Back to Journals » OncoTargets and Therapy » Volume 13
lncRNAHIF1A-AS2 Promotes Renal Carcinoma Cell Proliferation and Migration via miR-130a-5p/ERBB2 Pathway
Authors Zhu Y, Yang Z, Chen H, Pan Y, Gong L, Chen F, Jin X, Wen S, Li Y, Chen G
Received 8 May 2020
Accepted for publication 4 August 2020
Published 1 October 2020 Volume 2020:13 Pages 9807—9820
DOI https://doi.org/10.2147/OTT.S260191
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Federico Perche
Yunxiao Zhu, Ziyi Yang, Han Chen, Yang Pan, Lifeng Gong, Falin Chen, Xiaoxiang Jin, Shuang Wen, Yi Li, Gang Chen
Department of Urology, The First Affiliated Hospital of Chongqing Medical University, Chongqing 400010, People’s Republic of China
Correspondence: Gang Chen
Department of Urology, The First Affiliated Hospital of Chongqing Medical University, Chongqing 400010, People’s Republic of China
Tel +86 15823057887
Email [email protected]
Background: Long non-coding RNAs (lncRNAs) are essential for tumorigenesis and progression of diverse cancers. This study aims to investigate the roles of lncRNAs on renal carcinoma.
Methods: The expression of lncRNA HIF1A-AS2 in clear cell renal cell carcinoma (ccRCC) and adjacent non-cancer tissues was identified by quantitative real-time PCR (qRT-PCR). Investigations were performed on biological function of lncRNA HIF1A-AS2 on cell proliferation, cell cycle, apoptosis and invasion of ccRCC by overexpression and knockdown experiments. Further, luciferase reporter assay and Western blot were constructed to explore molecular mechanisms underlying the function of lncRNA HIF1A-AS2.
Results: HIF1A-AS2 was highly expressed in kidney cancer tissues and ccRCC cells. Interference of HIF1A-AS2 in vivo hindered cell proliferation, invasion and migration while accelerated cell apoptosis. Overexpression of HIF1A-AS2 presented an opposite effect that repressed the expression of miR-130a-5p, and miR-130a-5p inhibited the expression of HIF1A-AS2. Additionally, rescue experiments exhibited that oncogenic function of HIF1A-AS2 was partially dependent on the suppression of miR-130a-5p.
Conclusion: Our results indicated a critical role for the HIF1A-AS2-miR-130a-5p axis in renal carcinoma progression, which may act as a promising diagnostic biomarker and a pivotal therapeutic target for renal carcinoma cures.
Keywords: lncRNA HIF1A-AS2, miR-130a-5p, renal carcinoma cell, cell proliferation, cell migration
Introduction
As one of high mortality cancers, renal carcinoma presents little clinical symptoms in early stages. Being diagnosed at an advanced stage plus poor prognosis, renal carcinoma patients suffer from a high failure rate of chemotherapy.1 Novel and high efficient options are in urgent need to improve the therapeutic efficacy of this disease. In general, tumorigenesis and progression are multi-step and multi-stage processes involved in extensive genomic alterations and gene mutations.2 Mechanisms of tumorigenesis and progression have been figured out at a molecular level which provides addition of effective alternative for the treatment of renal malignancies.3
Recognized as a type of RNA transcripts exceeding 200 nt, lncRNAs regulate a broad array of processes both physiologically and pathologically in cancer, including cell cycles, chromatin modification, gene transcription and translation, cellular differentiation and oncogenic or tumor suppressive signals.4–6 Growing evidence has indicated a crucial role for lncRNAs in the initiation and progression of multiple cancer types. However, little has known about the function and mechanism of lncRNAs in renal carcinoma, and further explorations remain necessary.
Increasing investigations have suggested both the small endogenous single-stranded non-coding RNAs and microRNAs (miRNAs) are crucial players in renal carcinoma, while how miR-130a-5p regulates post-transcriptionally is still unclear. Growing evidence has indicated that lncRNA functions as a competitive endogenous RNA (ceRNA) by binding multiple miRNAs through competitions.7–9 Frequent interactions between lncRNAs and miRNAs fulfill respective biological functions in a physiological and pathological manner. An example of HOTAIR depletion mediated by RNAi has been reported significantly reduced the proliferation and survival of renal carcinoma cells both in vitro and in vivo.10 SOX2-OT has been considered to be another common driver lncRNA in kidney carcinoma cells, which participates in the regulation of oncogene SOX2.11 Unluckily, no complete narration has been made on the association of miR-130a-5p with lncRNAs. As an important target for cancer therapy, ERBB2 enhances the expression in cancers and involves in essential signaling processes.12
Our study aims to investigate the differentially expressed HIF1A-AS2 in renal carcinoma tissues in contrast to normal tissues. The disabled HIF1A-AS2 weakens the proliferation, migration, and invasion of the renal carcinoma cells. Importantly, HIF1A-AS2 serves as a ceRNA sponging miR-130a-5p and facilitates the expression of ERBB2 and it has been confirmed that HIF1A-AS2 accelerates renal carcinoma tumorigenesis and metastasis by suppressing miR-130a-5p. Furthermore, our findings are expected to present a fresh biomarker as well as a robust therapeutic target for renal carcinoma.
