Back to Journals » OncoTargets and Therapy » Volume 12
LncRNA SNHG6 promotes the migration, invasion, and epithelial-mesenchymal transition of colorectal cancer cells by miR-26a/EZH2 axis
Authors Zhang M, Duan W, Sun W
Received 7 December 2018
Accepted for publication 28 March 2019
Published 2 May 2019 Volume 2019:12 Pages 3349—3360
DOI https://doi.org/10.2147/OTT.S197433
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Federico Perche
Mingyuan Zhang, Wenbiao Duan, Weiliang Sun
Department of Gastrointestinal Surgery, Yinzhou People’s Hospital, Ningbo, Zhejiang 315000, People’s Republic of China
Objective: Colorectal cancer (CRC) is a leading cause of cancer-related deaths worldwide. Small nucleolar RNA host gene 6 (SNHG6) was reported to function as an oncogene in a number of cancers. Here, we aimed to further explore the roles and molecular mechanism of SNHG6 in CRC metastasis.
Methods: The expression levels of SNHG6, miR-26a, and enhancer of zeste homolog 2 (EZH2) mRNA were assessed by quantification real-time PCR in CRC tissues and cell lines. Western blot analysis was performed to determine the levels of E-cadherin, Snail, Vimentin, N-cadherin, and EZH2. Cell migration and invasion capacities were detected by transwell assay. Dual-luciferase reporter assay or RNA Immunoprecipitation assay was employed to verify the interaction between SNHG6 and miR-26a, or EZH2 and miR-26a.
Results: Our data indicated that SNHG6 and EZH2 mRNA were upregulated, and miR-26a was downregulated in CRC tissues and cell lines. SNHG6 knockdown suppressed the migration, invasion, and epithelial-mesenchymal transition (EMT) of CRC cells. Moreover, SNHG6 binded to miR-26a and repressed miR-26a expression. EZH2 was a direct target of miR-26a, and it was regulated by SNHG6/miR-26a. MiR-26a inhibitor undermined the effect of SNHG6 knockdown on cell migration, invasion, and EMT. Additionally, EZH2 antagonized the effect of miR-26a on cell migration, invasion, and EMT in CRC cells.
Conclusion: SNHG6 knockdown suppressed cell migration, invasion, and EMT at least partly by sponging miR-26a and regulating EZH2 expression in CRC cells, providing a strategy for blocking CRC metastasis.
Keywords: small nucleolar RNA host gene 6 (SNHG6), miR-26a, enhancer of zeste homolog 2 (EZH2), epithelial-mesenchymal transition (EMT)
Introduction
Colorectal cancer (CRC), one of the most common malignancies, is a leading cause of cancer-related deaths around the world.1 Although the development of diagnostic and therapeutic methods has improved the survival rate of CRC patients, the prognosis of patients with distant metastases is unfavorable.2 Therefore, a better understanding of the molecular mechanism associated with metastasis in CRC is an urgent need.
Long non-coding RNAs (lncRNAs) are defined as non-coding functional transcripts of >200 nucleotides in length that are recognized as major players in a multitude of pathways across species.3 Aberrant regulation of lncRNAs is demonstrated to implicate in a variety of human diseases, including cancers.4 Small nucleolar RNA host gene 6 (SNHG6) was reported to function as an oncogene in a number of cancers, including hepatocellular carcinoma,5 gastric cancer,6 glioma,7 and lung adenocarcinoma.8 SNHG6 knockdown was confirmed to repress the migration and epithelial-mesenchymal transition (EMT) of glioma cells,7 lung adenocarcinoma,8 and gastric cancer cells.9 Additionally, upregulation of SNHG6 was associated with poor prognosis of CRC patients.10 SNHG6 enhanced the proliferation of CRC cells through directly inhibiting p21 expression by recruiting enhancer of zeste homolog 2 (EZH2) to the p21 promoter.11 Moreover, SNHG6 sequestered miR-760 to promote CRC progression by regulating FOXC1, highlighting its role as a potential therapeutic target for CRC.12 Hence, in the present study, we aimed to further explore the roles and molecular mechanism of SNHG6 in CRC metastasis.
