Back to Journals » OncoTargets and Therapy » Volume 13
LncRNA SNHG11 Promotes Proliferation, Migration, Apoptosis, and Autophagy by Regulating hsa-miR-184/AGO2 in HCC
Authors Huang W, Huang F , Lei Z, Luo H
Received 3 November 2019
Accepted for publication 19 December 2019
Published 14 January 2020 Volume 2020:13 Pages 413—421
DOI https://doi.org/10.2147/OTT.S237161
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr XuYu Yang
Wei Huang, Feizhou Huang, Zhao Lei, Hongwu Luo
Department of Hepatopancreatobiliary Surgery, The Third Xiangya Hospital, Central South University, Changsha 410013, Hunan, People’s Republic of China
Correspondence: Hongwu Luo
Department of Hepatopancreatobiliary Surgery, The Third Xiangya Hospital, Central South University, Changsha 410013, Hunan, People’s Republic of China
Tel/Fax +86 731 8861 8576
Email [email protected]
Background: The most common malignant tumor of the digestive system is HCC. However, the mechanism and pathogenesis of HCC occurrence and progress are still unknown. LncRNA is closely related to the occurrence and progress of HCC. It is important to investigate the effect and role of lncRNA in HCC.
Materials and Methods: LncRNA microarray assay was used to screen the differential expression profile of lncRNA. SNHG11, miR-184 and GO2 expression was analyzed by RT-PCR. The ability of SNHG11 to serve as a sponge for miRNA and the fact that miR-184 directly targets mRNA were revealed by dual luciferase assay and RIP. Apoptosis and autophagy related proteins were detected by Western blot. Cell proliferation, invasion, migration, and apoptosis were detected by CCK-8 assay, wound healing assay, transwell assay, and flow cytometry.
Results: LncRNA microarray assay and RT-PCR results revealed that the expression of SNHG11 was increased in HCC tumor tissues and also upregulated in HCC cells. SNHG11 had a connection with poor survival rate in HCC. In addition, dual luciferase assay and RIP results revealed that SNHG11 serves as a sponge for miR-184 and miR-184 directly targets AGO2. Pearson correlation analysis showed that SNHG11 with miR-184 and miR-184 with AGO2 were negative correlations, and SNHG11 with AGO2 was a positive correlation. Cell function assay and Western blot showed SNHG4/miR-184/AGO2 regulatory loop was critical for HCC cell proliferation, migration, apoptosis, and autophagy.
Conclusion: Our study demonstrated that the expression of SNHG11 is higher in HCC; moreover, SNHG11 promotes proliferation, migration, apoptosis, and autophagy by regulating AGO2 via miR-184 in HCC. Our verification of the role of SNHG11 may provide a novel biomarker for the diagnosis, therapy, and prognosis of HCC.
Keywords: hepatocellular cancer, LncRNA SNHG11, apoptosis, biomarker
Introduction
Hepatocellular carcinoma (HCC) forms 75–85% of primary liver cancer. Globally, liver cancer has become the sixth most common cancer, comprising about 5% of new cases of cancer. It is also the fourth leading cause of cancer death and accounts for about 8% of cancer deaths in 2018.1 Therefore, it is necessary to understand the occurrence and development mechanisms of HCC. Finding new molecular targets is very important to HCC prevention and treatment.
Long non-coding RNA (lncRNA) is a type of RNA more than 200 nt in length. Due to the differential expression of lncRNAs in many tissues and cells, lncRNAs have become a “star gene” of high concern in recent years. LncRNAs have been recognized as being closely related to a variety of diseases, such as the occurrence, development, and prognosis of malignant tumors, cardiovascular diseases, and endocrine diseases, and have played important roles in regulating physiological processes such as cell proliferation, differentiation, and apoptosis.2–7 Therefore, further research and verification of the role of abnormal lncRNA expression in HCC may provide novel ideas for the treatment of HCC.