Materials and Methods
Clinical Tissue Samples
A total of 42 cases of RCC renal cancer and 42 paired non-cancerous renal tissues as control were collected from patients who underwent partial or radical nephrectomy in The First Affiliated Hospital of Chongqing Medical University. The tissues were immediately frozen in liquid nitrogen and stored at −80°C. The postoperative specimens were diagnosed as ccRCC by two pathologists. Patients were aged from 34 to 76 years old (27 males, 15 females). Clinical stages and T stages were determined according to the 2010 International Union against Cancer (UICC)/American Joint Committee on Cancer (AJCC) TNM classification guidelines.13 Five patients were in the TNM stage I/II and 33 patients were in the TNM stage III/IV. This study was approved by the Ethics Committee of The First Affiliated Hospital of Chongqing Medical University (approval number: 2019-S15) and informed consent was obtained from the patients.
Cell Culture
Renal carcinoma cell lines, namely, ACHN, OSRC-2, 786-O Caki-1, and human normal renal tubular epithelial cell line HK2 cells were obtained from the American Type Culture Collection (ATCC, USA). ACHN, OSRC-2, 786-O, and Caki-1 cells were cultured in DMEM (Gibco, USA) with 10% of fetal bovine serum (FBS) (Hyclone, USA) and 1% of penicillin/streptomycin (P/S) (Gibco, USA). HK-2 cells were cultured in keratinocyte medium (KM, ScienCell, USA) with 1% of keratinocyte growth supplement (KGS, ScienCell, USA) and 1% of penicillin/streptomycin (ScienCell, USA). All cells were cultivated with 5% CO2 at 37°C.
Cell Transfection and Vector Construction
The miR-130a-5p mimics, inhibitor and related negative controls (NCs) were purchased from the Guangzhou Ribobio Co. Ltd. (China). Lentiviral HIF1A-AS2-OE was purchased from the Chongqing Biomedicine Biotechnology Co., Ltd. (China). Both siRNAs and miRNA were transfected in ACHN and OSRC-2 cells by using Lipofectamine RNAiMAX (Invitrogen, USA) following the manufacturer’s instructions of use. The miR-130a-5p inhibitor or HIF1A-AS2-OE was transfected in ACHN and OSRC-2 cells. In brief, cells were cultured in a 6-well plate for fusion at 50–70% before transfection. Of 5 mL siRNA (20 mM) (or miRNA mimic (20 mM)) was diluted into 100 mL of Opti-MEM medium (GIBCO, USA), and 6 mL of Lipofectamine RNAiMAX was added into the prepared 100 mL of Opti-MEM medium. After 5 minutes, diluted siRNA (or miRNA mimic) was mixed with the diluted Lipofectamine RNAiMAX for 15 min. Then the mixture was administrated to the cells in an 800 mL of fresh medium, incubated for 6 hours before replacing of the medium. The transfected cells were incubated for 48 hours for further analysis. The siRNA sequences for lnc-HIF1A-AS2 silencing were as follows: siRNA-1: GGTTGGATCTAACACTAACTGT; siRNA-2: TATAGTCACTTTGCCAGCTCAA; and siRNA-3: CAGTTACTCATGGAATATATTC.
Cell Proliferation Assay
Renal carcinoma cells were planted in a 96-well plate and transfected with 100 nM HIF1A-AS2 siRNA or control siRNA. The proliferation rate was determined on days 0, 2, 4, 6 and 8 by MTT assay (Beyotime, China) according to the manufacturer’s instructions of use.
RNA-fluorescence in situ Hybridization (RNA-FISH)
Subcellular distribution of HIF1A-AS2 was subjected to RNA-FISH in renal carcinoma cells. Cells were fixed in 4% of polyformaldehyde at room temperature for 30 minutes, and dehydrated with ethanol at concentrations of 70% (twice), 80%, 95% (twice), and 100%, respectively. Specimens were incubated and hybridized with digoxigenin-labeled RNA-FISH probes at 37°C overnight, and dyed with anti-digoxigenin-FITC conjugated secondary antibody (Roche, Switzerland) for 2 h. The antisense probe was utilized as a negative control. The fluorescence was measured with a confocal microscope (Carl Zeiss, USA). Sequence of RNA-FISH probe was as follows: CCATTGCATTGCAGTAGCATC.