MicroRNAs (miRNAs), a class of small non-coding RNAs of 19–23 nucleotides, act as a negative regulator of gene expression by binding to the 3ʹ-untranslated region of their target mRNAs.13 Deregulation of miRNAs is involved in multiple biological processes in cancer, including metastasis.14 MiR-26a has been reported as a tumor suppressor miRNA in CRC,15 osteosarcoma,16 and hepatocellular carcinoma.17 Additionally, previous studies demonstrated that EZH2 played an important role in the progression and metastasis of CRC.18,19 In the present study, our data indicated that SNHG6 and EZH2 mRNA were upregulated, and miR-26a was downregulated in CRC tissues and cell lines. Furthermore, SNHG6 knockdown suppressed cell migration, invasion, and EMT by sponging miR-26a and regulating EZH2 expression in CRC cells. Collectively, our data suggested the SNHG6 might serve as a potential strategy for blocking CRC metastasis.
Materials and methods
Clinical specimens and cell culture
Twenty-nine pairs of CRC tissues and corresponding noncancerous tissues were obtained from CRC patients who had undergone surgical resection at Yinzhou People’s Hospital between March 2014 and May 2016. All clinical specimens were stored at −80°C until RNA extraction. No conventional therapy was performed at pre-operation. Prior written informed consent from all patients and Institutional Review Board approval was obtained from the Ethics Committee of Yinzhou People’s Hospital in accordance with the ethical guidelines of the Declaration of Helsinki. Human CRC cell lines (SW480, SW620, HCT8, and HT-29) and human normal colon mucosal epithelial cell line (NCM460), purchased from American Type Culture Collection (ATCC, Manassas, VA, USA), were cultured in DMEM medium (Invitrogen, Karlsruhe, Germany) containing 10% fetal bovine serum (FBS, Biochrom AG, Berlin, Germany), 1% penicillin/streptomycin (Invitrogen) at 37°C in a humidified atmosphere of 5% CO2.
Cell transfection
The modified miR-26a mimics, miRNA inhibitors (anti-miR-26a), siRNA targeting SNHG6 (si-SNHG6), and corresponding negative controls were chemically enhanced oligonucleotides designed and synthetized by Applied Biosystems (Foster city, CA, USA). SNHG6 and EZH2 overexpression plasmids (Vector-SNHG6 and Vector-EZH2) also were chemically synthetized by Applied Biosystems, and Vector was used as a control. Cells were transfected with 100 mM of the indicated oligonucleotide or 50 ng of plasmid using Lipofectamine 2000 transfection reagent (Invitrogen) according to the protocols of manufacturers.
Quantification real-time PCR (qRT-PCR)
The expression levels of SNHG6, EZH2 mRNA, miR-26a, and miR-16 were examined by the qRT-PCR assay. Briefly, total RNA from tissues and cells was extracted using Trizol (Invitrogen). Then, the integrity and quality of RNA extracts were determined using an Agilent BioAnalyzer 2100 (Agilent, Palo Alto, CA, USA). For SNHG6 and EZH2 mRNA, cDNA was reversely transcribed from RNA extracts with M-MLV reverse transcriptase (Promega, Madison, WI, USA) and qRT-PCR was performed with a LightCycler 480 Probes Master Kit (Roche Diagnostics, Mannheim, Germany) on a LightCycler 480 instrument (Roche Diagnostics). The expression levels of SNHG6 and EZH2 mRNA were calculated by the 2−ΔΔCT method with GAPDH as an endogenous control. For miR-26a and miR-16, miScript II RT kit (Qiagen, Hilden, Germany), miScript Primer Assays (Qiagen), and miScript SYBR Green PCR kit (Qiagen) were used. The U6 small nuclear RNA was used as an internal control. Primers used for quantitative PCR were listed as follows: SNHG6: 5ʹ-ATACTTCTGCTTCGTTACCT-3ʹ (forward) and 5ʹ-CTCATTTTCATCATTTGCT-3ʹ (reverse); EZH2 mRNA: 5ʹ-TTGTTGGCGAAGCGTGTAAAATC-3ʹ (forward) and 5ʹ-TCCCTAGTCCCGCGCAATGAGC-3ʹ (reverse); GAPDH: 5ʹ-TATGACTCTACCCACGGCAAG-3ʹ (forward) and 5ʹ-TACTCAGCACCAGCATCACC-3ʹ (reverse).
