s Hospital. All written informed consents were received from patients.
Cell Lines
Bladder cancer cell lines: UMUC3, SW780, 5637, T24 and one normal urothelial cell line: SVHUC-1 cells were obtained from the Tumor Cell Bank of the Chinese Academy of Medical Science (Shanghai, China). The UMUC3, T24 and SVHUC-1 cells were cultured in Dulbecco’s Modified Eagle Medium (Invitrogen, Carlsbad, CA, USA) plus 10% FBS and ampicillin and streptomycin at 37 °C and 5% CO2. The 5637 cells and SW780 cells were cultured in RPMI-1640 Medium (Invitrogen, Carlsbad, CA, USA) plus 10% FBS and ampicillin and streptomycin at 37 °C and 5% CO2.
CCK8 Assay
CCK8 assays were performed using CCK8 detection kit (7 sea biotech, Shanghai, China) according to the instruction of the Kit.
Cell Transfection
Small interfering RNA oligonucleotides (siRNA, 5ʹ-AUAAAAUGCCACACAACCA-3ʹ) that targeting BCAR4 and a negative control were purchased from GenePharma (Shanghai, China). miR-644a mimics (5ʹ-AGUGUGGCUUUCUUAGAGC-3ʹ), miR-control (5ʹ-AUCUGCGUAAGAUUCGAGUC-3ʹ), miR-644a inhibitors (5ʹ- GCTCTUUGUUUGCCUCUCT-3ʹ) and control inhibitors (5ʹUAACUAAUACAUCGGAUU-3ʹ) were purchased from GenePharma (Shanghai, China). Cells were transfected with 100 nM miR-644a mimics, miR-control, miR-644a inhibitors or control inhibitors using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions.
RNA Preparation and RT-PCR
Total RNA was extracted from tissues or cells using Trizol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. Then, total RNA was subsequently reverse-transcribed into cDNAs using the PrimeScript RT reagent Kit (Promega, Madison, WI, USA) according to the protocol of the manufacturer. RT-PCR was performed using SYBR Green PCR Master Mix reagents (Takara) in the 7300 Real-Time PCR System (Applied Biosystems). The primers were listed as below:
U6 forward, 5ʹ-CGCTTCGGCAGCACATATA C-3ʹ; U6 reverse, 5ʹ-TTCACGAATTTGCGTGTCAT-3ʹ; GAPDH forward, 5ʹ-CATGAGAAGTATGACAACAGCC-3ʹ; GAPDH reverse, 5ʹ-AGTCCTTCCACGATACCAAAGT-3ʹ; TLX1 forward, 5ʹ- GACCGGCTCCTACAACGTG-3ʹ; TLX1 reverse, 5ʹ- CTGCGGTTACTCTCCATCCAG-3ʹ; BCAR4 forward, 5ʹ- gaactgctatatgcatttgc-3ʹ; BCAR4 reverse, 5ʹ- acgaattaggctcttgattg-3ʹ; 18S forward, 5ʹ -cagccacccgagattgagca-3ʹ; 18S reverse, 5ʹ-tagtagcgacgggcggtgtg-3ʹ.
Transwell Migration and Invasion Assay
Transwell assay was performed as previously described.28 Briefly, migration or invasion chamber was coated with Matrigel (BD Biosciences, Cowley, United Kingdom). Cells were then seeded into the upper chamber with serum-free DMEM medium and the lower chamber was supplemented with DMEM containing 10% FBS. After 24h incubation, cells in the lower chamber were fixed with 4% paraformaldehyde and stained with 0.1% crystal violet for 30 min.
Luciferase Assay
pMIR-TLX1-3ʹUTR (WT or Mutant) or pMIR-BCAR4 (WT or Mutant) and miR-644a (mimics or inhibitor) were transfected into T24 or SW780 cells along with pRL-TK vectors (Promega, USA). After 24 hrs culturing, luciferase activities were measured using a dual Glo™ Luciferase Assay System (Promega) according to the manufacturer’s protocols.
