Back to Journals » Cancer Management and Research » Volume 12
LncRNA UCA1 Affects the Cell Proliferation, Migration, Invasion and Apoptosis of Hepatic Carcinoma Cells by Targeting MicroRNA-193a-3p
Authors Wang HZ, Liu L, Xu Y, Zhang GY, Wang YY
Received 20 July 2020
Accepted for publication 28 August 2020
Published 30 October 2020 Volume 2020:12 Pages 10897—10907
DOI https://doi.org/10.2147/CMAR.S270396
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Professor Seema Singh
Hong-Zhen Wang,1 Li Liu,2 Yan Xu,1 Guang-Ye Zhang,3 Yan-Yan Wang2
1Department of Oncology, Rizhao City Hospital of Traditional Chinese Medicine, Rizhao City, Shandong Province 276800, People’s Republic of China; 2Department of ENT, Rizhao City Hospital of Traditional Chinese Medicine, Rizhao City, Shandong Province 276800, People’s Republic of China; 3Department of Hepatology, Rizhao City Hospital of Traditional Chinese Medicine, Rizhao City, Shandong Province 276800, People’s Republic of China
Correspondence: Yan-Yan Wang
Department of ENT, Rizhao City Hospital of Traditional Chinese Medicine, Rizhao City, Shandong Province 276800, People’s Republic of China
Tel +86-13906331429
Email [email protected]
Objective/Background: Hepatic carcinoma (HCC) is the fourth lethal cancer in the world, but its relationship with lncRNA urothelial cancer-associated 1 (UCA1)/microRNA-193a-3p axis remains unclear, so this study would explore the relationship.
Methods: A real-time polymerase chain reaction (RT-PCR) assay was carried out to quantify lncRNA UCA1 and microRNA-193a-3p in HCC tissues and cells, and relevant overexpression or inhibition vectors were constructed to analyze the influences of lncRNA UCA1 and microRNA-193a-3p on HCC cells. A Transwell assay was used to measure invasion and migration of HCC cells, and a Western blot assay to quantify protein biomarkers of apoptosis, invasion, and migration, a MTT assay to determine cell viability, a flow cytometry to detect cell cycle, and a dual-luciferase reporter gene assay to analyze the correlation between lncRNA UCA1 and microRNA-193a-3p.
Results: LncRNA UCA1 was increased in HCC, while microRNA-193a-3p was decreased. Down-regulated lncRNA UCA1 could up-regulate microRNA-193a-3p, and down-regulated lncRNA UCA1 or up-regulated microRNA-193a-3p would strengthen cell apoptosis and weaken cell migration, invasion, and proliferation. Furthermore, lncRNA UCA1 could negatively regulate microRNA-193a-3p by binding to it.
Conclusion: LncRNA UCA1 promotes malignant hyperproliferation of HCC cells by repressing microRNA-193a-3p.
Keywords: hepatic carcinoma, lncRNA UCA1/microRNA-193a-3p, cell carcinogenesis
Introduction
Hepatic carcinoma (HCC) is the fourth lethal cancer in the world, with a relatively high metastatic rate of extrahepatic carcinoma.1,2 It can be roughly classified into hepatocellular carcinoma and intrahepatic cholangiocarcinoma according to differences in cancerous cells.3 The incidence of HCC in 2016 increased to 1.14 times that in 1990,4 and obesity, excessive drinking, hepatitis B/C virus, flagellin/lipopolysaccharide antibody, gallstones, and statins may be factors leading to the cancer.5–10 Currently, it is a mainstream of research to analyze tumorigenesis from gene regulation, which helps to improve the accuracy of tumor target therapy.
