Back to Journals » International Journal of General Medicine » Volume 13
Long Non-Coding RNA A2M-AS1 Promotes Breast Cancer Progression by Sponging microRNA-146b to Upregulate MUC19
Authors Liu Y, Zhang Q, Wu J, Zhang H, Li X, Zheng Z, Luo M, Li L, Xiang Y, Yang F, Wu L
Received 24 August 2020
Accepted for publication 15 October 2020
Published 27 November 2020 Volume 2020:13 Pages 1305—1316
DOI https://doi.org/10.2147/IJGM.S278564
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Scott Fraser
Yuncong Liu,* Qi Zhang,* Jing Wu, Hanqun Zhang, Xin Li, Zhaopeng Zheng, Min Luo, Libo Li, Yang Xiang, Feiyue Yang, Li Wu
Department of Oncology, Guizhou Provincial People’s Hospital, Guiyang 550002, Guizhou, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Feiyue Yang; Li Wu
Department of Oncology, Guizhou Provincial People’s Hospital, No. 83, Zhongshan East Road, Guiyang 550002 Guizhou, People’s Republic of China
Tel/Fax +86-0851-85937611
Email [email protected]; [email protected]
Background: Long non-coding RNA (lncRNA) A2M-AS1 has been indicated to be augmented in breast cancer (BC), with its specific function undetermined. Therefore, this study is designed to investigate the mechanism of lncRNA A2M-AS1 in BC.
Methods: The expression of A2M-AS1, microRNA (miR)-146b, and MUC19 in BC tissues and cells was measured. Then, the interaction among A2M-AS1, miR-146b, and MUC19 was detected. After A2M-AS1, miR-146b, and MUC19 expression were altered in BC cells, cell proliferation, invasion, and apoptosis were detected, and the protein levels of Hippo-related proteins (YAP and p-YAP) were evaluated. Tumor growth assay was also performed to validate the effects of A2M-AS1 and miR-146b in vivo.
Results: A2M-AS1 and MUC19 were highly expressed in BC, while miR-146b was poorly expressed. A2M-AS1 acts as a molecular sponge for miR-146b, which targeted and negatively modulated MUC19. A2M-AS1 accelerated BC cell proliferation, invasion, and colony formation and suppressed apoptosis via the miR-146b/MUC19/Hippo axis, which was confirmed in vivo.
Conclusion: Taken above together, an oncogenic role for A2M-AS1 in BC was elicited by acting as a miR-146b sponge to promote MUC19 expression. The findings will present some cues for a further approach to BC.
Keywords: breast cancer, long non-coding RNA A2M-AS1, microRNA-146b, Mucin19, Hippo signaling pathway
Introduction
Among the female population, breast cancer (BC) accounts for 11.6% of all cancers and 6.6% of cancer-related death in 2018 worldwide.1 Recently, there are great advances in therapeutic modalities, such as surgery, radiotherapy, and chemotherapy, while the therapeutic efficacy for BC remains unsatisfactory.2,3 It has been unraveled that there are multiple pathological mechanisms underlying breast tumorigenesis which are still not fully elucidated.4 Therefore, it is essential to determine the molecular mechanisms of BC occurrence and progression to advance BC therapies.
Long non-coding RNAs (lncRNAs) have been indicted as important molecules in tumorigenesis in recent years.5 A prior study has proved that lncRNA A2M-AS1 is involved in pancreatic cancer, with its specific role undetermined.6 Furthermore, A2M-AS1 elevation has been suggested to be related to poor recurrence-free and metastasis-free survival of BC patients, implying its possible prognostic and therapeutic role in BC.7 However, whether A2M-AS1 indeed modulates the progression of BC and how the mechanism works remain unsettled. LncRNAs act as competing endogenous RNAs (ceRNAs) by competitively interacting with shared microRNAs (miRNAs).8 miRNAs are a group of non-coding RNAs that play an important part in cell proliferation and survival as well as tumorigenesis by binding to target mRNAs, contributing to mRNA translational inhibition or degradation.9 Moreover, in our research, LNCIPEDIA (https://lncipedia.org/) predicted that lncRNA A2M-AS1 sponged miR-146b. miR-146b is shown to inhibit invasion and migration of BC cells in vitro, and to suppress lung metastasis in vivo.10 During the present study, TargetScan (http://www.targetscan.org/) predicted a targeting relationship between miR-146b and Mucin19 (MUC19). Mucins are secreted by epithelial cells of many normal human organs, which are of significance in pancreatic cancer treatment.11 More specifically, MUC6 expression was tightly linked to higher histologic grade, lymph node metastasis and HER2-positivity in breast ductal carcinomas.12 Also, the driving role of MUC2 in cell proliferation, apoptosis and metastasis has been indicated in BC.13 Intriguingly, MUC1-C suppressed the activation of the Hippo pathway through the dephosphorylation of the oncogenic YAP protein,14 indicating the possible association between Mucins and the Hippo pathway. The Hippo pathway, evolutionarily conserved from Drosophila to mammals, modulates tissue growth and organ size by restraining cell proliferation and migration and enhancing apoptosis.15 On the basis of the aforementioned evidence, we hypothesized that lncRNA A2M-AS1 had the potency to mediate BC cell function through the miR-146b/MUC19/Hippo axis in vitro and in vivo. Our studies might provide more insights into A2M-AS1 as a potential prognostic biomarker and therapeutic target for BC.