Transwell assay of migration and invasion
For migration assay, 1.0×105 cells resuspended in 200 µL of serum-free medium were seeded into the upper chamber of transwell plates in a 24-well format with 8 µm pore size (BD Falcon, Franklin Lakes, NJ, USA). For invasion assay, 200 µL of serum-free medium containing 1.0×105 cells was added into the upper chamber with Matrigel-coated membrance (BD Falcon). In both assays, 600 µL of medium containing 10% FBS was added to the lower chamber as a chemoattractant. Twenty-four hours later, migrated or invaded cells were fixed with 90% methanol (Sigma–Aldrich, St. Louis, MO, USA) and stained with 0.1% crystal violet (Sigma–Aldrich). Images were captured, and the cells numbers were determined by Image J software (National Institutes of Health, Bethesda, Maryland, USA) under a microscope (Leica, Wetzlar, Germany) in random fields.
Western blot
Cells were lysed in lysis buffer comprised 50 mM Tris-HCl, pH=7.5, 0.1% SDS, 150 mM NaCl, 1% NP-40, 0.5% sodium-deoxycholate and protease inhibitor cocktail (Roche Diagnostics). The concentration of protein extracts was measured with the BCA Protein Assay Kit (Thermo Fisher Scientific, Waltham, MA, USA). Twenty micrograms of proteins was separated by gel electrophoresis on 10% gels, and then transferred to PVDF membranes (Roche Diagnostics). Blocked by 5% non-fat milk in Tween-20, the membranes were blotted with anti-E-cadherin (1:1,000, Cell Signaling Technology, Danvers, MA, USA), anti-Snail (1:1,000, Cell Signaling Technology), anti-Vimentin (1:500, Abcam, Cambridge, UK) and anti-N-cadherin (1:500, Abcam), anti-EZH2 (1:1,000, Cell Signaling Technology), and anti-β-actin (1:1,000, Cell Signaling Technology). Following the incubation with horseradish peroxidase-conjugated secondary antibodies (1:500, Abcam), the protein bands were visualized by using the enhanced chemiluminescence system (GE Healthcare, Chicago, IN, USA).
Dual-luciferase reporter assay
Online software Starbase v2.0 was performed to predict the target miRNAs of SNHG6. The wild-type SNHG6 reporter plasmid containing the potential binding sites of miR-26a (SNHG6-WT) and its mutant-type SNHG6-MUT was constructed by Applied Biosystems. For the luciferase assay, SNHG6-WT or SNHG6-MUT constructs were transiently transfected into SW480 and SW620 cells together with miR-26a mimics or miR-NC mimics. After 36 hrs post-transfection, the relative luciferase activities were determined using a Dual Luciferase Assay System (Promega).
RNA immunoprecipitation (RIP) assay
RIP assay was performed to examine the potentially endogenous interaction between SNHG6 and miR-26a, or EZH2 and miR-26a using Magna RIPTM RNA Immunoprecipitation Kit (Millipore, Bedford, MA, USA). Briefly, cells were transfected with miR-26a mimics, Vector-SNHG6, Vector-EZH2, or respective controls, and then were lysed in lysis buffer. Subsequently, cell lysates were incubated with anti-Ago2 (Abcam) or anti-IgG (Abcam) and protein A/G magnetic beads. The magnetic bead-bound complexes were purified by Dnase and Proteinase K (Applied Biosystems). Lastly, qRT-PCR assays were used to determine the relative enrichment of SNHG6, EZH2 mRNA, miR-26a, and miR-16 with a LightCycler 480 Probes Master Kit on a LightCycler 480 instrument.
Statistical analysis
All data were analyzed using SPSS 19.0 software (SPSS Inc., Chicago, IL, USA) and were presented as mean ± standard deviation (SD) of three independent experiments. The differences between the two groups were compared by the Student’s t-test. The one-way variance analysis (ANOVA) was used to analyze the differences between multiple groups. P-values less than 0.05 were considered statistically significant. *P<0.05, **P<0.01 or ***P<0.001.