Cell Cycle Analysis
After the collection and transfection, cells were fixed in cold ethanol at 70% 4°C overnight. The fixed samples were washed twice in fresh cold PBS and incubated with RNase A (50 mg/mL, Beyotime) and propidium iodide (PI) (100 mg/mL, Beyotime) at 37°C for 30 min. FACSCalibur (BD Biosciences, USA) was used to collect cells, and cell cycle distribution was analyzed by FlowJo software7.6.1 (Tree Star, USA).
Apoptosis Assay
Annexin V-FITC Apoptosis Detection Kit I (BD, USA) was utilized for apoptosis assays following the manufacturer’s instructions of use. Apoptotic cells were evaluated and analyzed by FACSCalibur and FlowJo software 7.6.1, respectively.
Transwell Invasion and Migration Assay
To perform an invasion assay, cells were suspended amid 250 µL of culture medium with 1% of FBS and embedded into the upper chamber of a 24-well transwell insert (poresize:8 μm, BD356230, Corning, USA) which was pre-coated with 25 µL of growth factor declined matrigel (diluted to three volumes with serum-free culture medium). As for the migration assay, cells were settled in the upper chamber of the 24-well insert directly without any process of pre-treatment. A 500 µL of fresh culture medium with 10% FBS was added into the lower chamber for both assays. After 24 or 36 hours, staining with 0.1% crystal violet was carried out on the invaded cells and migrated cells, respectively. Ten pictures of each specimen were randomly photographed. The stained crystal violet was resolved in a 200 µL of 50% ethanol containing 0.05 mM sodium citrate and 0.05 mM citric acid and measured at OD570 using an ELISA reader (Molecular Devices).
Wound Healing Assay
Wound healing assay was performed when ACHN and OSRC-2 cells were seeded in 6-well plates with fresh medium containing 10% FBS. When fused monolayer cells were formed, the membrane was scratched with a 200-µL sterile pipette. The cell culture medium was refreshed and wound photographs were taken at different time points (0 and 12 h after scratching). The coverage of the intermediate space was measured at three positions for each replicate, repeated twice.
Western Blot Analysis
Western blot analysis was performed as per the manufacturer’s instructions of use. Primary antibodies included E-Cadherin (ab219332; Abcam, US), SNAIL (ab53519; Abcam), and ERBB2 (A2071; Abclonal). The horseradish peroxidase (HRP)-conjugated secondary antibodies were purchased from the Zhongshan Golden Bridge Biotechnology Company (ZDR5306 or ZDR5307, China).
Plasmid Construction and Luciferase Reporter Assay
HIF1A-AS2 cDNA fragments were amplified by RT-PCR from the complementary DNA of renal carcinoma, which comprised a miR-130a-5p binding site, inserted between Sgf I and Pme I restriction sites in the reporter plasmid psiCHECK-2 (Promega, USA). For constructing HIF1A-AS2 reporter gene plasmids with a mutant miR-130a-5p binding site, the Site-Directed Mutagenesis System (Beyotime, China) was utilized in terms of the manufacturer’s instructions of use. The luciferase reporter assay was conducted as per the manufacturer’s instructions of use.
Statistical Analysis
All statistical analyses were made on the SPSS 17.0 platform (SPSS; USA). Differences between both groups were analyzed using Student’s t-test and differences among multiple groups were analyzed by adopting one-way ANOVA LSD t-test. P < 0.05 was considered as significant differences. Correlations between two variables were analyzed by linear regression.
Results
HIF1A-AS2 Was Highly Expressed in ccRCC Tissues and RCC Cell Lines
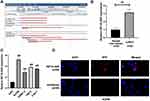
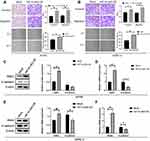
HIF1A-AS2 was located at chromosome 14 (hg19, 62,213,757–62,215,807) (Figure 1A). The expression of HIF1A-AS2 in these ccRCC tissues was notably increased compared to that in the paired non-cancer tissues (Figure 1B). Meanwhile, the qRT-PCR analysis demonstrated a remarkably elevated expression of HIF1A-AS2 in multiple human RCC cell lines than that in the human normal renal tubular epithelial cell line HK2 cells (Figure 1C). These findings exhibited a role of HIF1A-AS2 as an oncogene in renal carcinoma. By determining subcellular location of HIF1A-AS2, we noticed distribution of HIF1A-AS2 was allocated both in nucleus and in cytoplasm of ACHN cells by RNA-FISH (Figure 1D).