Materials and Methods
Ethics Statement
This project was authorized by the Institutional Review Board of Guizhou Provincial People’s Hospital (Approval date: 2017.05.26) and informed consent was obtained from all patients. All clinic operations followed principles embodied in the Declaration of Helsinki. All animal studies were performed under the approval of the Ethics Committee of Guizhou Provincial People’s Hospital (Approval date: 2019.01.21). The report of animal experiments is in accordance with the ARRIVE guidelines.
Study Subjects
From September 2017 to September 2018, BC tissues and its matched adjacent normal tissues (>5 cm from tumor tissues) of 26 BC patients (27–69 years with a median age of 49 years) treated in Guizhou Provincial People’s Hospital were collected. The inclusion criteria were as follows: (1) patients without severe pulmonary or renal impairment; (2) patients who agreed to be followed-up; (3) patients without a history of severe cardiovascular and immune system disease; and (4) patients without chronic or acute infectious diseases. Patients with preoperative adjuvant chemotherapy were excluded. The tissue samples were stored in liquid nitrogen at −80°C. All patients were diagnosed and confirmed pathologically. Detailed clinicopathological characteristics of the patients are listed in Table 1.
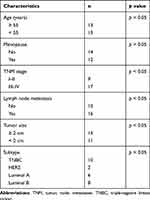 |
Table 1 The Clinicopathological Characteristics of Patients Included |
Cell Culture and Grouping
BC cells MCF-7 (ZQ0071), MDA-MB-231 (ZQ0118), MDA-MB-468 (ZQ0373) (Shanghai Zhong Qiao Xin Zhou Biotechnology Co., Ltd., Shanghai, China) and normal mammary epithelial cells MCF-10A (American Type Culture Collection, Manassas, VA, USA) were maintained in Dulbecco’s modified Eagle’s medium (DMEM) comprised of 10% fetal bovine serum (FBS), maintained with 5% CO2 at 37°C. After adherence, cells were detached by 0.25% trypsin (Hyclone, Marlborough, MA, USA) for the experiments.
Cells were transfected with miR-146b mimic, A2M-AS1 overexpression plasmid (oe-A2M-AS1), shRNA targeting A2M-AS1 (sh-A2M-AS1), MUC19 overexpression plasmid (oe-MUC19), miR-146b mimic + oe-A2M-AS1, miR-146b mimic + oe-MUC19 or sh-A2M-AS1 + oe-MUC19 as well as their respective controls. The overexpression vectors were packaged with adenovirus (Genomeditech, Shanghai, China, http://www.genomeditech.com). After transfection, cells were maintained in an incubator (37°C, 5% CO2, saturated humidity) for 6 h. The medium containing the transfection solution was replaced by the DMEM with 10% FBS (Santa Cruz Biotechnology Inc., Santa Cruz, CA, USA) for another 48-h incubation.
5-Ethynyl-2ʹ-Deoxyuridine (EdU) Assay
Cells were seeded into 24-well plates with three replicates in each group. Cells were incubated with EdU (10 µmol/L, Thermo Fisher, Waltham, MA, USA) for 2 h. The cells were fixed and stained based on the instructions of the EdU kits. After being observed and photographed under a fluorescence microscope (FM-600, Pudan Optical Instrument, Shanghai, China), cells in 6–10 random fields of view were counted, and the EdU-positive rate was calculated.
Colony Formation Assay
The cells were seeded in 75-mm plates (800 cells/plate), cultured for 9 days, fixed in methanol for 20 min, stained with 10% Giemsa for 20 min, washed with deionized water, and dried. The number of colonies with more than 20 cells per plate was counted under the anatomical microscope, and the colony formation rate was calculated.