Results
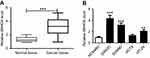
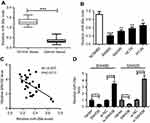
Upregulation of SNHG6 in CRC tissues and cell lines
Firstly, we detected the expression levels of SNHG6 in CRC tissues and adjacent noncancerous tissues by qRT-PCR. The data revealed a significant upregulation of SNHG6 expression in CRC tissues compared to normal control (Figure 1A). Then, SNHG6 expression was assessed in CRC cell lines. Results indicated that except HCT8 cells, SNHG6 expression was highly elevated in CRC cell lines (SW480, SW620, and HT-29) compared with normal cell line NCM460 (Figure 1B).
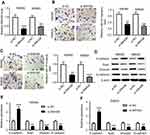
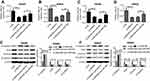
SNHG6 knockdown resulted in decreased migration, invasion and EMT of SW480 and SW620 cells
Then, loss-of-function experiments were performed to explore the function of SNHG6 on CRC metastasis by transfecting with si-SNHG6 into SW480 and SW620 cells. As shown in Figure 2A, si-SNHG6 transfection in SW480 and SW620 cells led to a drastic reduction of SNHG6 expression compared with negative control. Subsequently, transwell assays demonstrated that compared to respective controls, SNHG6 knockdown significantly suppressed the migration and invasion in SW480 and SW620 cells (Figure 2B and C). Further, western blot results revealed that SNHG6 knockdown resulted in a decrease of Snail, Vimentin, and N-cadherin levels, as well as an increase of E-cadherin expression, indicating an inhibition of EMT in SW480 and SW620 cells (Figure 2D–F).
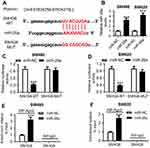
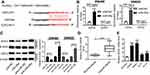
SNHG6 directly binded to miR-26a in SW480 and SW620 cells
To further explore the molecular mechanism by which SNHG6 knockdown inhibited the migration, invasion, and EMT of CRC cells, online software Starbase v2.0 was performed to predict the target miRNAs of SNHG6. Of interest, there existed several potential complementary sites between SNHG6 and miR-26a (Figure 3A). To confirm the prediction, dual-luciferase reporter assays were performed by transfecting with SNHG6-WT or SNHG6-MUT constructs into SW480 and SW620 cells with miR-26a mimics. As shown in Figure 3B, in comparison to control, transfection of miR-26 mimics resulted in about 3.25-fold increase of miR-26 expression in SW480 cells, and 4.23-fold increase in SW620 cells. Moreover, these results showed that compared with homologous control, miR-26a overexpression strikingly attenuated the luciferase activity of SNHG6-WT constructs, while it failed to affect the luciferase activity of SNHG6-MUT constructs (Figure 3C and 3D). RIP assay was used to examine the potentially endogenous interaction between SNHG6 and miR-26a. These data presented that SNHG6 was substantially enriched by miR-26a overexpression with anti-Ago2 in SW480 and SW620 cells (Figure 3E and 3F). Also, the association between SNHG6 and miR-26a in SW480 and SW620 cells was supported in Figure S1A and B.
miR-26a expression was downregulated in CRC and was suppressed by SNHG6
Further, we observed miR-26a expression in CRC tissues and cell lines. qRT-PCR results presented that compared to the corresponding counterpart, miR-26a expression was highly downregulated in CRC tissues and cell lines (Figure 4A and B). Moreover, miR-26a expression was inversely correlated with SNHG6 level in CRC tissues (Figure 4C). Additionally, SW480 and SW620 cells were transfected with Vector-SNHG6 or si-SNHG6, followed by the detection of the miR-26a level. Results revealed that compared to their counterpart, miR-26a level was significantly decreased by Vector-SNHG6 introduction, while it was markedly increased in the presence of si-SNHG6 (Figure 4D).