Influence of HIF1A-AS2 on RCC Cell Proliferation, Cell Cycle, and Apoptosis
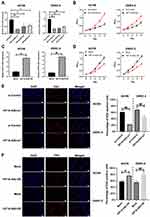
For a further investigation on the exact biological roles of HIF1A-AS2, the ACHN and OSRC-2 cells were selected for subsequent experimental use. The expression of HIF1A-AS2 was determined by qRT-PCR and the interfered HIF1A-AS2-si1 exhibited the best results (Figure 2A). Findings of MTT assay revealed that the interfered HIF1A-AS2 expression retarded the growth of ACHN and OSRC-2 cells (Figure 2B). Overexpression HIF1A-AS2 in ACHN and OSRC-2 cells (Figure 2C) with results of MTT assay indicating that overexpressed HIF1A-AS2 remarkably enhanced cell growth compared to the negative control cells (Figure 2D). Accordant with the MTT assay results, similarly inhibited proliferation was observed in EdU assay after transfection with HIF1A-AS2 siRNA and enhanced proliferation was noted in overexpressed-HIF1A-AS2 cells (Figure 2E and F).
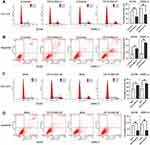
To ascertain the association of HIF1A-AS2 with cycle of renal carcinoma cells, flow cytometry was performed to determine the percentage of cells in phase S, with results indicating that HIF1A-AS2 silence drastically decreased the cell percentage in phase S, while an increase exhibited in phase G2 (Figure 3A). Results of apoptosis assay revealed that ACHN and OSRC-2 cells transfected with HIF1A-AS2 siRNA presented a higher percentage of cell apoptosis in both early and late phases in contrast to the control group (Figure 3B). Notably, HIF1A-AS2 silence increased percentage of G2 phase in ACHN cells. It is implied that ACHN cells were arrest in the G2 phase. The opposite result was displayed in ACHN and OSRC-2 cells transfected with HIF1A-AS2-OE (Figure 3C and D). As previously described results, HIF1A-AS2 promoted cell proliferation of renal carcinoma by accelerating cell cycles and hindering cellular apoptosis.
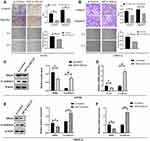
HIF1A-AS2 Promoted the Invasion and Migration in RCC Cell Lines
For further exploring biological effects of HIF1A-AS2 in renal malignancy development, a transwell assay and a scratch wound were performed to identify how HIF1A-AS2 affects invasion and migration abilities of kidney cancer cells. Results exhibited that HIF1A-AS2 silencing dramatically alleviated cellular invasion and migration in ACHN and OSRC-2 cells (Figure 4A and B), whereas overexpressed HIF1A-AS2 was confirmed producing a role in promotion of the invasion and migration in ACHN and OSRC-2 cells (Figure 5A and B). In the wound healing assay, cell motility was observed linking to ability of migration. HIF1A-AS2 silencing disabled ACHN and OSRC-2 cell migration after being scratched (Figure 4A and B), and overexpressed HIF1A-AS2 displayed a diminished migratory capacity in ACHN and OSRC-2 cells (Figure 5A and B). Moreover, we also identified levels of epithelial marker E-cadherin and mesenchymal marker SNAIL in ACHN and OSRC-2 cells. Interference of HIF1A-AS2 obviously boosted protein and mRNA levels of E-cadherin and minimized that of SNAIL (Figure 4C–F). Conversely, overexpression of HIF1A-AS2 drastically reduced levels of E-cadherin while increased levels of SNAIL (Figure 5C–F). Our study assumed HIF1A-AS2 might serve as a trigger for epithelial-mesenchymal transition in renal oncology.