In our study, LncRNA SNHG11 was increased in HCC tumor tissues and in 4 HCC cells. Overexpression of SNHG11 led to poor survival rate in HCC. To investigate the effect and role of SNHG11 in HCC, bioinformatics analysis, and luciferase reporter assay found that the binding sites of SNHG11 are miR-184 and miR-184 directly targets argonaute RISC catalytic component 2(AGO2), SNHG11 with AGO2 was positively correlation. Further, our investigations revealed that the SNHG4/miR-184/AGO2 regulatory loop was critical for HCC cell proliferation, migration, apoptosis, and autophagy.
Materials and Methods
Tumor Tissues and Normal Tissues
From 2017 to 2018, HCC tumor tissues (n=57) and matched normal samples (n=57) were obtained from Third Xiangya Hospital, Central South University. Before surgery, none of them received radiotherapy or chemotherapy. During surgery, all tissues were stored in liquid nitrogen for further research. The study was approved by the Ethics Committee of Xiangya Third Hospital, and written informed consent was signed with the patients before surgery.
Cell Culture and Cell Transfection
HL-7702, SK-HEP-1, Hep G2, HuH-7, and Li-7 were obtained from the Cell Collection Committee of the Chinese Academy of Sciences (Shanghai, China) and cultured at 37°C in a 5% CO2 humidified incubator.
LncRNA SNHG11 cDNA was synthesized and cloned into vector (Biotech, China). Plasmids were transfected into SK-HEP-1 and Hep G2 cells with Lipofectamine 2000 (Invitrogen, USA) according to the manufacture’s protocol.
RNA Extraction and lncRNA Microarray Analysis
Total RNA was extracted from HCC tissues and cells by Trizol (Invitrogen, USA). According to Low Input Quick Amp WT Labeling Kit (Agilent, USA) and standard operation procedure, the qualified samples of total RNA were amplified by cDNA. SBC human lncRNA microarray (Shanghai Biotechnology Corporation, China) was used to screen the expression profile of lncRNA.
Real-Time PCR Analysis
SNHG11, miR-184, and AGO2 expression were measured by real-time qPCR with the CFX96Tm Real-Time System (Bio-Rad, USA). Sangon Biotech (Shanghai, China) provided the primers and GAPDH/U6 acted as the internal reference. The following primers were used: SNHG11-Forward: TGGGAGTTGTCATGTTGGGA, SNHG11-Reverse: ACTCGTCACTCTTG -GTCTGT; miR-184-Forward: TGGACGGAGAACTGAUAAGGGT, miR-184-Reverse: GTGCA -GGGTCCGAGGT; AGO2-Forward: AGTGCCCGAGGAGAGTTAAC, AGO2-Reverse: AGAT -GCGATCCTTGCCTTCT; U6-Forward: CTCGCTTCGGCAGCACA, U6-Reverse: AACGCT -TCACGAATTTGCGT; GAPDH-Forward: ATGTTCGTCATGGGTGTGAA, GAPDH-Reverse: CAGTGATGGCATGGACTGT. Data were analyzed using the comparative Ct(2−ΔΔCt) method.
CCK8 Analysis
Aliquots of 100 μL with a concentration of 5 × 105/mL cells were seeded into a 96-well plate for 24 h, then 10 μL CCK-8 reagent was added and incubated at 37°C for 2 h in the dark. The absorbance values of each group were measured with a microplate detector (EnSpire 2300, PerkinElmer, USA) at 450 nm, and repeated three times in each group.
Flow Cytometry Assay
SK-HEP-1, Hep G2 cells transfected with the plasmids were stained with propidium iodide (PI) and Annexin V-FITC Apoptosis Detection Kit I (BD, USA) according to manufacturer’s protocol, then the cells were measured by FACS (BD, USA) for apoptosis and Cell Quest Research Software (BD, USA) was used to analyze data.
Wound Healing and Transwell Assay
For this assay, 5 × 105 cells were inoculated into 6-well plates for 24 h, 200 μL tips were used to draw three lines on 6-well plate, then cells were incubated at 37°C for 48 h and observed with an optical microscope (Nikon, Japan).