Transwell Assay
The Matrigel (356,234, BD Biosciences, NJ, USA) was placed at 4°C overnight, and cells were diluted by serum-free medium with 1:3. Matrigel (50 μL) was added on the transwell apical chamber and solidified in the incubator for 30 min. Cells were suspended in serum-free medium (1 × 105 cells/mL), incubated for 12 h. The basolateral chamber was supplemented with a medium containing 10% FBS. After the addition of 100 μL cell suspension, the chamber was cultured for 1 d at 37°C. The non-invaded cells in the Matrigel membrane were wiped off by cotton swabs. Cells were fixed with methanol, stained by toluidine blue (Sigma-Aldrich, MO, USA) and photographed under the inverted microscope (CarlZeiss, Oberkochen, Germany). The number of permeating cells was counted manually in five randomly selected areas.
Flow Cytometry
Flow cytometry was conducted using Annexin V-FITC/PI apoptosis detection kit (MA0220, Dalian Meilun Biotech Co., Ltd., Dalian, China). Cells (2–5 × 105 cell/mL) were subjected to a 5-mins centrifugation at 500 g. Cells were resuspended with 195 μL binding buffer, incubated avoiding light exposure with 5 μL Annexin V-FITC for 10 min and with 10 μL PI (20 μg/mL) for 5 mins and detected by a flow cytometer.
Dual-Luciferase Reporter Gene Assay
LNCIPEDIA (https://lncipedia.org/) and TargetScan (http://www.targetscan.org/) were used to predict the binding sites between lncRNA A2M-AS1 and miR-146b as well as between miR-146b and MUC19, which were verified by dual-luciferase reporter gene assay. The PGLO-A2M-AS1-WT, PGLO-A2M-AS1-MUT, PGLO-MUC19-WT, and PGLO-MUC19-MUT were constructed and co-transfected with miR-146b mimic into HEK293T cells for 48 h. The cells were lysed to obtain the supernatant using Trans Detect Double-Luciferase Reporter Assay Kit (FR201-01, TransGen Biotech, Beijing, China). Dual-luciferase reporter gene assay system (E1910, Promega Corporation, Madison, WI, USA) was utilized to test luciferase activity. The relative luciferase activity = firefly luciferase activity/renilla luciferase activity.
RNA-Fluorescence in situ Hybridization (FISH)
FISH was conducted using a specific probe of A2M-AS1 sequence. In short, the Cy5-labeled probe was specific for A2M-AS1. DAPI was utilized to stain nuclei. The subcellular localization of A2M-AS1 was detected by an RNA-FISH Probe Kit (Shanghai GenePharma Co., Ltd., Shanghai, China). Fluorescence images were captured by a Zeiss LSM880 NLO (2 + 1 with BIG) confocal scanning microscope (Leica Microsystems, Mannheim, Germany).
RNA Immunoprecipitation (RIP) Assay
Using RIP kit (Millipore, Billerica, MA, USA), the cell extract supernatant with the treated beads for 6 h was subjected to washing with RIP buffer (6 times). The purified RNA was used to assess A2M-AS1 expression by RT-qPCR.
RNA Pull-Down Assay
The 50 nM biotin-labeled WT-bio-LncRNA A2M-AS1 and MUT-bio-LncRNA A2M-AS1 were transfected into MCF-7 cells. After 48 h, cells were maintained in the specific lysate (Ambion, Austin, TX, USA) for 10 min, and centrifuged at 14,000 g to obtain the supernatant. The remaining lysate was co-cultured with M-280 streptavidin magnetic beads (S3762, Sigma) that have been pre-coated by RNase-free and yeast tRNA (TRNABAK-RO, Sigma) for 3 h, then rinsed twice with cool lysis buffer, three times with low salt buffer and once with high salt buffer. The total RNA was extracted to determine A2M-AS1 expression with RT-qPCR.