Statistical Analysis
A Student’s t-test was used to analyze the differences between two groups. A one-way ANOVA followed by a Tukey’s post-hoc test was used for multiple comparisons. The overall survival was analyzed by Kaplan-Meier analysis and a Log rank test. GraphPad Prism 6 software was used to analyze all results. The results were expressed as mean ± SD. P < 0.05 was considered to be significant.
Results
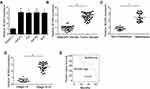
LncRNA BCAR4 Is Associated with Bladder Cancer Development
LncRNA BCAR4 was proved to participate in the regulation of many cancers. However, the function of BCAR4 in the regulation of bladder cancer progression remains unknown. To explore the role of BCAR4 in bladder cancer, we firstly examined the expression of BCAR4 in bladder cancer cell lines and normal bladder cells. Through RT-PCR assay, we found that BCAR4 was highly expressed in bladder cancer cell lines (Figure 1A). Moreover, we also collected 38 bladder cancer samples and found that the expression of BCAR4 in bladder cancer samples was higher than paired healthy bladder tissues (Figure 1B). Through RT-PCR analysis, we discovered that the expression of BCAR4 was associated with bladder cancer metastasis and advanced stages (Figure 1C and D). To explore the overall survival rate of bladder cancer patients with different BCAR4 expression, we performed Ker-Mein analysis. We divided the 38 bladder cancer patients into 2 groups based on the median expression of BCAR4. We found that lower BCAR4 expression group possessed better overall survival while higher BCAR4 expression group possessed worse overall survival (Figure 1E). Collectively, we showed that the expression of BCAR4 was associated with bladder cancer development.
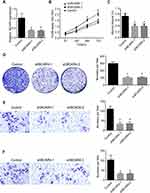
Knockdown of BCAR4 Decreased Bladder Cancer Progression
To find the function of BCAR4 in the regulation of bladder cancer progression, we performed a series of functional assays. Firstly, we constructed two shRNAs specifically targeted to BCAR4 sequence (Figure 2A). Then, we performed CCK8 assays and found that the cell viability of bladder cells were significantly decreased after knockdown of BCAR4 (Figure 2B). Moreover, colony formation assay also proved that BCAR4 knockdown decreased the proliferation of bladder cancer cells (Figure 2D). Finally, we performed transwell assays and wound healing assays to examine the migration and invasive abilities of bladder cancer cells. We found that the knockdown of BCAR4 remarkably inhibited the migration and invasive abilities of bladder cancer cells (Figure 2C, E and F). Taken together, these data showed that BCAR4 promoted the proliferation, migration and invasive of bladder cancer cells.
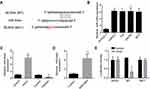
BCAR4 Binds to miR-644a Directly
To find the molecular mechanism that BCAR4 regulated bladder cancer development, we performed bioinformatics analysis. We found that BCAR4 can form complementary base pairing with miR-644a directly (Figure 3A). Besides, we also discovered that miR-644a was highly expressed in bladder cancer cell lines compared to normal bladder cell line (Figure 3B). To illustrate the molecular regulation between BCAR4 and miR-644a, we constructed miR-644a overexpression plasmid and miR-644a inhibitor (Figure 3C). We found that the expression of miR-644 was significantly elevated after knockdown of BCAR4 (Figure 3D). Moreover, we also performed luciferase assay. We discovered that BCAR4 can bind to miR-644a directly and overexpression of miR-644a significantly decreased the expression of BCAR4 (Figure 3E). Besides, miR-644a cannot regulate the expression of BCAR4 mutant (Figure 3E). Collectively, we showed that BCAR4 can bind to miR-644a directly and regulate the expression of miR-644a.