Aliquots of 100μL with a concentration of 1×105/mL cells were inoculated in the upper chambers, and to the bottom chamber was added medium containing 10% FBS for about 24 h. Then the chamber was fixed in anhydrous methanol for 20 min, dyed with 0.1% crystal violet for 20 min, before washing and drying. An inverted microscope (Nikon, Japan) was used to measure the number of migrating cells.
Luciferase Reporter Assay
SNHG11 wild-type (WT), AGO2 WT, and SNHG11 mutant-type (MUT), AGO2 MUT were constructed. The miRNA mimics and negative control were obtained from Ribobio (Guangzhou, China). The luciferase activity and normalized Renilla activity were measured by dual luciferase reporter assay system (Promega, Madison, WI) following the manufacturer’s protocol.
Pull-Down Assay with Biotinylated miRNA
The pull-down assay with biotinylated miRNA was performed as described previously,8 then the RNAs were purified using RNeasy Mini Kit (QIAGEN), and SNHG11 and miR-184 were detected by qRT-PCR.
Western Blot Assay
Protein was extracted with RIPA reagent from SK-HEP-1, Hep G2 cells, and the concentration of protein was tested by BCA Protein Concentration Assay Kit (Thermo Scientific, USA) following the manufacturer’s protocol. According to the experimental design, the primary antibodies we selected were: AGO2, LC3 I/LC3 II, Beclin-1, cleaved caspase3, and GAPDH (Cell Signaling Technology, USA). The chemiluminescence system (Bio-Rad, USA) imaging system was used to measure the protein samples.
In vitro Tumor Xenografts
About 1×106 cells were injected into the upper back of 4–8-week BALB/c Nude mice. The tumor size was measured as follows: 0.5×W2×L every week. One month later, the BALB/c Nude mice were killed and the tumor size was measured. The laboratory animals were approved by the medical laboratory animal ethics committee of Third Xiangya Hospital, Central South University. Instructions with respect to caring for laboratory animals (released by the Ministry of Science and Technology of the People’s Republic of China on September 30, 2006) were followed for the welfare of the animals.
Statistical Analysis
All data were analyzed by SPSS 20.0 and shown as means ± standard; the differences between group data were analyzed by ANOVA and post hoc Tukey’s test. Survival analysis was performed by KaplanMeier curves and log rank test and Pearson correlation coefficient analysis was used to analysis SNHG11, miR-184, AGO2. *P<0.05, #P<0.05 were considered statistically significant.
Results
SNHG11 Was Upregulated in HCC Patients and HCC Cell Lines
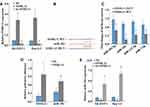
LncRNA microarray assay was used to screen the differential expression profile of lncRNA and detected thousands of lncRNA abnormal expressions in HCC tumor tissues compared with normal samples (Figure 1A). The highest expression levels of lncRNAs were selected and tested by RT-PCR; lncRNA SNHG11 expression was higher in HCC tumor tissues compared with normal samples (Figure 1B). Kaplan–Meier curve and log rank test showed that higher expression of SNHG11 had poor survival rate in HCC (Figure 1C). SNHG11 was increased in HCC cells compared to normal cell lines (Figure 1D).
SNHG11 Serves as a Sponge for miR-184 in HCC
To investigate the function and role of SNHG11 in HCC, sh-SNHG11, SNHG11 overexpression, and control groups were transfected into SK-HEP-1 and Hep G2 cells. SNHG11 expression had a higher level when treated with SNHG11 overexpression and had a lower level when treated with sh-SNHG11 in SK-HEP-1 and Hep G2 (Figure 2A). Bioinformatics analysis found that SNHG11 may have 4 miRNAs (miR-183-5p, miR-184, miR-122-5p, miR-7-5p) directly binding sites. The luciferase reporter assay found that miR-184 mimic could significantly decrease luciferase activity was significant reduced by WT-SNHG11 (Figure 2B and C). RIP assay showed that miR-184 interacts with SNHG11 (Figure 2D), miR-184 expression was increased in the sh-SNHG11 group and decreased in the SNHG11 overexpression group compared with the control group in SK-HEP-1 and Hep G2 (Figure 2E).