LncRNA UCA1 is a long-chain noncoding RNA on human chromosome 19. Currently, it is generally accepted that long-chain non-coding RNA does not directly participate in the coding of genetic information, but it can regulate the post-transcriptional level of genes by binding to mRNA responsible for transmission. LncRNA UCA1 is a common cancer factor, which can affect cell carcinogenesis by regulating various downstream target genes. For example, it promotes the diffusion of oncolytic vaccinia virus among ovarian cancer cells by activating the CDC42 pathway, and finally improves the oncolytic efficiency of the virus,11 and it enhances the viability of bladder cancer cells by modulating the microRNA-582-5p/ATG7 axis.12 In addition, it also enhances the viability of cancer cells by the microRNA-28-5p/HOXB3 pathway,13 and enhances the sensitivity of breast cancer cells to tamoxifen via the E2H2/p21 pathway and PI3K/Akt pathway.14 It is worth mentioning that lncRNA UCA1 is abnormally up-regulated in cases with HCC and is strongly linked to the carcinogenesis of normal hepatic cells. According to previous studies, lncRNA UCA1, as a molecular sponge, can regulate the biological function of HCC cells through non-coding RNA such as miR-216b, thus promoting the development and progression of HCC.15–18 In addition to lncRNA, miRNA is also an important member of gene epigenetic regulation. Many studies have revealed that microRNA-193a and its spliceosomes are involved in the progression of HCC.19,21
Given the abnormal expression of LncRNA UCA1 and microRNA-193a-3p in HCC and the existence of sequence fragments binding to microRNA-193a-3p on lncRNA UCA1,22 we suspected that microRNA-193a-3p and lncRNA UCA1 may affect the development of HCC through a certain regulatory relationship. Although some studies have confirmed the roles of lncRNA UCA1 and miR-193a-3p in HCC, there is no study on how the lncRNA UCA1/microRNA-193a-3paxis affects hepatocarcinogenesis, so this study would explore a possible mechanism of hepatocarcinogenesis.
Materials and Methods
Patients with HCC
Seventy-seven HCC tissue specimens and sixty-four corresponding adjacent normal tissues were sampled from patients with HCC in our hospital. The inclusion criteria of the patients: Patients diagnosed as HCC according to clinical characteristics or pathological sections, and those without mental diseases. The exclusion criteria of them: Patients with other comorbid tumors, patients with previous treatment history of HCC, and those unwilling to cooperate for the treatment. The patients were informed of the information about the study during the whole study process, and the study was carried out under the permission of the Ethics Committee of Rizhao City Hospital of Traditional Chinese Medicine. Tissue samples were cut into sections, and stored in −80°C for future analysis.
Cell Transfection
Human normal hepatic cell (THLE-3) and HCC cell lines including Hep3B, SNU-398 and SNU-449 purchased from the American Type Culture Collection (ATCC) were cultured in the dulbecco’s modified eagle medium (DMEM) (Hyclone Company) supplemented with 10% fetal bovine serum (FBS) solution (Gibco Company), and 1% penicillin/streptomycin solution (100x, Solarbio Company). Afterwards, the cells were transferred to a cell culture flask (Thermo Fisher Company), and 5 mL medium preheated to 37°C was added into the culture flask. The cells were cultured in a 5%CO2 animal cell incubator (German Binder Company) at 37°C until they were in a good growth condition. Before transfection, the medium was replaced with a FBS-free medium, and at transfection, the cells were seeded into a 6-well plate at 1×105cells/well. LncRNA UCA1 siRNA, microRNA-193a-3p inhibitor mimics and NC vectors were all purchased from Shanghai Sangon Biotech Co., Ltd. (China). The cell lines were transfected with a Lipofectamine 2000 transfection kit (Invitrogen, the United States) in accordance with the kit instructions. The concentration of lncRNA UCA1 siRNA, microRNA-193a-3p inhibitor mimics, and NC vectors was all 1μg/mL. After 8 hours of transfection, the culture medium was replaced, and after 24 hours of transfection, the cells could be used for subsequent extraction of RNA, protein or other biological function experiments.