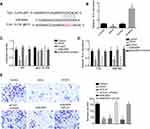
BCAR4 Promotes the Expression of TLX1 by Regulating the Expression of miR-644a
To find the potential gene that regulated by BCAR4 and miR-644a, we performed bioinformatics analysis again using TargetScan7 software. We showed that TLX1 was a potential target that can bind to miR-644a directly (Figure 4A). We found that the expression of TLX1 was elevated after inhibiting miR-644a expression while the expression of TLX1 was decreased after overexpression of miR-644a (Figure 4B). Through luciferase assay, we showed that miR-644a bind s to WT TLX1 other than MUT TLX1 (Figure 4C). Besides, we also found that overexpression of miR-644a or knockdown of BCAR4 significantly decreased TLX1 expression while inhibition of miR-644a can rescue the decreased BCAR4 expression caused by knockdown of BCAR4 (Figure 4D). Consistently, the migration abilities of bladder cancer cells were decreased after knockdown of BCAR4, TLX1 or overexpression of miR-644a. And also, inhibition of miR-644a coupled with knockdown of TLX1 or overexpression of miR-644a coupled with knockdown of BCAR4 can rescued the numbers of migration bladder cancer cells (Figure 4E). Collectively, we showed that TLX1 was targeted by BCAR4 and miR-644a to regulate the migration of bladder cancer.
Discussion
In this study, we showed that the expression of BCAR4 was much higher in bladder cancer cell lines and bladder cancer samples. Knockdown of BCAR4 significantly decreased the proliferation, migration and invasion abilities of bladder cancer cells. Moreover, we discovered that BCAR4 can bind to miR-644a directly and regulate the expression of TLX1. The expression of miR-644a was also proved to be higher in bladder cancer cell lines. Besides, we showed that knockdown of BCAR4 can increase the expression of miR-644a and inhibition of miR-644a can increase the expression of TLX1. So, we found that overexpression of TLX1 significantly promote the expression of TLX1. Finally, through transwell migration assays, we showed that overexpression of TLX1 or inhibition of miR-644a increased the migration abilities of bladder cancer cells decreased by knockdown of BCAR4, TLX1 or overexpression of miR-644a.
LncRNAs were shown to play vital functions in the regulation of bladder cancer development.9 Pan J and colleagues found that lncRNA UCA1 can promotes the cisplatin or gemcitabine resistance in bladder cancer through modulating miR-196a-5p.29 Ye T and colleagues showed that linc00346 is upregulated in bladder cancer tissues and knockdown of linc00346 inhibits bladder cancer progression.30 Though many lncRNAs were proved to participate in the regulation of bladder cancer development.31,32 However, the function of BCAR4 in the regulation of bladder cancer have not been characterized.
TLX1 is a transcription factor that controls the cell fate specification and organ expansion during spleen development.33 Tlx1 deletion in mice causes asplenia.34 Moreover, many studies have showed that TLX1 was associated with T-cell acute lymphoblastic leukemia.35,36 However, the function of TLX1 in the regulation of bladder cancer remains unknown. In our study, we firstly showed that BCAR4 and miR-644a can regulate the expression of TLX1 and the expression of TLX1 was associated with bladder cancer progression.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Martinez Rodriguez RH, Buisan Rueda O, Ibarz L. Bladder cancer: present and future. Med Clin (Barc). 2017;149:449–455. doi:10.1016/j.medcli.2017.06.009