HIF1A-AS2 Acted as a ceRNA by Sponging miR-130a-5p to Facilitate ERBB2
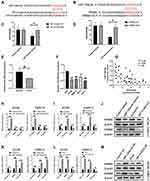
A further investigation was carried out to explore the mechanism of HIF1A-AS2 on cell growth of renal carcinoma. miRNAs with complementary base pairing with HIF1A-AS2 were searched by taking advantage of NCBI BLAST algorithm (https://blast.ncbi.nlm.nih.gov/Blast.cgi). Importantly, miR-130a-5p was found possessing conserved complementary base pairings with HIF1A-AS2 (Figure 6A). Cloned HIF1A-AS2 Wild-type (WT) and mutation (MUT) structures containing miR-130a-5p binding sites were inserted into psiCHECK2 dual luciferase reporter plasmids, and transfected into 293T cells together with control miRNA mimics and miR-130a-5p mimics, respectively, through which the one regulated by HIF1A-AS2 was figured out. Results of overexpressed miR-130a-5p exhibited robust diminution of luciferase activity in wild-type HIF1A-AS2 constructs (Figure 6C). Wild-type structures were mutated as depicted in Figure 6A and transfected into 293T cells with control miRNA mimics and miR-130a-5p mimics. These results of our assay demonstrated that mutation of binding sties muted the effects of miR-130a-5p on luciferase activity of HIF1A-AS2 (Figure 6C). Subsequently, qRT-PCR assay substantiated that expression of miR-130a-5p was lowered greatly in the RCC cell lines and ccRCC tissues than their control groups (Figure 6E and F). Meanwhile, there was a negative correlation between HIF1A-AS2 and miR-130a-5p in the mRNA level of the ccRCC tissues (Figure 6G). Additionally, ectopic expression of miR-130a-5p efficiently decreased the expression level of HIF1A-AS2 and HIF1A-AS2 interference showed an increase in miR-130a-5p expression in ACHN and OSRC-2 cells (Figure 6H and I). Conversely, the miR-130a-5p inhibitor largely promoted HIF1A-AS2 expression in ACHN and OSRC-2 cells compared with the inhibitor control and the overexpression of HIF1A-AS2 attenuated the expression of miR-130a-5p (Figure 6K and L).
To figure out whether HIF1A-AS2 served as a sponge for miR-130a-5p, we performed dual-luciferase reporter, qRT-PCR and Western blot assays to evaluate the expression of ERBB2 as a target of miR-130a-5p. Luciferase reporter assay showed that miR-130a-5p overexpression substantially repressed activity of the reporter with WT rather than MUT 3ʹUTR of ERBB2 (Figure 6B and D). Western blot and qRT-PCR confirmed that transfected ACHN cells and OSRC-2 with miR-130a-5p mimics or HIF1A-AS2 siRNA efficiently diminished the expression of ERBB2 (Figure 5H–J). On the contrary, weakening miR-130a-5p or enhancing HIF1A-AS2 levels displayed an increase in ERBB2 expression (Figure 6K–M). Based on these results, we argued that knockdown of HIF1A-AS2 released the repression to miR-130a-5p, thereby promoting the expression of ERRB2.
HIF1A-AS2 Enhanced RCC Cell Proliferation and Aggressive Behavior Through Sponging miR-130a-5p
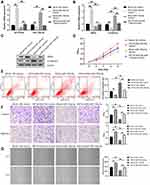
We detected the effects of miR-130a-5p on RCC cells and determined whether miR-130a-5p overexpression could inhibit the proliferation and progression of renal cancer cells induced by HIF1A-AS2 overexpression. As illustrated in Figure 7A, miR-130a-5p and HIF1A-AS2 were successfully overexpressed in ACHN cells, respectively. However, when ACHN cells were treated with both miR-130a-5p mimics and HIF1A-AS2-OE, neither expressions of miR-130a-5p mimics and HIF1A-AS2-OE exhibited apparent difference compared to mock + NC mimics group. Observation results revealed that ACHN cells with high miR-130a-5p expression declined the SNAIL levels but increased the E-cadherin levels. Meanwhile, overexpression of miR-130a-5p could weaken the regulation of HIF1A-AS2 to SNAIL and E-cadherin (Figure 7B and C). The proliferation, migration and invasion of cells were repressed while apoptosis was promoted in ACHN cells by upregulating miR-130a-5p expression. Furthermore, the phenotypes caused by HIF1A-AS2-OE were also identified and partly rescued by miR-130a-5p mimics subjected to MTT, cell apoptosis, Transwell, and wound healing assays (Figure 7D–G).