RT-PCR Assay
Total RNA of tissues and cells was extracted using the Trizol method. The optical density (OD) of the total RNA at 260–280 nm was measured using an ultraviolet spectrophotometer, and the RNA with OD 260/OD 280>1.8 was used for subsequent qPCR assay. The RNA was quantified and analyzed using a FastKing one-step reverse transcription-fluorescence quantitative kit (Tiangen Biotech (Beijing) Co., Ltd.) and ABI PRISM 7000 (Applied Biosystems, the United States). The primers of lncRNA UCA1 and microRNA-193a-3p were designed and synthesized by Shanghai Sangon Biotech Co., Ltd. For lncRNA UCA1, F: 5′- CTC TCC TAT CTC CCT TCA CTG A-3′, R: 5-CTT TGG GTT GAG GTT CCT GT −3ʹ; for microRNA-193a-3p, F: 5ʹ- TGG GTC TTT GCG GGC −3ʹ, R: 5-GAA TAC CTC GGA CCC TGC-3ʹ. A qPCR assay was carried out under a reaction system consisting of 50 μL total volume containing 1.25 μL of upstream primer, 1.25 μL downstream primer, 1.0 uL probe, 10 pg/μg RNA template, 5 μL 50×ROX Reference Dye ROX, and RNase-Free ddH2O added to adjust the total volume. The reaction process included reverse transcription at 50°C for 30 min (one cycle) and pre-denaturation at 95°C for 3 min (one cycle) followed by 40 cycles of denaturation at 95°C for 15 s, and annealing at 60°C for 30 s. The results were analyzed using an ABI PRISM 7000 instrument, and the data were normalized using the 2–ΔΔCt method, with U6 and GAPDH as internal references.
Western Blotting Assay
Both protease inhibitor (Solarbio Company) and 20 mMTris-HCl solution (pH7.5, Solarbio Company) were used to prepare cell protein extract, and adherent cells were trypsinized, and prepared into cell suspension. The suspension was added with 1 mL cell protein extract, followed by repeated pipetting until the cells were fully lysed. Then, the solution was centrifuged in a pre-cooling centrifuge at 1.6×104×g for 20 min at 4°C to take the supernatant, and the protein concentration in the supernatant was determined using the bicinchoninic acid (BCA) method. The protein was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and the separated protein was transferred to a nitrocellulose (NC) membrane, and let to stand at room temperature for 1 h (blocked with 5% skim milk – PBS solution). Subsequently, cleaved Caspase 3, cleaved Caspase 9, Bax, E-cadherin, N-cadherin, β-catenin, Bcl-2 and β-actin primary antibodies (Abcam Company) were added into the suspension, and let to stand at 4°C overnight. The NC membrane was washed with PBS solution three times, and then added with goat anti-rabbit secondary antibody (HRP conjugant, Abcam Company), and let to stand at indoor temperature for 1 hour. Finally, the NC membrane was washed with PBS solution, and visualized by electrochemiluminescence (ECL) reagent. The internal reference protein was β-actin, and the relative expression of the protein to be detected was recorded as the gray value of the band to be detected/the gray value of β-actin protein band.
Transwell Assay
The cells were transferred to the upper compartment containing 200 μL mixed solution supplemented with 10% FBS +1% DMEM at 2×104 cells/well. The lower compartment was added with 500 μL DMEM supplemented with 10% FBS. The transwell insert was cultured under 5%CO2 at 37°C for 24 h. After 2 hours, the fluid in the upper compartment was removed, and the cells on the microporous membrane were wiped off. The cells on the other side of the microporous membrane were immobilized for 20 min, and the membrane was stained with crystal violet for 15min, and then cleaned with PBS solution. Photographs about cell migration were obtained under a 200-fold microscope. The cells in 3 randomly selected fields were counted, and the values were averaged as the number of cells penetrating the membrane. The experiment was repeated three times. Invasion was detected in the same way as above steps after 8% matrix gel was laid on the membrane, and the number of cells per well was changed to 5×104.
MTT Assay
The cells were seeded into three wells in each 96-well plate (four plates in total) at 5×103cells/100 μL in each well. One plate was taken out at 24h, 48 h, 72 h, and 96 h after cells were seeded, respectively. Each plate was added with 5 mg/mL MTT solution dissolved in dimethylsulfoxide (DMSO) (Solarbio Company) at 10 μL/well, and then cultured continuously for 1 h. Subsequently, the medium was taken out, and the OD at 570 nm was measured using an enzyme mark instrument.
Flow Cytometry
The cells were prepared into suspension through enzymolysis, and the number of cells was controlled to 1×106. The cells were immobilized in 70% ice-cold ethanol solution at ambient temperature of 4°C for 30 min, and then the ethanol solution was removed. A mixed solution of propidium iodide (50 ng/mL)/RNase (0.2 mg/mL)/0.1% Triton X-100 solution was added into the cells, and the cells were incubated at room temperature for 30 min. Subsequently, the FACScan flow cytometer (Becton Dickinson Company, the United States) was adopted to analyze apoptosis.