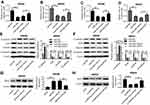
miR-26a inhibitor undermined the effect of SNHG6 knockdown on migration, invasion, and EMT of SW480 and SW620 cells
To provide further mechanistic insight into the link between SNHG6 and miR-26a on CRC metastasis, SW480, and SW620 cells were cotransfected with si-SNHG6 and anti-miR-26a. Transwell assays revealed that transfection of anti-miR-26a evidently antagonized si-SNHG6-mediated anti-migration and anti-invasion effects in SW480 and SW620 cells (Figure 5A–D). In parallel, the anti-EMT effect of si-SNHG6 was significantly abolished by anti-miR-26a transfection in SW480 and SW620 cells (Figure 5E and F).
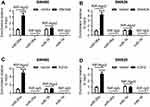
EZH2 was a direct target of miR-26a
Next, online software Starbase v2.0 was used to search for the targets of miR-26a. The predicted data presented that EZH2 was a potential target of miR-26a (Figure 6A). Thus, RIP assay was performed by transfecting with miR-26a mimics into SW480 and SW620 cells. Results presented that compared to negative control, the introduction of miR-26a mimics resulted in an abundant enrichment of EZH2 mRNA in SW480 and SW620 cells (Figure 6B). Moreover, the association between EZH2 and miR-26a was confirmed in Figure S1C and D. Further, we determined whether miR-26a regulated EZH2 expression in CRC cells. As expect, EZH2 expression was significantly reduced by – miR-26a mimics transfection, while it was highly elevated when introduced with anti-miR-26a in SW480 and SW620 cells (Figure 6C).
Subsequently, EZH2 mRNA expression was determined in CRC tissues and adjacent noncancerous tissues, and CRC cell lines and NCM460 cells. These data revealed a significant upregulation of EZH2 mRNA expression in CRC tissues and cell lines compared to their counterparts (Figure 6D and E).
EZH2 antagonized the effect of miR-26a on cell migration, invasion, and EMT, and it was regulated by SNHG6/miR-26a
Lastly, we investigated whether miR-26a exerting a regulatory function on CRC metastasis was mediated by EZH2. SW480 and SW620 cells were transfected with miR-26a mimics alone or together with Vector-EZH2. Transwell results indicated that transfection of miR-26a mimics in SW480 and SW620 cells led to a dramatical suppression of the migration and invasion compared to their counterparts (Figure 7A–D). Moreover, miR-26a mimics transfection significantly inhibited the EMT of SW480 and SW620 cells (Figure 7E and F). Further, our results presented that Vector-EZH2 transfection antagonized the effect of miR-26a on migration, invasion, and EMT in SW480 and SW620 cells (Figure 7A–F).
Further, we observed how did SNHG6/miR-26a affect EZH2 expression in CRC cells. These results revealed that compared with respective controls, EZH2 expression was strikingly increased by Vector-SNHG6 transfection in SW480 cells, while it was significantly decreased in the presence of si-SNHG6 in SW620 cells (Figure 7G and 7H). Additionally, the regulatory effect of SNHG6 on EZH2 expression was abrogated in response to the alteration of miR-26a expression in SW480 and SW620 cells (Figure 7G and 7H).
Discussion
CRC is a leading cause of tumor-related deaths worldwide, with the majority attributable to distant metastasis.20 Increasing evidences have suggested that some lncRNAs play promotional roles in CRC metastasis. For example, upregulation of TUG1 enhanced the migration and invasion of CRC cells in vitro and promoted their metastasis in vivo.21 LncRNA-ATB overexpression was tightly associated with lymph node metastasis and hematogenous metastasis of CRC.22 Also, 91H was verified as an independent distant metastasis indicator and 91H silencing repressed the migration and invasion capacities of CRC cells.23
Previous studies demonstrated that the dysregulation of SNHG6 was closely involved in various cancers metastasis. For instance, Chang et al,5 reported that upregulation of SNHG6 accelerated tumor growth and metastasis through inducing EMT in hepatocellular carcinoma. Wang et al,24 verified that SNHG6 overexpression enhanced the migration and invasion via miR-125b/NUAK1 axis, and promoted EMT by upregulating Snail1/2 expression in bladder cancer cells. Yan and colleague9 found that SNHG6 overexpression was associated with invasion depth, distant metastasis, and TNM stage, and SNHG6 knockdown inhibited EMT processes of gastric cancer cells. In the present study, our data indicated that SNHG6 was upregulated in CRC tissues and cells, consistent with previous studies.10,11 We also found that SNHG6 knockdown repressed the migration and invasion of CRC cells, similar to a recent document.12 Moreover, we firstly manifested that SNHG6 knockdown resulted in decreased EMT in CRC cells. All these results hinted that SNHG6 might contribute to the metastasis of CRC.