Discussion
As a necessary class of non-protein-coding RNAs, lncRNAs are closely involved in the development and progression of cancer.14–16 When compared with studies on miRNAs, little description has been recorded about lncRNAs associated with renal carcinoma. Since the natural antisense transcript of HIF1A was characterized in 1999,17 it encodes proteins to stabilize protein p53 and has been recognized as a transcription factor for hypoxia inducible genes during hypoxia. HIF1A-AS2 is complementary to the 3ʹ untranslated region of HIF1α messenger RNA and is strikingly overexpressed specifically in nonpapillary ccRCC.17–19 Bertozzi et al (2011) demonstrate that HIF1A-AS2 is expressed in several renal cancers, suggesting that they may have a role in cancer development in patients.20 Analysis of GSE96574 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE96574) showed that the expression of HIF1A-AS2 was dramatically higher in ccRCC tissues compared to paired noncancerous renal tissues, which was consistent with the result of TCGA KIRC DataSets analyzed by TANRIC (https://ibl.mdanderson.org/tanric/_design/basic/query.html). Besides, HIF1A-AS2 was found to be dysregulated in several tumors, such as colorectal cancer, breast cancer, bladder cancer and gastric cancer.21–24 In previously described studies, much evidence has confirmed that HIF1A-AS2 promotes cell proliferation, migration, and invasion. Our study demonstrated that lncRNA HIF1A-AS2 was overexpressed in ccRCC tissues and RCC cell lines. Additionally, HIF1A-AS2 could suppress renal carcinoma cellular proliferation, invasion, and migration and promote apoptosis. Overexpression of HIF1A-AS2 elevated levels of SNAIL and reduced levels of E-cadherin. SNAIL roles as a strong repressor of transcription of the E-cadherin gene, which inhibits cancer progression.25,26 Furthermore, HIF1A-AS2 forms a reciprocal repression feedback loop with miR-130a-5p and the phenotypes caused by HIF1A-AS2 overexpression can be partly rescued by miR-130a-5p overexpression. Our findings defined a functional role of HIF1A-AS2 in ccRCC and technically disclosed that HIF1A-AS2 might form a regulatory axis with miR-130a-5p/ERBB2.
OncomiR (http://www.oncomir.org/) is an online resource for exploring miRNA dysregulation in cancer, aligned and normalized miRNA-seq and RNA-seq data were obtained from TCGA DataSets.27 Analysis of OncomiR shows that miR-130a-5p upregulated in normal tissue compared to ccRCC tissues, which was consistent with our qRT-PCR results. Luciferase assays showed that miR-130a-5p could depress ERBB2 and HIF1A-AS2 expression through direct binding to their promoter. ERBB2 is implicated and overexpressed in the development of various cancers.28 Inhibitors targeting the ERBB2 selective tyrosine kinase inhibit tumor growth including RCC.12 Analysis of ERBB2 expression revealed that HIF1A-AS2 competitively binds miR-130a-5p to enhance ERBB2 expression, thereby promoting the progression of renal tumors. Previous studies have proved that some protein-coding genes work as direct targets of miR-130a-5p.29,30 Our future studies are scheduled to perform a deeper screening of targets below the HIF1A-AS2-miR-130a-5p axis associated with renal cancer development and progression. Findings have proved that promoter hypermethylation elicits miR-130a-5p downregulation. In line with extensive reports on the relationship between miR-130a-5p expression and its promoter methylation of renal carcinomas, miR-130a-5p CpG methylation may be a promising diagnostic marker for colorectal cancer, pancreatic cancer, breast cancer, kidney cancer, urothelial cancer, and soft tissue sarcoma. What we have discovered in this study provides novel evidence in post-transcriptional regulation of miR-130a-5p. As reported by several laboratories, members of the miR-130a-5p are direct targets of p5331,32 which produce certain roles in HIF1A-AS2 regulation.33–35 Consequently, we determined levels of HIF1A-AS2 and miR-130a-5p in wide-type renal carcinoma cells, with findings demonstrated that the deletion of p53 dramatically suppressed the expression of miR-130a-5p by elevating the level of HIF1A-AS2 indicating that p53 functioned as a potential upstream regulator of the HIF1A-AS2-miR-130a-5p/c axis.
Massive evidence has revealed that miR-130a-5p roles as more than a regulator in the processes of cellular proliferation, apoptosis, invasion and migration of tumors, it functions as a modulator of cancer cellular stemness and chemoresistance as well. Thence, the biological effects of HIF1A-AS2 on renal carcinoma remain to be under exploration, including properties and resistance of cancer stem cells.
Conclusions
In short, our study has demonstrated that HIF1A-AS2 plays an oncogenic role in renal carcinoma, whose function links to the regulation of miR-130a-5p/ERBB2 pathway. HIF1A-AS2 may also work as a novel marker for kidney cancer diagnosis and a promising target of therapeutic intervention on renal carcinoma cures.
Data Sharing Statement
All data generated or analyzed during this study are included in this published article.
Funding
This work was supported by the General project of Chongqing Healthy and Family Planning commission (20142001), the General project of Chongqing Educational Commission (KJ1400233), the project of Chongqing Science and Technology Commission (CSTC2015SHMSZX120067) and High-level Medical Reserved Personnel Training Project of Chongqing.
Disclosure
The authors declare that they have no conflicts of interest for this work.
References
1. Zhou Y, Zhang R, Ding Y, et al. Prognostic nomograms and Aggtrmmns scoring system for predicting overall survival and cancer-specific survival of patients with kidney cancer. Cancer Med. 2020;9:2710–2722.