Dual-Luciferase Reporter Gene Assay
The cells were incubated on a 96-well plate, and a dual luciferase assay was carried out to the cells when they grew well. The GLO-UCA1-wt and GLO-UCA1-mut vectors were constructed, and co-transfected with miR-152 mimics and NC mimics into cells, respectively. After 48 hours of transfection, their luciferase activity was detected using the dual-luciferase reporter gene assay system (Promega).
Statistical Analyses
In this study, the data were statistically analyzed using SPSS20.0 (Asia Analytics Formerly SPSS, China), and visualized into figures using GraphPad Prism 8.0. The measurement data were expressed as the mean ± standard deviation (mean ± SD), and comparison between tumor tissues and corresponding tumor-adjacent tissues, between the NC siRNA group and the UCA1 siRNA group, between the NC mimics group and the miR mimics group, and between the UCA1 siRNA group and the UCA1 si+miR inhibitor group was carried out using the independent-samples t-test, and comparison among various cells was carried out by the one-way ANOVA. Post hoc pairwise comparison was carried out using the LSD t-test, and Pearson’s correlation analysis was carried out to study the correlation between lncRNA UCA1 and microRNA-193a-3p. All data were analyzed using the two-tailed test, and 95% was used as the confidence interval. P<0.05 indicates a statistical difference.
Results
Increased LncRNA UCA1 and Decreased MicroRNA-193a-3p in Cases HCC
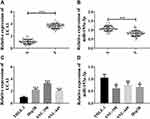
Firstly, we collected 77 HCC tissues and 69 adjacent normal tissues, and quantified lncRNA UCA1 and microRNA-193a-3p in those specimens, finding that compared with adjacent normal tissues, HCC tissues showed increased lncRNA UCA1 and decreased microRNA-193a-3p (Figure 1A and B). We also quantified lncRNA UCA1 and microRNA-193a-3p in human normal hepatic cells and HCC cells, finding that lncRNA UCA1 was up-regulated in HCC cells and microRNA-193a-3p was down-regulated in them (Figure 1C and D). The above results implied that the increase of lncRNA UCA1 expression and decrease of microRNA-193a-3p expression might play a role in hepatocarcinogenesis. Since the highest expression of lncRNA UCA1 was found in SUN-398 cells, we selected SUN-398 cells as research objects to study the influences of lncRNA UCA1 and microRNA-193a-3p on HCC (Figure 1).
Effects of LncRNA UCA1 on HCC Cells
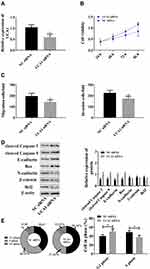
Since the abnormal up-regulation of lncRNA UCA1 is related to HCC, we down-regulated lncRNA UCA1 to explore its role in hepatocarcinogenesis. It came out that UCA1 siRNA could strongly down-regulate lncRNA UCA1 (Figure 2A), and down-regulation of lncRNA UCA1 could reduce cell viability, migration and invasion (Figure 2B and C). In addition, it was also found that down-regulated lncRNA UCA1 could increase cleaved Caspase 3, cleaved Caspase 9, E-cadherin, and Bax, and decrease N-cadherin, β-catenin, and Bcl2; and reduce cells in S phase and increase cells in G1phase. The above results implied that down-regulation of lncRNA UCA1 could give rise to cells apoptosis via cleaved Caspase 3, cleaved Caspase 9, Bax, Bcl2, and inhibit proliferation and migration via E-cadherin, N-cadherin and β-catenin, which suggested that lncRNA UCA1 could promote malignant hyperproliferation of HCC cells (Figure 2).
MicroRNA-193a-3p Inhibited the Malignant Hyperproliferation of HCC Cells
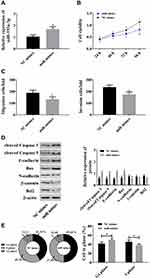
Since the down-regulation of microRNA-193a-3p may be linked to HCC cells, we up-regulated microRNA-193a-3p to analyze its effects on HCC cells. It was turned out that up-regulated microRNA-193a-3p suppressed the viability and proliferation of cells, weakened migration and invasion of them, up-regulated cleaved Caspase 3, cleaved Caspase 9, E-cadherin, and Bax, and down-regulated N-cadherin, Bcl2, and β-catenin. The above results implied that microRNA-193a-3p, as a tumor suppressor, could inhibit the malignant hyperproliferation of HCC cells (Figure 3).