The competing endogenous RNA hypothesis proposed that lncRNA might act as natural miRNA sponges to inhibit intracellular miRNA function.25 Thus, online software Starbase v2.0 was performed to predict the target miRNAs of SNHG6. Among these candidates, miR-26a was selected for further research due to its role as a tumor suppressor miRNA in various cancers, such as nasopharyngeal carcinoma,26 breast cancer,27 and gastric cancer.28 Moreover, downregulation of miR-26a was associated with lymph node metastasis and miR-26a repressed the metastasis in gastric cancer.28 MiR-26a was also demonstrated to function as an anti-metastasis miRNA in nasopharyngeal carcinoma,29 triple negative breast cancer30, and hepatocellular carcinoma.31 Additionally, miR-26a repressed CRC progression by hindering the binding of hnRNP A1-CDK6 mRNA and inducing apoptosis.32 High expression of miR-26a led to an inhibition of migration behavior of CRC cells and fucosyltransferase 4 expression, a protein associated with invasion and metastatic properties of CRC.15 Further, we first confirmed that SNHG6 binded to miR-26a and repressed miR-26a expression in CRC cells. Our data also indicated a significant downregulation of miR-26a expression in CRC tissues and cells, in accordance with former work.15 Furthermore, we validated that SNHG6 knockdown exerted its anti-migration, anti-invasion, and anti-EMT effects through sponging miR-26a in CRC cells.
Then, online software Starbase v2.0 was used to search for the targets of miR-26a. Among these candidates, EZH2 was chosen for follow-up experiments because it played vital roles in the development, progression, and metastasis of CRC.18,19,33 Moreover, EZH2 upregulation was a mark of advanced and metastasis tumors including breast and prostate cancer.34,35 In addition, EZH2 overexpression epigenetically silenced a number of miRNAs, and thus negatively regulated the metastasis of hepatocellular carcinoma.36 EZH2 depletion induced by H19 promoted bladder cancer metastasis via inhibition of E-cadherin expression.37 In this study, we verified that EZH2 was upregulated in CRC tissues and cell lines. Also, we manifested that EZH2 was a direct target of miR-26a in CRC cells, which was consistent with previous reports.29,38 Moreover, EZH2 antagonized the effect of miR-26a on cell migration, invasion, and EMT, and it was regulated by SNHG6/miR-26a in CRC cells.
Conclusion
In conclusion, our data indicated that SNHG6 knockdown repressed the migration, invasion, and EMT at least partly by sponging miR-26a and regulating EZH2 expression in CRC cells. Target SNHG6 might be a potential strategy for blocking CRC metastasis.
Ethical statement
Prior written informed consent from all patients and Institutional Review Board approval was obtained at Yinzhou People’s Hospital.