2. Zhou W, Yang F, Xu Z, et al. Comprehensive analysis of copy number variations in kidney cancer by single-cell exome sequencing. Front Genet. 2019;10:1379. doi:10.3389/fgene.2019.01379
3. Niziol J, Sunner J, Beech I, et al. Localization of metabolites of human kidney tissue with infrared laser-based selected reaction monitoring mass spectrometry imaging and silver-109 nanoparticle-based surface assisted laser desorption/ionization mass spectrometry imaging. Anal Chem. 2020;92(6):4251–4258.
4. Zheng H, Li BH, Liu C, Jia L, Liu FT. Comprehensive analysis of lncRNA-mediated ceRNA crosstalk and identification of prognostic biomarkers in Wilms’ tumor. Biomed Res Int. 2020;2020:4951692. doi:10.1155/2020/4951692
5. Cui WW, Sun YL, Chen C, et al. LncRNA CRNDE promotes the development of Wilms’ tumor by regulating microRNA-424. Eur Rev Med Pharmacol Sci. 2020;24(3):1088–1097. doi:10.26355/eurrev_202002_20159
6. Zhang X, Yan Z, Wang L, Zhang S, Gao M. STAT1-induced upregulation of lncRNA RHPN1-AS1 predicts a poor prognosis of hepatocellular carcinoma and contributes to tumor progression via the miR-485/CDCA5 axis. J Cell Biochem. 2020.
7. Wu P, Mo Y, Peng M, et al. Emerging role of tumor-related functional peptides encoded by lncRNA and circRNA. Mol Cancer. 2020;19(1):22.
8. Chen Y, Zhang X, An Y, Liu B, Lu M. LncRNA HCP5 promotes cell proliferation and inhibits apoptosis via miR-27a-3p/IGF-1 axis in human granulosa-like tumor cell line KGN. Mol Cell Endocrinol. 2020;503:110697. doi:10.1016/j.mce.2019.110697
9. Shi S, Li D, Li Y, Feng Z, Du Y, Nie Y. LncRNA CR749391 acts as a tumor suppressor to upregulate KLF6 expression via interacting with miR-181a in gastric cancer. Exp Ther Med. 2020;19(1):569–578. doi:10.3892/etm.2019.8226
10. Chen Z, Lin S, Li JL, et al. CRTC1-MAML2 fusion-induced lncRNA LINC00473 expression maintains the growth and survival of human mucoepidermoid carcinoma cells. Oncogene. 2018;37(14):1885–1895. doi:10.1038/s41388-017-0104-0
11. Zhou W, Zhao Z, Wang R, et al. Identification of driver copy number alterations in diverse cancer types and application in drug repositioning. Mol Oncol. 2017;11(10):1459–1474. doi:10.1002/1878-0261.12112
12. Nagasawa J, Mizokami A, Koshida K, Yoshida S, Naito K, Namiki M. Novel HER2 selective tyrosine kinase inhibitor, TAK-165, inhibits bladder, kidney and androgen-independent prostate cancer in vitro and in vivo. Int J Urol. 2006;13(5):587–592. doi:10.1111/j.1442-2042.2006.01342.x
13. Edge SB, Compton CC. The American Joint Committee on Cancer: the 7th edition of the AJCC cancer staging manual and the future of TNM. Ann Surg Oncol. 2010;17(6):1471–1474. doi:10.1245/s10434-010-0985-4
14. Zhou K, Li S, Du G, et al. LncRNA XIST depletion prevents cancer progression in invasive pituitary neuroendocrine tumor by inhibiting bFGF via upregulation of microRNA-424-5p. Onco Targets Ther. 2019;12:7095–7109. doi:10.2147/OTT.S208329
15. Lin X, Yang F, Qi X, et al. LncRNA DANCR promotes tumor growth and angiogenesis in ovarian cancer through direct targeting of miR-145. Mol Carcinog. 2019;58(12):2286–2296. doi:10.1002/mc.23117
16. Akbari M, Yassaee F, Aminbeidokhti M, Abedin-Do A, Mirfakhraie R. LncRNA SRA1 may play a role in the uterine leiomyoma tumor growth regarding the MED12 mutation pattern. Int J Women's Health. 2019;11:495–500. doi:10.2147/IJWH.S211632
17. Thrash-Bingham CA, Tartof KD. aHIF: a natural antisense transcript overexpressed in human renal cancer and during hypoxia. J Natl Cancer Inst. 1999;91(2):143–151. doi:10.1093/jnci/91.2.143