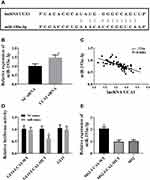
Inhibition of LncRNA UCA1 on MicroRNA-193a-3p in a Targeted Manner
It was predicted through miRcode website that lncRNA UCA1 had a sequence fragment that can be used to bind to microRNA-193a-3p (Figure 4A). It was also found that down-regulation of lncRNA UCA1 could give rise to up-regulation of microRNA-193a-3p (Figure 4B), and lncRNA UCA1 was negatively correlated with microRNA-193a-3p (Figure 4C). We constructed lncRNA UCA1 wt and lncRNA UCA1 mut vectors, and co-transfected them with microRNA-193a-3p mimics into HCC cells, respectively, finding that when microRNA-193a-3p mimics and lncRNA UCA1 wt were co-transfected, they were paired and bound, resulting in a decrease in luciferase activity. The above results indicated that lncRNA UCA1 inhibited the microRNA-193a-3p expression by binding to it (Figure 4).
LncRNA UCA1 Regulated HCC Cell Line via MiR-193a-3p
UCA1 siRNA and UCA1 siRNA+microRNA-193a-3p inhibitor were transfected into SUN-398 cells, respectively, to analyze the changes of cell biological behaviors, finding that inhibition of microRNA-193a-3p could offset the decline in cell proliferation, migration, invasion, increase in apoptosis, and down-regulation of microRNA-193a-3p caused by UCA1 (Figure 5). The above results suggested that lncRNA UCA1 could promote malignant proliferation of HCC cells by down-regulating microRNA-193a-3p.
Discussion
In this study, firstly, we found that lncRNA UCA1 in cases with HCC increased, while microRNA-193a-3p in them decreased, which was in consistent with results in other studies.15–18,23–25 Therefore, we speculated that lncRNA UCA1 and microRNA-193a-3p were involved in the development of HCC. Down-regulation of lncRNA UCA1 resulted in up-regulated microRNA-193a-3p, and sequence matching based on the miRcode website showed that there were sequence fragments that can bind to microRNA-193a-3p at the 3ʹUTR of lncRNA UCA1. In order to verify the existence of these pairing loci, we constructed lncRNA UCA1 wt and lncRNA UCA1 mut vectors, and co-transfected them with microRNA-193a-3pmimics into HCC cells, respectively. The results showed that when microRNA-193a-3pmimics and lncRNA UCA1 wt were co-transfected, they were paired and bound, resulting in a decrease in luciferase activity. The above results indicate that lncRNA UCA1 may regulate the survival of HCC cells through targeted inhibition of microRNA-193a-3p.
Afterwards, based on the above viewpoints, we carried out a series of assays to study the role of lncRNA UCA1/microRNA-193a-3p axis in HCC. We regulated lncRNA UCA1 siRNA and microRNA-193a-3pmimics vectors in HCC cells. It came out that down-regulated lncRNA UCA1 or up-regulated microRNA-193a-3p inhibited cell migration, proliferation and invasion, and strengthened cell apoptosis, and down-regulation of both lncRNA UCA1 and microRNA-193a-3p intensified cell proliferation, migration, and invasion, and weakened apoptosis. The above results implied that lncRNA UCA1 could promote proliferation, migration, and invasion of cells and inhibit apoptosis of them by down-regulating microRNA-193a-3p.
MicroRNA-193a-3p is a mature spliceosome of microRNA-193a, with a length of about 22bp. MicroRNA-193a-3p can regulate the development and progression of tumors by changing the biological processes of breast cancer cells, prostate cancer cells, colorectal cancer cells, HCC cells and other cells.19–26 According to the results of this study, up-regulation of microRNA-193a-3p could cause the increase in cleaved Caspase 3, cleaved Caspase 9, E-cadherin and Bax, and the decrease in N-cadherin, β-catenin, and Bcl2. Increase in cleaved Caspase 3, cleaved Caspase 9, Bax and decrease in Bcl2 can promote cell apoptosis and inhibit cell proliferation, while increase in E-cadherin and decrease in N-cadherin andβ-catenin can suppress cell migration and invasion. Therefore, lncRNA UCA1 promotes the over-proliferation and malignant expansion of HCC cells through the microRNA-193a-3p-based regulation mechanism, and finally induces the formation and metastasis of HCC. Furthermore, the results of this study also suggested that down-regulation of lncRNA UCA1 or up-regulation of microRNA-193a-3p may be beneficial to the treatment of HCC patients.