Disclosure
All authors declare that they have no conflicts of interest in this work.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30. doi:10.3322/caac.21387
2. Siegel RL, Miller KD, Fedewa SA, et al. Colorectal cancer statistics, 2017. CA Cancer J Clin. 2017;67(3):177–193. doi:10.3322/caac.21395
3. Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet. 2014;15(1):7–21. doi:10.1038/nrg3606
4. Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Mol Cancer. 2011;10:38. doi:10.1186/1476-4598-10-93
5. Chang L, Yuan Y, Li C, et al. Upregulation of SNHG6 regulates ZEB1 expression by competitively binding miR-101-3p and interacting with UPF1 in hepatocellular carcinoma. Cancer Lett. 2016;383(2):183–194. doi:10.1016/j.canlet.2016.09.034
6. Li Y, Li D, Zhao M, et al. Long noncoding RNA SNHG6 regulates p21 expression via activation of the JNK pathway and regulation of EZH2 in gastric cancer cells. Life Sci. 2018;208:295–304. doi:10.1016/j.lfs.2018.07.032
7. Meng Q, Yang BY, Liu B, Yang JX, Sun Y. Long non-coding RNA SNHG6 promotes glioma tumorigenesis by sponging miR-101-3p. Int J Biol Markers. 2018;33(2):148–155. doi:10.1177/1724600817747524
8. Liang R, Xiao G, Wang M, et al. SNHG6 functions as a competing endogenous RNA to regulate E2F7 expression by sponging miR-26a-5p in lung adenocarcinoma. Biomed Pharmacother. 2018;107:1434–1446. doi:10.1016/j.biopha.2018.08.099
9. Yan K, Tian J, Shi W, Xia H, Zhu Y. LncRNA SNHG6 is associated with poor prognosis of gastric cancer and promotes cell proliferation and EMT through epigenetically silencing p27 and sponging miR-101-3p. Cell Physiol Biochem. 2017;42(3):999–1012. doi:10.1159/000478682
10. Li M, Bian Z, Yao S, et al. Up-regulated expression of SNHG6 predicts poor prognosis in colorectal cancer. Pathol Res Pract. 2018;214(5):784–789. doi:10.1016/j.prp.2017.12.014
11. Li Z, Qiu R, Qiu X, Tian T. SNHG6 Promotes Tumor Growth via Repression of P21 in Colorectal Cancer. Cell Physiol Biochem. 2018;49(2):463–478. doi:10.1159/000492986
12. Zhu Y, Xing Y, Chi F, Sun W, Zhang Z, Piao D. Long noncoding RNA SNHG6 promotes the progression of colorectal cancer through sponging miR-760 and activation of FOXC1. Onco Targets Ther. 2018;11:5743–5752. doi:10.2147/OTT.S170246
13. Hammond SM. An overview of microRNAs. Adv Drug Deliv Rev. 2015;87:3–14. doi:10.1016/j.addr.2015.05.001
14. Reddy KB. MicroRNA (miRNA) in cancer. Cancer Cell International. 2015;15(1):38. doi:10.1186/s12935-015-0185-1
15. Li Y, Sun Z, Liu B, Shan Y, Zhao L, Jia L. Tumor-suppressive miR-26a and miR-26b inhibit cell aggressiveness by regulating FUT4 in colorectal cancer. Cell Death Dis. 2017;8(6):e2892. doi:10.1038/cddis.2017.518
16. Wang Z, Wang Z, Liu J, Yang H. Long non-coding RNA SNHG5 sponges miR-26a to promote the tumorigenesis of osteosarcoma by targeting ROCK1. Biomed Pharmacother. 2018;107:598–605. doi:10.1016/j.biopha.2018.08.025
17. Li Y, Guo D, Zhao Y, et al. Long non-coding RNA SNHG5 promotes human hepatocellular carcinoma progression by regulating miR-26a-5p/GSK3beta signal pathway. Cell Death Dis. 2018;9(9):888. doi:10.1038/s41419-018-0882-5
18. Tang J, Zhong G, Wu J, Chen H, Jia Y. Long noncoding RNA AFAP1-AS1 facilitates tumor growth through enhancer of zeste homolog 2 in colorectal cancer. American Journal of Cancer Research. 2018;8(5):892–902.
19. Li X, Xing J, Wang H, Yu E. The SLC34A2-ROS-HIF-1-induced upregulation of EZH2 expression promotes proliferation and chemo-resistance to apoptosis in colorectal cancer. Bioscience Reports. 2018.
20. Chaffer CL, Weinberg RA. A perspective on cancer cell metastasis. Science. 2011;331(6024):1559–1564. doi:10.1126/science.1203543
21. Sun J, Ding C, Yang Z, et al. The long non-coding RNA TUG1 indicates a poor prognosis for colorectal cancer and promotes metastasis by affecting epithelial-mesenchymal transition. J Transl Med. 2016;14:42. doi:10.1186/s12967-016-0786-z
22. Iguchi T, Uchi R, Nambara S, et al. A long noncoding RNA, lncRNA-ATB, is involved in the progression and prognosis of colorectal cancer. Anticancer Res. 2015;35(3):1385–1388.