2. Smith AB. Recent developments in the management of bladder cancer: introduction. Urol Oncol. 2018;36:95–96. doi:10.1016/j.urolonc.2017.10.026
3. Moriello G, Terpstra ME, Earl J. Outcomes following physical therapy incorporating hippotherapy on neuromotor function and bladder control in children with down syndrome: a case series. Phys Occup Ther Pediatr. 2019;1–14. doi:10.1080/01942638.2019.1615601
4. Dobruch J, Daneshmand S, Fisch M, et al. Gender and bladder cancer: a collaborative review of etiology, biology, and outcomes. Eur Urol. 2016;69:300–310. doi:10.1016/j.eururo.2015.08.037
5. Fung C, Pandya C, Guancial E, et al. Impact of bladder cancer on health related quality of life in 1476 older Americans: a cross-sectional study. J Urol. 2014;192:690–695. doi:10.1016/j.juro.2014.03.098
6. Smith AB, Jaeger B, Pinheiro LC, et al. Impact of bladder cancer on health-related quality of life. BJU Int. 2018;121:549–557. doi:10.1111/bju.14047
7. Robertson AG, Kim J, Al-ahmadie H, et al. Comprehensive molecular characterization of muscle-invasive bladder cancer. Cell. 2017;171:540–556 e525. doi:10.1016/j.cell.2017.09.007
8. Lv M, Zhong Z, Huang M, et al. lncRNA H19 regulates epithelial-mesenchymal transition and metastasis of bladder cancer by miR-29b-3p as competing endogenous RNA. Biochim Biophys Acta Mol Cell Res. 2017;1864:1887–1899. doi:10.1016/j.bbamcr.2017.08.001
9. Martens-uzunova ES, Böttcher R, Croce CM, et al. Long noncoding RNA in prostate, bladder, and kidney cancer. Eur Urol. 2014;65:1140–1151. doi:10.1016/j.eururo.2013.12.003
10. Hua Q, Lv X, Gu X, et al. Genetic variants in lncRNA H19 are associated with the risk of bladder cancer in a Chinese population. Mutagenesis. 2016;31:531–538. doi:10.1093/mutage/gew018
11. Wang H, Niu L, Jiang S, et al. Comprehensive analysis of aberrantly expressed profiles of lncRNAs and miRNAs with associated ceRNA network in muscle-invasive bladder cancer. Oncotarget. 2016;7:86174–86185. doi:10.18632/oncotarget.13363
12. Ouyang S, Zhou X, Chen Z, et al. LncRNA BCAR4, targeting to miR-665/STAT3 signaling, maintains cancer stem cells stemness and promotes tumorigenicity in colorectal cancer. Cancer Cell Int. 2019;19:72. doi:10.1186/s12935-019-0784-3
13. Wang L, Chunyan Q, Zhou Y, et al. BCAR4 increase cisplatin resistance and predicted poor survival in gastric cancer patients. Eur Rev Med Pharmacol Sci. 2017;21:4064–4070.
14. van Agthoven T, Dorssers LCJ, Lehmann U, et al. Breast Cancer Anti-Estrogen Resistance 4 (BCAR4) drives proliferation of IPH-926 lobular carcinoma cells. PLoS One. 2015;10:e0136845. doi:10.1371/journal.pone.0136845
15. Wei L, Yi Z, Guo K, Long X. Long noncoding RNA BCAR4 promotes glioma cell proliferation via EGFR/PI3K/AKT signaling pathway. J Cell Physiol. 2019;234:23608–23617. doi:10.1002/jcp.28929
16. Siddique H, Al-ghafari A, Choudhry H, et al. Long noncoding RNAs as prognostic markers for colorectal cancer in Saudi patients. Genet Test Mol Biomarkers. 2019;23:509–514. doi:10.1089/gtmb.2018.0308
17. Mohr AM, Mott JL. Overview of microRNA biology. Semin Liver Dis. 2015;35:3–11. doi:10.1055/s-0034-1397344
18. Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;6:376–385. doi:10.1038/nrm1644
19. Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597–610. doi:10.1038/nrg2843
20. Zhou X, Yang PC. MicroRNA: a small molecule with a big biological impact. MicroRNA. 2012;1:1. doi:10.2174/2211536611201010001