This study has explored the relationship between the lncRNA UCA1/microRNA-193a-3p axis and HCC cells from the perspective of molecular biology. The results showed that microRNA-193a-3p mediated the regulation of lncRNA UCA1 on the life process of HCC cells (Figure 6). However, this study still has certain limitations. LncRNA UCA1 and miR-193a-3p can be used as biomarkers of HCC, but this study has not explored it, so the possibility of the two as diagnostic substances for HCC will be discussed in further research. This study has also not discussed the target gene of miR-193a-3p, so the downstream target genes of miR-193a-3p will also be fully discussed in subsequent studies. At the same time, in addition, the relationship between lncRNA UCA1/miR-193a-3p axis and drug resistance of HCC also needs further research.
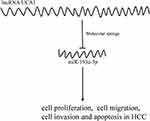 |
Figure 6 MiR-193a-3p, as an intermediate link, mediates the regulation of lncRNA UCA1 on the life procession of HCC cells. |
To sum up, lncRNA UCA1 inhibits microRNA-193a-3p by binding to it, and inhibition of microRNA-193a-3p leads to over-proliferation of HCC cells and reduction of apoptosis of them. The role of lncRNA UCA1/microRNA-193a-3p axis in HCC implies that down-regulation of lncRNA UCA1 or up-regulation of microRNA-193a-3p can be used for HCC treatment. In addition, lncRNA UCA1 may also have early diagnostic value for HCC.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Petrick JL, McGlynn KA. The changing epidemiology of primary liver cancer. Curr Epidemiol Rep. 2019;6(2):104–111. doi:10.1007/s40471-019-00188-3
2. Elmoghazy W, Ahmed K, Vijay A, et al. Hepatocellular carcinoma in a rapidly growing community: epidemiology, clinico-pathology and predictors of extrahepatic metastasis. Arab J Gastroenterol. 2019;20(1):38–43.
3. Ma L, Hernandez MO, Zhao Y, et al. Tumor cell biodiversity drives microenvironmental reprogramming in liver cancer. Cancer Cell. 2019;36(4):418–430. doi:10.1016/j.ccell.2019.08.007
4. Liu Z, Jiang Y, Yuan H, et al. The trends in incidence of primary liver cancer caused by specific etiologies: results from the global burden of disease study 2016 and implications for liver cancer prevention. J Hepatol. 2019;70(4):674–683.
5. Tran KT, Cardwell CR, Thrift AP, et al. Statin use and risk of liver cancer: evidence from two population‐based studies. Int J Cancer. 2019. doi:10.1002/ijc.32506
6. Wang Y, Xie L-F, Lin J. Gallstones and cholecystectomy in relation to risk of liver cancer. Eur J Cancer Prev. 2019;28(2):61–67. doi:10.1097/CEJ.0000000000000421
7. Yang B, Petrick JL, Thistle JE, et al. Bacterial translocation and risk of liver cancer in a Finnish cohort. Cancer Epidemiol Biomarkers Prev. 2019;28(4):807–813. doi:10.1158/1055-9965.EPI-18-0240
8. Pang Y, Kartsonaki C, Turnbull I, et al. Adiposity in relation to risks of fatty liver, cirrhosis and liver cancer: a prospective study of 0.5 million Chinese adults. Sci Rep. 2019;9(1):785. doi:10.1038/s41598-018-36460-7
9. Saitta C, Pollicino T, Raimondo G. Obesity and liver cancer. Ann Hepatol. 2019;18(6):810–815. doi:10.1016/j.aohep.2019.07.004
10. Grewal P, Viswanathen VA. Liver cancer and alcohol. Clin Liver Dis. 2012;16(4):839–850. doi:10.1016/j.cld.2012.08.011
11. Horita K, Kurosaki H, Nakatake M, et al. lncRNA UCA1-mediated Cdc42 signaling promotes oncolytic vaccinia virus cell-to-cell spread in ovarian cancer. Mol Ther Oncolytics. 2019;13:35–48. doi:10.1016/j.omto.2019.03.003
12. Wu J, Li W, Ning J, et al. Long noncoding RNA UCA1 targets miR-582-5p and contributes to the progression and drug resistance of bladder cancer cells through ATG7-mediated autophagy inhibition. Onco Targets Ther. 2019;12:495. doi:10.2147/OTT.S183940
13. Cui M, Chen M, Shen Z, et al. LncRNA‐UCA1 modulates progression of colon cancer through regulating the miR‐28‐5p/HOXB3 axis. J Cell Biochem. 2019;120(5):6926–6936. doi:10.1002/jcb.27630
14. Li Z, Yu D, Li H, et al. Long non-coding RNA UCA1 confers tamoxifen resistance in breast cancer endocrinotherapy through regulation of the EZH2/p21 axis and the PI3K/AKT signaling pathway. Int J Oncol. 2019;54(3):1033–1042.