18. Qiu JJ, Lin XJ, Zheng TT, Tang XY, Hua KQ. Natural antisense transcript of hypoxia-inducible factor 1 regulates hypoxic cell apoptosis in epithelial ovarian cancer. Onco Targets Ther. 2018;11:9101–9110. doi:10.2147/OTT.S173816
19. Uchida T, Rossignol F, Matthay MA, et al. Prolonged hypoxia differentially regulates hypoxia-inducible factor (HIF)-1alpha and HIF-2alpha expression in lung epithelial cells: implication of natural antisense HIF-1alpha. J Biol Chem. 2004;279(15):14871–14878. doi:10.1074/jbc.M400461200
20. Bertozzi D, Iurlaro R, Sordet O, Marinello J, Zaffaroni N, Capranico G. Characterization of novel antisense HIF-1α transcripts in human cancers. Cell Cycle. 2011;10(18):3189–3197. doi:10.4161/cc.10.18.17183
21. Chen WM, Huang MD, Kong R, et al. Antisense long noncoding RNA HIF1A-AS2 is upregulated in gastric cancer and associated with poor prognosis. Dig Dis Sci. 2015;60(6):1655–1662. doi:10.1007/s10620-015-3524-0
22. Lin J, Shi Z, Yu Z, He Z. LncRNA HIF1A-AS2 positively affects the progression and EMT formation of colorectal cancer through regulating miR-129-5p and DNMT3A. Biomed Pharmacother. 2018;98:433–439. doi:10.1016/j.biopha.2017.12.058
23. Chen M, Zhuang C, Liu Y, et al. Tetracycline-inducible shRNA targeting antisense long non-coding RNA HIF1A-AS2 represses the malignant phenotypes of bladder cancer. Cancer Lett. 2016;376(1):155–164. doi:10.1016/j.canlet.2016.03.037
24. Wang Y, Zhang G, Han J. HIF1A-AS2 predicts poor prognosis and regulates cell migration and invasion in triple-negative breast cancer. J Cell Biochem. 2019;120(6):10513–10518. doi:10.1002/jcb.28337
25. Cano A, Pérez-Moreno MA, Rodrigo I, et al. The transcription factor snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nat Cell Biol. 2000;2(2):76–83. doi:10.1038/35000025
26. Gheldof A, Berx G. Cadherins and epithelial-to-mesenchymal transition. Prog Mol Biol Transl Sci. 2013;116:317–336.
27. Li J, Han L, Roebuck P, et al. TANRIC: an interactive open platform to explore the function of lncRNAs in cancer. Cancer Res. 2015;75(18):3728–3737. doi:10.1158/0008-5472.CAN-15-0273
28. Zhang H, Berezov A, Wang Q, et al. ErbB receptors: from oncogenes to targeted cancer therapies. J Clin Invest. 2007;117(8):2051–2058. doi:10.1172/JCI32278
29. Sun Z, Gao S, Xuan L, Liu X. Long non-coding RNA FEZF1-AS1 induced progression of ovarian cancer via regulating miR-130a-5p/SOX4 axis. J Cell Mol Med. 2020;24(7):4275–4285. doi:10.1111/jcmm.15088
30. Wang W, Wu D, He X, et al. CCL18-induced HOTAIR upregulation promotes malignant progression in esophageal squamous cell carcinoma through the miR-130a-5p-ZEB1 axis. Cancer Lett. 2019;460:18–28. doi:10.1016/j.canlet.2019.06.009
31. Zhang D, Yan B, Yu S, et al. Coenzyme Q10 inhibits the aging of mesenchymal stem cells induced by D-galactose through Akt/mTOR signaling. Oxid Med Cell Longev. 2015;2015:867293. doi:10.1155/2015/867293
32. Park KW, Kundu J, Chae IG, et al. Carnosol induces apoptosis through generation of ROS and inactivation of STAT3 signaling in human colon cancer HCT116 cells. Int J Oncol. 2014;44(4):1309–1315. doi:10.3892/ijo.2014.2281
33. Idogawa M, Nakase H, Sasaki Y, Tokino T. Prognostic effect of long noncoding RNA NEAT1 expression depends on p53 mutation status in cancer. J Oncol. 2019;2019:4368068.
34. Idogawa M, Nakase H, Sasaki Y, Tokino T. Corrigendum to “Prognostic effect of long noncoding RNA NEAT1 expression depends on p53 mutation status in cancer”. J Oncol. 2019;2019:4757046.
35. Pereira T, Brito JAR, Guimaraes ALS, et al. MicroRNA profiling reveals dysregulated microRNAs and their target gene regulatory networks in cemento-ossifying fibroma. J Oral Pathol Med. 2018;47(1):78–85. doi:10.1111/jop.12650
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.