RT-qPCR
TRIZOL reagents (15596–018, Solarbio, Beijing, China) were first applied for total RNA extraction from cells. BC and adjacent normal tissues were added to tubes (100 mg/mL) with TRIzol, broken up with an ultrasonic breaker and incubated on ice for 10 min. After centrifugation at 12000 g at 4°C for 10 min, 700 μL supernatant was incubated on ice with chloroform for 10 min, centrifuged again at 12000 g at 4°C for 10 min, and the supernatant was discarded. Cells in each tube were added with 1 mL freshly prepared 75% ethanol, centrifuged at 4°C at 12,000 g for 15 min, and air-dried. Afterwards, 30 mL diethyl pyrocarbonate (DEPC) was added to dissolve the RNA. RNA concentration was determined by NanoDrop ND-2000 (Thermo Fisher scientific), and RNA quality was assessed using formaldehyde denaturing gels. Next, reverse transcription was implemented using TaqMan MicroRNA Assays Reverse Transcription primer (4427975, Applied Biosystems, Foster City, CA, USA)/PrimeScript RT reagent Kit (RR047A, Takara). Reverse transcription system was prepared as follows: 5 μg total RNA, 1 μL Oligo(dT)20, 1 μL 10 mmol/L dNTP, 3 μL 0.1mol/L DTT and 10 μL DEPC. The reaction cycle was pre-denaturation at 92°C for 2 min, denaturation at 95°C for 20 s, annealing at 60°C for 40 s, and extension at 72°C for 2 min for a total of 35 cycles. Fluorescence quantification of cDNA was performed by SYBR Premix Ex Taq II Kit (Takara) with a PCR system of 2 μL cDNA, 12.5 μL SYBR premix, 1 μL Primer and 9.5 μL DEPC. TaKaRa was authorized to synthesize the primers (Table 2) used in the assay. ABI 7500 system (ABI, CA, USA) was employed for RT-qPCR. The target product was sequenced at Sangon Biological Engineering Technology & Services Co., Ltd. (Shanghai, China). The specificity of the gene and the origin of the product species were verified by the Blast program (NCBI, USA), and the primers and gene product were shown to be human-specific sequences in the Blast program. The relative expression of genes was determined by 2−ΔΔCt method. Glyceraldehyde phosphate dehydrogenase (GAPDH) and U6 were, respectively, used as normalization controls. The amplification efficiencies of the target and reference genes were calculated by relative standard curves which were close to 100%, and the efficiency deviation was within 5%.
 |
Table 2 Primer Sequences for RT-qPCR |
Immunoblotting
Total protein was extracted using radioimmunoprecipitation assay lysis buffer (R0010, Solarbio), lysed for 15 min at 4°C, followed by a centrifugation (12000 rpm, 15 min). Bicinchoninic acid (20201ES76, Yisheng Biotechnology Co., Ltd., Shanghai, China) was utilized to assess the protein concentration. After separation by means of 8%–14% sodium dodecyl sulfate-polyacrylamide gel electrophoresis, the isolated proteins (15 μg) were transferred to the polyvinylidene fluoride membrane, which was blocked for a period of 1 h with the use of 5% BSA. The membrane was subjected to an incubation overnight at a temperature of 4°C with diluted primary antibodies to MUC19 (ab123813, 1:1000, Abcam, Cambridge, UK), p-YAP (ab62751, 1:1000, Abcam), YAP (ab81183, 1:1000, Abcam), and GAPDH (ab8245, 1:1000, Abcam). The membrane was subjected to additional incubation with horseradish peroxidase (HRP)-labelled goat anti-rabbit IgG (ab205718, 1:20000, Abcam) or goat anti-mouse (ab6789, 1:5000, Abcam) for a period of 1 h. After that, the membrane was visualized by chemiluminescence (ECL). Grayscale analysis was implemented with the use of ImageJ 1.48 u Software (National Institutes of Health). The relative ratio was calculated with GAPDH deemed as the internal reference.
Xenograft Studies
Twenty 6-week-old female nude mice (weighing 16–18 g) provided by Hunan SJA Laboratory Animal Co., Ltd. (Hunan, China) were housed in specific-pathogen-free conditions at 24°C–26°C with the relative humidity of 40%~60%. MCF-7 cells stably transfected with sh-NC, sh-A2M-AS1, sh-A2M-AS1 + mimic NC, sh-A2M-AS1 + miR-146b mimic plasmids were dispersed into single-cell suspension, and 0.1 mL cell suspension (1 × 106 cells/mL MCF-7 cells) was subcutaneously injected into each nude mouse. Volume changes in nude mice were recorded on the basis of the formula: V = (A × B2)/2 (A was long diameter, B was short diameter, mm3). After 35 days, the nude mice received euthanasia by CO2, followed by the removal of tumors, photograph and weighing.
Statistical Analysis
The data were represented as mean ± standard deviation, which were implemented by means of unpaired t-test between two groups or one-way analysis of variance (ANOVA) or two-way ANOVA for more than two groups. The values were considered significantly different at ap < 0.05 using SPSS 21.0 software (IBM Corp. Armonk, NY, USA).
Results
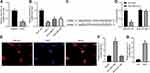
LncRNA A2M-AS1 Expression is Elevated in BC Tissues and Cells
The quantitation by RT-qPCR showed that A2M-AS1 expression in BC tissues was higher than that in adjacent normal tissues (p < 0.05) (Figure 1A). Consistently, A2M-AS1 expression was elevated in BC cells (MCF-7, MDA-MB-231, and MDA-MB-468) compared with MCF-10 cells (p < 0.05) (Figure 1B).