SNHG11 as a ceRNA Binding to miR-184 and Targets AGO2 in HCC Cells
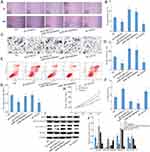
Bioinformatics analysis revealed that miR-184 and AGO2 3ʹ-UTR may have binding sites (Figure 3A). Future, luciferase reporter assay showed that luciferase activity was significantly inhibited by co-transfected WT-AGO2 and miR-184 (Figure 3B). To investigate whether SNHG11 regulated the expression of AGO2 via targeting miR-184, we compared AGO2 mRNA expression mediated by sh-SNHG11, sh-SNHG11+ miR-184 inhibitor, miR-184 inhibitor, sh-AGO2, and negative control by RT-PCR. AGO2 mRNA expression was increased in sh-SNHG11+miR-184 inhibitor and miR-184 inhibitor group and significantly reduced in sh-SNHG11 and sh-AGO2 group (Figure 3C and D).
The expression of miR-184 and AGO2 was measured by RT-qPCR in 57 pairs of HCC tumor tissues and matched normal samples; miR-184 expression was decreased and AGO2 expression was increased in HCC tumor tissues (Figure 3E and F). In addition, Pearson correlation analysis showed that SNHG11 with miR-184 and miR-184 with AGO2 were negative correlations and SNHG11 with AGO2 was a positive correlation. Above all, SNHG11 directly binds miR-184 to target AGO2 in HCC (Figure 3G).
SNHG4/miR-184/AGO2 Regulatory Loop Is Critical for HCC Cell Proliferation, Migration, Apoptosis, and Autophagy
In vitro, cell proliferation, invasion, migration, and apoptosis w detected by CCK-8 assay, wound healing assay, transwell assay, and flow cytometry; sh-SNHG11 and sh-AGO2 groups could significantly suppress cell viability, invasion, migration, and promoted apoptosis, while sh-SNHG11+miR-184 inhibitor and miR-184 inhibitor groups could significantly enhance proliferation, invasion, and migration, and inhibited apoptosis in Hep G2 (Figure 4A–G). When SNHG11 and AGO2 expression were decreased, the cell activity, invasion, and migration decreased and apoptosis increased. When miR-184 expression was reduced, it could neutralize the inhibition of cell viability, invasion, and migration, and promoted apoptosis in HCC. In addition, in vivo, the tumor average volume of overexpression of the SNHG11 groups was significantly larger than the control group. However, the tumor average volume contrasted with knockdown SNHG11 expression groups (Figure 4H). Western blot detected the expression of apoptosis and autophagy related protein: AGO2, Beclin-1, LC3 II/LC3 I, cleaved Caspase3, and AGO2, Beclin-1, and LC3 II/LC3 I expression was decreased and cleaved Caspase3 expression was increased in the sh-SNHG11, sh-AGO2 group, while the opposite result was found in sh-SNHG11+miR-184 inhibitor and miR-184 inhibitor groups (Figure 4I and J). In conclusion, SNHG11 downregulated the expression of miR-184 and upregulated AGO2 expression, indicating that SNHG11 acts as a ceRNA, binding miR-184 and targeting AGO2 to enhance cell viability, invasion, migration, apoptosis, and autophagy in HCC.
Discussion
HCC is a disease that seriously endangers human health. It is important to explore the pathogenesis of HCC and find effective early diagnostic markers and biological treatment targets. More and more evidence shows that lncRNA is closely related to the occurrence and progress of HCC.9,10 In our study, LncRNA microarray assay and RT-PCR results showed that SNHG11 had higher expression in HCC tumor tissues compared with normal samples. SNHG11 was also increased in HCC cells than normal cell lines. Bioinformatics analysis revealed that SNHG11 had poor survival rate in HCC.