21. Li Y, Yan X, Ren L, Li Y. miR-644a inhibits cellular proliferation and invasion via suppression of CtBP1 in gastric cancer cells. Oncol Res. 2018;26:1–8. doi:10.3727/096504016X14772410356982
22. Ebron JS, Shankar E, Singh J, et al. MiR-644a disrupts oncogenic transformation and warburg effect by direct modulation of multiple genes of tumor-promoting pathways. Cancer Res. 2019;79:1844–1856. doi:10.1158/0008-5472.CAN-18-2993
23. Matullo G, Naccarati A, Pardini B. MicroRNA expression profiling in bladder cancer: the challenge of next-generation sequencing in tissues and biofluids. Int J Cancer. 2016;138:2334–2345. doi:10.1002/ijc.29895
24. Shegay PV, Zhavoronkov AA, Gaifullin NM, et al. Potentialities of MicroRNA diagnosis in patients with bladder cancer. Bull Exp Biol Med. 2017;164:106–108. doi:10.1007/s10517-017-3935-3
25. Ueno Y, Fujisaki K, Hosoda S, et al. Transcription factor Tlx1 marks a subset of lymphoid tissue organizer-like mesenchymal progenitor cells in the neonatal spleen. Sci Rep. 2019;9:20408. doi:10.1038/s41598-019-56984-w
26. Vanden Bempt M, Demeyer S, Broux M, et al. Cooperative enhancer activation by TLX1 and STAT5 drives development of NUP214-ABL1/TLX1-positive T cell acute lymphoblastic leukemia. Cancer Cell. 2018;34:271–285 e277. doi:10.1016/j.ccell.2018.07.007
27. Riz I, Hawley TS, Luu TV, Lee NH, Hawley RG. TLX1 and NOTCH coregulate transcription in T cell acute lymphoblastic leukemia cells. Mol Cancer. 2010;9:181. doi:10.1186/1476-4598-9-181
28. Liu W, Ma W, Yuan Y, Zhang Y, Sun S. Circular RNA hsa_circRNA_103809 promotes lung cancer progression via facilitating ZNF121-dependent MYC expression by sequestering miR-4302. Biochem Biophys Res Commun. 2018;500:846–851. doi:10.1016/j.bbrc.2018.04.172
29. Pan J, Li X, Wu W, et al. Long non-coding RNA UCA1 promotes cisplatin/gemcitabine resistance through CREB modulating miR-196a-5p in bladder cancer cells. Cancer Lett. 2016;382:64–76. doi:10.1016/j.canlet.2016.08.015
30. Ye T, Ding W, Wang N, et al. Long noncoding RNA linc00346 promotes the malignant phenotypes of bladder cancer. Biochem Biophys Res Commun. 2017;491:79–84. doi:10.1016/j.bbrc.2017.07.045
31. Wu J, Li W, Ning J, et al. Long noncoding RNA UCA1 targets miR-582-5p and contributes to the progression and drug resistance of bladder cancer cells through ATG7-mediated autophagy inhibition. Onco Targets Ther. 2019;12:495–508. doi:10.2147/OTT.S183940
32. Dai L, Chai C-M, Shen T-Y, et al. LncRNA ITGB1 promotes the development of bladder cancer through regulating microRNA-10a expression. Eur Rev Med Pharmacol Sci. 2019;23:6858–6867. doi:10.26355/eurrev_201908_18725
33. Lenti E, Farinello D, Yokoyama KK, et al. Transcription factor TLX1 controls retinoic acid signaling to ensure spleen development. J Clin Invest. 2016;126:2452–2464. doi:10.1172/JCI82956
34. Nakahara R, Kawai Y, Oda A, et al. Generation of a Tlx1(CreER-Venus) knock-in mouse strain for the study of spleen development. Genesis. 2014;52:916–923. doi:10.1002/dvg.22829
35. Durinck K, Van Loocke W, Van der Meulen J, et al. Characterization of the genome-wide TLX1 binding profile in T-cell acute lymphoblastic leukemia. Leukemia. 2015;29:2317–2327. doi:10.1038/leu.2015.162
36. Liu Y, Easton J, Shao Y, et al. The genomic landscape of pediatric and young adult T-lineage acute lymphoblastic leukemia. Nat Genet. 2017;49:1211–1218. doi:10.1038/ng.3909
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.