A2M-AS1 Knockdown Restrains BC Cell Malignant Phenotype
MCF-7 cells with the highest expression of A2M-AS1 were treated with A2M-AS1 knockdown, and MDA-MB-231 cells with the poorest expression of A2M-AS1 were treated with A2M-AS1 overexpression. RT-qPCR validated that A2M-AS1 expression was elevated in MDA-MB-231 cells transfected with expression vectors containing A2M-AS1, and decreased in MCF-7 cells transfected with sh-A2M-AS1 (p < 0.05) (Figure 2A).
EdU, Transwell, colony formation assay, and flow cytometry verified that overexpressed A2M-AS1 promoted MDA-MB-231 cell proliferation, invasion and colony formation and delayed the apoptosis, while the opposite results were observed in MCF-7 cells which were treated by sh-A2M-AS1 (p < 0.05) (Figure 2B–E).
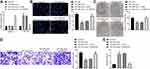
LncRNA A2M-AS1 Sponges miR-146b in BC Cells
The quantitation by RT-qPCR showed that miR-146b expression in BC tissues decreased (p < 0.05) (Figure 3A). Consistently, miR-146b expression decreased in BC cells (MCF-7, MDA-MB-231, and MDA-MB-468) compared with MCF-10 cells (p < 0.05) (Figure 3B).
LNCIPEDIA (https://lncipedia.org/) predicted the binding relationship between A2M-AS1 and miR-146b (Figure 3C). A dual-luciferase reporter assay (Figure 3D) displayed that elevated miR-146b reduced the A2M-AS1-WT luciferase activity (p < 0.05), while the A2M-AS1-MUT luciferase activity showed no obvious difference (p > 0.05). RNA-FISH showed that A2M-AS1 localized to the cytoplasm (Figure 3E). RIP demonstrated that Ago2 was able to enrich A2M-AS1 compared with IgG (Figure 3F). In addition, the RNA pull-down assay showed that WT-miR-146b significantly increased the enrichment of A2M-AS1 (p < 0.05) (Figure 3G).
A2M-AS1 Accelerates BC Cell Proliferation, Invasion, and Colony Formation and Suppresses Apoptosis by Competitively Binding to miR-146b
In order to study the function of A2M-AS1 and miR-146b in BC, we upregulated miR-146b and/or A2M-AS1 in MCF-7 cells. RT-qPCR (Figure 4A) displayed that compared with MCF-7 cells transfected with mimic NC, miR-146b expression elevated (p < 0.05), and A2M-AS1 expression showed no obvious difference (p > 0.05) in MCF-7 cells treated with miR-146b mimic. Simultaneous overexpression of A2M-AS1 and miR-146b increased A2M-AS1 expression and reduced miR-146b expression in MCF-7 cells in comparison to miR-146b restoration alone.
It was verified that MCF-7 cell proliferation, invasion and colony formation abilities were repressed, and the apoptosis was facilitated by the treatment of miR-146b mimic. Moreover, simultaneous restoration of A2M-AS1 and miR-146b accelerated MCF-7 cell proliferation, invasion and colony formation, and inhibited apoptosis in comparison to miR-146b restoration alone (p < 0.05) (Figure 4B–E). Overall, upregulation of miR-146b reversed the promoting effects of A2M-AS1 in BC cell activity.
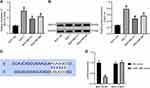
miR-146b Targets and Negatively Regulates MUC19
The results of RT-qPCR and immunoblotting exhibited that MUC19 expression was elevated in BC cells (MCF-7, MDA-MB-231, and MDA-MB-468) compared with MCF-10A cells (p < 0.05) (Figure 5A and B).
TargetScan (http://www.targetscan.org/) predicted the targeting relationship between miR-146b and MUC19 (Figure 5C). Moreover, the dual-luciferase reporter assay was used to verify the binding relationship between miR-146b and MUC19 (Figure 5D). miR-146b mimic reduced the luciferase activity of MUC19-3ʹ-UTR-WT (p < 0.05), while the MUC19-3ʹ-UTR-MUT luciferase activity showed no evident difference (p > 0.05). Therefore, the obtained data suggested that miR-146b could target MUC19 and inhibited MUC19 expression.
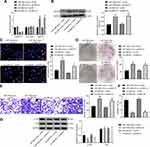
A2M-AS1 Inhibition Prevents the BC Progression in vitro via the miR-146b/MUC19/Hippo Axis
MCF-7 cells treated with miR-146b mimic or A2M-AS1 knockdown were transfected with MUC19 overexpression, respectively. MUC19 expression increased in MCF-7 cells received miR-146b mimic + oe-MUC19 relative to MCF-7 cells treated with miR-146b mimic + oe-NC. Similar upregulation of MUC19 was observed in MCF-7 cells treated with sh-A2M-AS1 + oe-MUC19 relative to sh-A2M-AS1+ oe-NC (p < 0.05) (Figure 6A and B).