lncRNA can act as a ceRNA to interact with miRNA and participate in the regulation of targeted gene expression. SNHG11 promotes proliferation and metastasis by targeting the Hippo pathway in colorectal cancer and may act as diagnostic markers for the detection of pancreatic cancer and prostate cancer.11–13 Recent studies have shown that lncRNA regulates cell proliferation, apoptosis, metastasis, invasion, angiogenesis, lipid metabolism, EMT, and many other processes, and has an impact on tumor size, TNM grade, postoperative survival, and prognosis of HCC.8,14–16 Bioinformatics analysis and luciferase reporter assay found that the binding site of SNHG11 is miR-184 and miR-184 directly targets AGO2. In this study, SNHG11 downregulated the expression of miR-184 and upregulated AGO2 expression, and thereby regulated cell viability, invasion, migration, and promoted apoptosis. Recent studies have shown that AGO2 might be involved in carcinogenesis, cell proliferation, migration, invasion, arrested cell cycle, and induced apoptosis, or recurrence of diffuse glioblastoma multiform, HCC, and hypopharyngeal cancer.17–19
Caspase-3 is currently recognized as an apoptosis-related gene, which can reflect apoptosis. Beclin-1 is currently recognized as genes involved in autophagy and promotes the occurrence of autophagy.20–23 Western blot revealed that sh-SNHG11 could downregulate Beclin-1 expression, while with AGO2, cleaved Caspase3 expression was increased, indicating that SNHG11 acts as a ceRNA binding miR-184 and targeting AGO2 to regulate autophagy and promoted apoptosis in HCC.
Conclusion
Our study demonstrated that SNHG11 is upregulated in HCC tissues and cell lines, and bioinformatics analysis revealed that SNHG11 has a connection with poor survival rate in HCC. Moreover, SNHG11 serves as a sponge for miR-184 and regulates AGO2 expression; the SNHG4/miR-184/AGO2 regulatory loop is critical for HCC cell proliferation, migration, apoptosis, and autophagy. SNHG11 may provide a novel biomarker for the diagnosis, therapy, prognosis of HCC.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.v68.6
2. Pant T, Dhanasekaran A, Bai X, et al. Genome-wide differential expression profiling of lncRNAs and mRNAs associated with early diabetic cardiomyopathy. Sci Rep. 2019;9(1):15345. doi:10.1038/s41598-019-51872-9
3. Wu D, Zhou Y, Fan Y, et al. LncRNA CAIF was downregulated in end-stage cardiomyopathy and is a promising diagnostic and prognostic marker for this disease. Biomarkers. 2019;24:1–4.
4. Xin Y, He X, Zhao W, et al. LncRNA PCAT6 increased cholangiocarcinoma cell proliferation and invasion via modulating miR-330-5p. Am J Transl Res. 2019;11(9):6185–6195.
5. Yu X, Zhe Z, Tang B, et al. α-Asarone suppresses the proliferation and migration of ASMCs through targeting the lncRNA-PVT1/miR-203a/E2F3 signal pathway in RSV-infected rats. Acta Biochim Biophys Sin (Shanghai). 2017;49(7):598–608. doi:10.1093/abbs/gmx048
6. Rochet E, Appukuttan B, Ma Y, Ma Y, Ashander LM, Smith JR. Expression of long non-coding RNAs by human retinal muller glial cells infected with clonal and exotic virulent toxoplasma gondii. NORAD. 2019;5(4):48.
7. Xie Q, Chen S, Tian R, et al. Long noncoding RNA ITPRIP-1 positively regulates the innate immune response through promotion of oligomerization and activation of MDA5. J Virol. 2018;92(17). doi:10.1128/JVI.00507-18.
8. Li H, Han Q, Chen Y, et al. Upregulation of the long non-coding RNA FOXD2-AS1 is correlated with tumor progression and metastasis in papillary thyroid cancer. Am J Transl Res. 2019;11(9):5457–5471.