23. Deng Q, He B, Gao T, et al. Up-regulation of 91H promotes tumor metastasis and predicts poor prognosis for patients with colorectal cancer. PLoS One. 2014;9(7):e103022. doi:10.1371/journal.pone.0103022
24. Wang C, Tao W, Ni S, Chen Q. Upregulation of lncRNA snoRNA host gene 6 regulates NUAK family SnF1-like kinase-1 expression by competitively binding microRNA-125b and interacting with Snail1/2 in bladder cancer. J Cell Biochem. 2019;120(1):357–367. doi:10.1002/jcb.27387
25. Kartha RV, Subramanian S. Competing endogenous RNAs (ceRNAs): new entrants to the intricacies of gene regulation. Front Genet. 2014;5:8. doi:10.3389/fgene.2014.00008
26. Lu J, He ML, Wang L, et al. MiR-26a inhibits cell growth and tumorigenesis of nasopharyngeal carcinoma through repression of EZH2. Cancer Res. 2011;71(1):225–233. doi:10.1158/0008-5472.CAN-10-1850
27. Gao J, Li L, Wu M, et al. MiR-26a inhibits proliferation and migration of breast cancer through repression of MCL-1. PLoS One. 2013;8(6):e65138. doi:10.1371/journal.pone.0065138
28. Deng M, Tang HL, Lu XH, et al. miR-26a suppresses tumor growth and metastasis by targeting FGF9 in gastric cancer. PLoS One. 2013;8(8):e72662. doi:10.1371/journal.pone.0072662
29. Yu L, Lu J, Zhang B, et al. miR-26a inhibits invasion and metastasis of nasopharyngeal cancer by targeting EZH2. Oncol Lett. 2013;5(4):1223–1228. doi:10.3892/ol.2013.1173
30. Liu P, Tang H, Chen B, et al. miR-26a suppresses tumour proliferation and metastasis by targeting metadherin in triple negative breast cancer. Cancer Lett. 2015;357(1):384–392. doi:10.1016/j.canlet.2014.11.050
31. Yang X, Liang L, Zhang XF, et al. MicroRNA-26a suppresses tumor growth and metastasis of human hepatocellular carcinoma by targeting interleukin-6-Stat3 pathway. Hepatology. 2013;58(1):158–170. doi:10.1002/hep.26305
32. Konishi H, Fujiya M, Ueno N, et al. microRNA-26a and −584 inhibit the colorectal cancer progression through inhibition of the binding of hnRNP A1-CDK6 mRNA. Biochem Biophys Res Commun. 2015;467(4):847–852. doi:10.1016/j.bbrc.2015.10.055
33. Liu X, Cui L, Hua D. Long non-coding RNA XIST regulates miR-137-EZH2 axis to promote tumor metastasis in colorectal cancer. Oncol Res. 2018. doi:10.3727/096504018X15195193936573
34. Yoo KH, Hennighausen L. EZH2 methyltransferase and H3K27 methylation in breast cancer. Int J Biol Sci. 2012;8(1):59–65.
35. Sreekumar A, Poisson LM, Rajendiran TM, et al. Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature. 2009;457(7231):910–914. doi:10.1038/nature07762
36. Au SL, Wong CC, Lee JM, et al. Enhancer of zeste homolog 2 epigenetically silences multiple tumor suppressor microRNAs to promote liver cancer metastasis. Hepatology. 2012;56(2):622–631. doi:10.1002/hep.25679
37. Luo M, Li Z, Wang W, Zeng Y, Liu Z, Qiu J. Long non-coding RNA H19 increases bladder cancer metastasis by associating with EZH2 and inhibiting E-cadherin expression. Cancer Lett. 2013;333(2):213–221. doi:10.1016/j.canlet.2013.01.033
38. Zhang B, Liu XX, He JR, et al. Pathologically decreased miR-26a antagonizes apoptosis and facilitates carcinogenesis by targeting MTDH and EZH2 in breast cancer. Carcinogenesis. 2011;32(1):2–9. doi:10.1093/carcin/bgq209
Supplementary material
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.