EdU, Transwell, colony formation assay, and flow cytometry confirmed that both upregulating miR-146b and MUC19 accelerated cell proliferation, invasion and colony formation and diminished the apoptosis compared with MCF-7 cells treated with miR-146b mimic + oe-NC. The treatment of sh-A2M-AS1+ oe-MUC19 contributed to the consistent results in comparison to MCF-7 cells treated with sh-A2M-AS1 + oe-NC (p < 0.05) (Figure 6C–F).
Immunoblotting (Figure 6G) presented that YAP expression showed no evident difference (p > 0.05) and p-YAP expression reduced (p < 0.05) in MCF-7 cells received miR-146b and MUC19 upregulation relative to MCF-7 cells received miR-146b mimic + oe-NC. Compared with MCF-7 cells treated with sh-A2M-AS1 + oe-NC, A2M-AS1 knockdown and overexpressed MUC19 exerted no effects on YAP expression (p > 0.05), while reduced p-YAP expression (p < 0.05). This evidence shows that A2M-AS1 regulated miR-146b/MUC19/Hippo axis, thus promoting BC progression.
Overexpression of A2M-AS1 Promotes the Tumor Growth in vivo by Downregulating miR-146b
Finally, we tested the effect of A2M-AS1 and miR-146b in tumorigenesis of BC cells in nude mice. Results (Figure 7A–C) displayed that A2M-AS1 upregulation potently increased the subcutaneous tumor volume and weight, which was reversed by miR-146b mimic (p < 0.05). In conclusion, A2M-AS1 overexpression contributed to tumorigenesis of BC cells in vivo, while elevated miR-146b exerted opposite effects on BC development.
Discussion
BC, the most prevalent malignant tumor in women around the world, remains the second most common factor for cancer-associated death in women despite the decreasing mortality rates.16 Existing literature has confirmed that lncRNAs function as a ceRNA to affect the BC progression by regulating miRNAs.17 Presently, lncRNA A2M-AS1 and MUC19 expression were much higher in BC tissues and cells, whereas miR-146b expression was lower. Downregulated A2M-AS1 suppressed BC cell proliferation, colony formation and invasion, and facilitated apoptosis. Moreover, A2M-AS1 was identified to be bound to miR-146b, and miR-146b expression could be conversely regulated by A2M-AS1 in BC cells. Dual-luciferase reporter assay presented that MUC19 3ʹ-UTR was targeted by miR-146b in BC cells. A2M-AS1 and miR-146b regulated the activity of the Hippo pathway via MUC19.
Our initial observations revealed that lncRNA A2M-AS1 was highly expressed in BC, and downregulated A2M-AS1 restrained BC cell proliferation and invasion and promoted apoptosis. LncRNA A2M-AS1 is demonstrated to influence the progression of pancreatic cancer.6 Interestingly, Fang et al have elucidated that lncRNA A2M-AS1 was also enriched in BC, which facilitated BC cell proliferation, invasion, and migration,7 while the roles of lncRNA A2M-AS1 in BC remain to be further explored. Following the evaluation of the functional relevance of lncRNA A2M-AS1 to BC cell function, this study demonstrated that lncRNA A2M-AS1 regulated BC cell biological activity by sponging miR-146b. LncRNAs mediated gene expression at transcriptional, translational, and post-translational levels.18 Since the proposal of ceRNA hypothesis in 2011,19 emerging evidence indicated that there are communications among all classes of RNA transcripts to modulate the expression patterns of each other via competing for shared sequences in miRNAs.20 In our study, dual-luciferase, RNA-FISH, RIP, and RNA pull-down assays verified that A2M-AS1 bound to miR-146b and downregulated miR-146b expression. Similar to our observations, Lnc-AL445665 promoted the progression of multiple uterine leiomyomas by sponging miR-146b-5p.21 Meanwhile, lncRNA SOX2-OT played as a tumor-supporting gene in nasopharyngeal carcinoma progression by interacting with miR-146b-5p.22 miR-146b expression was reduced in multiple human cancers, such as glioma and hepatocellular carcinoma, which was correlated with repressed cell invasion, migration, and viability.23,24 Consistently, miR-146b expression was decreased in BC in the current study. Hurst et al have found that miR-146b was downregulated in BC, and its overexpression limited BC cell migration and invasion to delay the BC development.10 Therefore, these findings support that lncRNA A2M-AS1 has the potential to function as a possible biomarker to hamper BC development by sponging miR-146b.