9. Xu W, Zhou G, Wang H, et al. Circulating lncRNA SNHG11 as a novel biomarker for early diagnosis and prognosis of colorectal cancer. Int J Cancer. 2019. doi:10.1002/ijc.32747
10. Ye G, Guo L, Xing Y, Sun W, Yuan M. Identification of prognostic biomarkers of prostate cancer with long non-coding RNA-mediated competitive endogenous RNA network. Exp Ther Med. 2019;17(4):3035–3040. doi:10.3892/etm.2019.7277
11. Jiang W, Cheng X, Wang T, Song X, Zheng Y, Wang L. LINC00467 promotes cell proliferation and metastasis by binding with IGF2BP3 to enhance the mRNA stability of TRAF5 in hepatocellular carcinoma. J Gene Med. 2019;e3134.
12. Yan PH, Wang L, Chen H, et al. LncRNA RUNX1-IT1 inhibits proliferation and promotes apoptosis of hepatocellular carcinoma by regulating MAPK pathways. Eur Rev Med Pharmacol Sci. 2019;23(19):8287–8294. doi:10.26355/eurrev_201910_19139
13. Zhang H, Liao Z, Liu F, et al. Long noncoding RNA HULC promotes hepatocellular carcinoma progression. Aging (Albany NY). 2019;11(20):9111.
14. Fu C, Xu X, Lu W, Nie L, Yin T, Wu D. Increased expression of long non-coding RNA CCAT2 predicts poorer prognosis in patients with hepatocellular carcinoma. Medicine (Baltimore). 2019;98(42):e17412. doi:10.1097/MD.0000000000017412
15. Song W, Zhang J, Zhang J, Sun M, Xia Q. Overexpression of lncRNA PIK3CD-AS1 promotes expression of LATS1 by competitive binding with microRNA-566 to inhibit the growth, invasion and metastasis of hepatocellular carcinoma cells. Cancer Cell Int. 2019;19:150. doi:10.1186/s12935-019-0857-3
16. Zhang JJ, Xu WR, Chen B, et al. The up-regulated lncRNA DLX6-AS1 in colorectal cancer promotes cell proliferation, invasion and migration via modulating PI3K/AKT/mTOR pathway. Eur Rev Med Pharmacol Sci. 2019;23(19):8321–8331. doi:10.26355/eurrev_201910_19143
17. Zhang Y, Wang B, Chen X, Li W, Dong P. AGO2 involves the malignant phenotypes and FAK/PI3K/AKT signaling pathway in hypopharyngeal-derived FaDu cells. Oncotarget. 2017;8(33):54735–54746. doi:10.18632/oncotarget.18047
18. Cheng N, Li Y, Han ZG. Argonaute2 promotes tumor metastasis by way of up-regulating focal adhesion kinase expression in hepatocellular carcinoma. Hepatology. 2013;57(5):1906–1918. doi:10.1002/hep.26202
19. Li Z, Hu C, Zhen Y, Pang B, Yi H, Chen X. Pristimerin inhibits glioma progression by targeting AGO2 and PTPN1 expression via miR-542-5p. Biosci Rep. 2019;39(5):BSR20182389.
20. Chen C, Wang K, Wang Q, Wang X. LncRNA HULC mediates radioresistance via autophagy in prostate cancer cells. Braz J Med Biol Res. 2018;51(6):e7080. doi:10.1590/1414-431x20187080
21. Zheng Y, Tan K, Huang H. Long noncoding RNA HAGLROS regulates apoptosis and autophagy in colorectal cancer cells via sponging miR-100 to target ATG5 expression. J Cell Biochem. 2019;120(3):3922–3933. doi:10.1002/jcb.27676
22. Xu DH, Chi GN, Zhao CH, Li DY. Long noncoding RNA MEG3 inhibits proliferation and migration but induces autophagy by regulation of Sirt7 and PI3K/AKT/mTOR pathway in glioma cells. J Cell Biochem. 2018.
23. Wang K, Yang C, Shi J, Gao T. Ox-LDL-induced lncRNA MALAT1 promotes autophagy in human umbilical vein endothelial cells by sponging miR-216a-5p and regulating Beclin-1 expression. Eur J Pharmacol. 2019;858:172338. doi:10.1016/j.ejphar.2019.04.019
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.