15. Wang F, Ying H-Q, He B-S, et al. Upregulated lncRNA-UCA1 contributes to progression of hepatocellular carcinoma through inhibition of miR-216b and activation of FGFR1/ERK signaling pathway. Oncotarget. 2015;6(10):7899. doi:10.18632/oncotarget.3219
16. Zhao B, Lu Y, Cao X, et al. MiRNA-124 inhibits the proliferation, migration and invasion of cancer cell in hepatocellular carcinoma by downregulating lncRNA-UCA1. Onco Targets Ther. 2019;12:4509. doi:10.2147/OTT.S205169
17. Cui X, Zhao C, Yao X, et al. SND1 acts as an anti-apoptotic factor via regulating the expression of lncRNA UCA1 in hepatocellular carcinoma. RNA Biol. 2018;15(10):1364–1375. doi:10.1080/15476286.2018.1534525
18. Xiao J-N, Yan T-H, Yu R-M, et al. Long non-coding RNA UCA1 regulates the expression of snail2 by miR-203 to promote hepatocellular carcinoma progression. J Cancer Res Clin Oncol. 2017;143(6):981–990. doi:10.1007/s00432-017-2370-1
19. Zhou HL, Zhou YF, Feng ZT. Long noncoding RNA ZFAS1 promotes hepatocellular carcinoma proliferation by epigenetically repressing miR-193a-3p. Eur Rev Med Pharmacol Sci. 2019;23:9840–9847.
20. Ma K, He Y, Zhang H, et al. DNA methylation-regulated miR-193a-3p dictates resistance of hepatocellular carcinoma to 5-fluorouracil via repression of SRSF2 expression. J Biol Chem. 2012;287(8):5639–5649. doi:10.1074/jbc.M111.291229
21. Salvi A, Conde I, Abeni E, et al. Effects of miR-193a and sorafenib on hepatocellular carcinoma cells. Mol Cancer. 2013;12(1):162. doi:10.1186/1476-4598-12-162
22. Nie W, Ge H-J, Yang X-Q, et al. LncRNA-UCA1 exerts oncogenic functions in non-small cell lung cancer by targeting miR-193a-3p. Cancer Lett. 2016;371(1):99–106. doi:10.1016/j.canlet.2015.11.024
23. Xie SD, Qin C, Jin LD, et al. Long noncoding RNA SNHG14 promotes breast cancer cell proliferation and invasion via sponging miR-193a-3p. Eur Rev Med Pharmacol Sci. 2019;23(6):2461–2468.
24. Chen D, Lu X, Yang F, et al. Circular RNA circHIPK3 promotes cell proliferation and invasion of prostate cancer by sponging miR-193a-3p and regulating MCL1 expression. Cancer Manag Res. 2019;11:1415.
25. Zhu Z, Du S, Yin K, et al. Knockdown long noncoding RNA nuclear paraspeckle assembly transcript 1 suppresses colorectal cancer through modulating miR‐193a‐3p/KRAS. Cancer Med. 2019;8(1):261–275. doi:10.1002/cam4.1798
26. Yu H-M, Wang C, Yuan Z, et al. LncRNA NEAT1 promotes the tumorigenesis of colorectal cancer by sponging miR‐193a‐3p. Cell Prolif. 2019;52(1):e12526. doi:10.1111/cpr.12526
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.