In addition, this study demonstrated that miR-146b targeted MUC19 and miR-146b elevation consequently suppressed its expression. Mucins, high molecular weight and multifunctional glycoproteins, are reported to protect and lubricate the luminal surfaces of epithelium-lined ducts in the human body.25 In addition, the secreted and transmembrane mucins are being recognized as effectors of carcinogenesis.26 It was noted that MUC2 are upregulated in BC, which could drive the cancer progression.13 A recent study has investigated that MUC4 conferred Trastuzumab resistance to HER2-positive BC cells.27 Also, MUC1 was also proved to predict the BC metastasis.28 Under the context of MUC19, MUC19 expression was notably increased in BC tissues and cells, and MUC19 was found to be targeted by miR-593.29 Furthermore, the Hippo pathway is known as a tumor inhibitor to participate in BC development.30 Hippo signaling pathway is involved in proliferation, migration, invasion, and stemness to affect BC progression.31 However, the linkage between miR-146b and the Hippo signaling pathway has not yet been explored. Our data provided that overexpression of MUC19 abrogated the stimulative role of miR-146b mimic or sh-A2M-AS1 on the Hippo signaling pathway by reducing the extent of YAP phosphorylation. Thus, overexpressed miR-146b activated Hippo signaling, which was negatively regulated by MUC19, thereby inhibiting cell proliferation, invasion, and colony formation and promoting apoptosis in BC.
Conclusions
To briefly conclude, the present study determined that lncRNA A2M-AS1 accelerated BC cell proliferation, invasion, and colony formation and delayed apoptosis by sponging miR-146b to upregulate MUC19 and disrupt Hippo signaling pathway. The result could open a new treatment target in BC treatment field. Further investigation into the specific regulatory mechanisms of the lncRNA A2M-AS1/miR-146b/MUC19/Hippo axis in BC will be summarized in our next study due to the limited reports on the relationship between lncRNA A2M-AS1 and the miR-146b/MUC19/Hippo axis.
Funding
This work was supported by Gui Zhou Provincial Science and Technology Bureau Joint Foundation (Gui Zhou LH [2016] 7197).
Disclosure
The authors declare that they have no conflicts of interest.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.21492
2. Ahmed S, Pati S, Le D, Haider K, Iqbal N. The prognostic and predictive role of 21-gene recurrence scores in hormone receptor-positive early-stage breast cancer. J Surg Oncol. 2020;122(2):144–154. doi:10.1002/jso.25952
3. Ferreira Almeida C, Oliveira A, Joao Ramos M, Fernandes PA, Teixeira N, Amaral C. Estrogen receptor-positive (ER(+)) breast cancer treatment: are multi-target compounds the next promising approach? Biochem Pharmacol. 2020;177:113989. doi:10.1016/j.bcp.2020.113989
4. Annaratone L, Cascardi E, Vissio E, et al. The multifaceted nature of tumor microenvironment in breast carcinomas. Pathobiology. 2020;87(2):125–142. doi:10.1159/000507055
5. Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Mol Cancer. 2011;10:38. doi:10.1186/1476-4598-10-38
6. Giulietti M, Righetti A, Principato G, Piva F. LncRNA co-expression network analysis reveals novel biomarkers for pancreatic cancer. Carcinogenesis. 2018;39(8):1016–1025. doi:10.1093/carcin/bgy069
7. Fang K, Caixia H, Xiufen Z, Zijian G, Li L. Screening of a novel upregulated lncRNA, A2M-AS1, that promotes invasion and migration and signifies poor prognosis in breast cancer. Biomed Res Int. 2020;2020:9747826. doi:10.1155/2020/9747826
8. Yamamura S, Imai-Sumida M, Tanaka Y, Dahiya R. Interaction and cross-talk between non-coding RNAs. Cell Mol Life Sci. 2018;75(3):467–484.
9. Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16(3):203–222. doi:10.1038/nrd.2016.246
10. Hurst DR, Edmonds MD, Scott GK, Benz CC, Vaidya KS, Welch DR. Breast cancer metastasis suppressor 1 up-regulates miR-146, which suppresses breast cancer metastasis. Cancer Res. 2009;69(4):1279–1283. doi:10.1158/0008-5472.CAN-08-3559
11. Torres MP, Chakraborty S, Souchek J, Batra SK. Mucin-based targeted pancreatic cancer therapy. Curr Pharm Des. 2012;18(17):2472–2481. doi:10.2174/13816128112092472
12. Do SI, Kim K, Kim DH, et al. Associations between the expression of mucins (MUC1, MUC2, MUC5AC, and MUC6) and clinicopathologic parameters of human breast ductal carcinomas. J Breast Cancer. 2013;16(2):152–158. doi:10.4048/jbc.2013.16.2.152
13. Astashchanka A, Shroka TM, Jacobsen BM. Mucin 2 (MUC2) modulates the aggressiveness of breast cancer. Breast Cancer Res Treat. 2019;173(2):289–299. doi:10.1007/s10549-018-4989-2
14. Alam M, Bouillez A, Tagde A, et al. MUC1-C represses the crumbs complex polarity factor CRB3 and downregulates the Hippo pathway. Mol Cancer Res. 2016;14(12):1266–1276. doi:10.1158/1541-7786.MCR-16-0233
15. Zhou X, Wang S, Wang Z, et al. Estrogen regulates Hippo signaling via GPER in breast cancer. J Clin Invest. 2015;125(5):2123–2135. doi:10.1172/JCI79573
16. Matamala N, Vargas MT, Gonzalez-Campora R, et al. Tumor microRNA expression profiling identifies circulating microRNAs for early breast cancer detection. Clin Chem. 2015;61(8):1098–1106. doi:10.1373/clinchem.2015.238691
17. Crudele F, Bianchi N, Reali E, Galasso M, Agnoletto C, Volinia S. The network of non-coding RNAs and their molecular targets in breast cancer. Mol Cancer. 2020;19(1):61.
18. Amelio I, Bernassola F, Candi E. Emerging roles of long non-coding RNAs in breast cancer biology and management. Semin Cancer Biol. 2020. doi:10.1016/j.semcancer.2020.06.019
19. Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146(3):353–358. doi:10.1016/j.cell.2011.07.014
20. Abdollahzadeh R, Daraei A, Mansoori Y, Sepahvand M, Amoli MM, Tavakkoly-Bazzaz J. Competing endogenous RNA (ceRNA) cross talk and language in ceRNA regulatory networks: a new look at hallmarks of breast cancer. J Cell Physiol. 2019;234(7):10080–10100. doi:10.1002/jcp.27941
21. Yang E, Xue L, Li Z, Yi T. Lnc-AL445665. 1–4 may be involved in the development of multiple uterine leiomyoma through interacting with miR-146b-5p. BMC Cancer. 2019;19(1):709. doi:10.1186/s12885-019-5775-1
22. Zhang E, Li X. LncRNA SOX2-OT regulates proliferation and metastasis of nasopharyngeal carcinoma cells through miR-146b-5p/HNRNPA2B1 pathway. J Cell Biochem. 2019;120(10):16575–16588. doi:10.1002/jcb.28917
23. Katakowski M, Buller B, Zheng X, et al. Exosomes from marrow stromal cells expressing miR-146b inhibit glioma growth. Cancer Lett. 2013;335(1):201–204. doi:10.1016/j.canlet.2013.02.019
24. Li C, Miao R, Liu S, et al. Down-regulation of miR-146b-5p by long noncoding RNA MALAT1 in hepatocellular carcinoma promotes cancer growth and metastasis. Oncotarget. 2017;8(17):28683–28695. doi:10.18632/oncotarget.15640
25. Mukhopadhyay P, Chakraborty S, Ponnusamy MP, Lakshmanan I, Jain M, Batra SK. Mucins in the pathogenesis of breast cancer: implications in diagnosis, prognosis and therapy. Biochim Biophys Acta. 2011;1815(2):224–240.
26. Kufe DW. Mucins in cancer: function, prognosis and therapy. Nat Rev Cancer. 2009;9(12):874–885. doi:10.1038/nrc2761
27. Mercogliano MF, De Martino M, Venturutti L, et al. TNFalpha-induced mucin 4 expression elicits trastuzumab resistance in HER2-positive breast cancer. Clin Cancer Res. 2017;23(3):636–648. doi:10.1158/1078-0432.CCR-16-0970
28. Ideo H, Hinoda Y, Sakai K, et al. Expression of mucin 1 possessing a 3ʹ-sulfated core1 in recurrent and metastatic breast cancer. Int J Cancer. 2015;137(7):1652–1660. doi:10.1002/ijc.29520
29. Song L, Xiao Y. Downregulation of hsa_circ_0007534 suppresses breast cancer cell proliferation and invasion by targeting miR-593/MUC19 signal pathway. Biochem Biophys Res Commun. 2018;503(4):2603–2610. doi:10.1016/j.bbrc.2018.08.007
30. Wu L, Yang X. Targeting the Hippo pathway for breast cancer therapy. Cancers (Basel). 2018;10(11):422. doi:10.3390/cancers10110422
31. Shi P, Feng J, Chen C. Hippo pathway in mammary gland development and breast cancer. Acta Biochim Biophys Sin (Shanghai). 2015;47(1):53–59. doi:10.1093/abbs/gmu114
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.