Back to Journals » Neuropsychiatric Disease and Treatment » Volume 17
LncRNA SAMMSON Overexpression Suppresses Vascular Smooth Muscle Cell Proliferation via Inhibiting miR-130a Maturation to Participate in Intracranial Aneurysm
Authors Pan W, Gao Y, Wan W, Xiao W, You C
Received 17 March 2021
Accepted for publication 21 May 2021
Published 4 June 2021 Volume 2021:17 Pages 1793—1799
DOI https://doi.org/10.2147/NDT.S311499
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Taro Kishi
Wen Pan,1 Yuan Gao,2 Weifeng Wan,3 Wenfeng Xiao,4 Chao You5
1Brain Center, Shougang Shuigang General Hospital, Liupanshui City, Guizhou Province, 553000, People’s Republic of China; 2Department of Neurosurgery, Liupanshui People’s Hospital, Liupanshui City, Guizhou Province, 553001, People’s Republic of China; 3Department of Neurosurgery, Affiliated Hospital of Southwest Medical University, Luzhou City, Sichuan Province, 646000, People’s Republic of China; 4Department of Neurosurgery, Sichuan Mianyang 404 Hospital, Mianyang City, Sichuan Province, 510700, People’s Republic of China; 5Department of Neurosurgery West China Hospital, Sichuan University, Chengdu Province, 610041, People’s Republic of China
Correspondence: Chao You
Department of Neurosurgery West China Hospital, Sichuan University, 37 Guoxue Lane, Wuhou District, Chengdu Province, 610041, People’s Republic of China
Email [email protected]
Background: MiR-130a is a recently identified critical player in vascular smooth muscle cell (VSMC) proliferation, which participates in intracranial aneurysm (IA). However, the involvement of miR-130a in IA and its upstream regulator are unknown. Our preliminary sequencing analysis revealed a close correlation between miR-130a and lncRNA SAMMSON across IA samples. Therefore, we further studied the crosstalk between SAMMSON and miR-130a in IA.
Methods: SAMMSON and miR-130a expression were measured using RT-qPCR. SAMMSON subcellular location was analyzed with nuclear fractionation assay. Their direct interaction was explored with RNA pull-down assay. The role of SAMMSON in miR-130a maturation was studied with overexpression analysis. VSMC cell proliferation was analyzed with BrdU assay.
Results: SAMMSON and premature miR-130a were deregulated in IA, while mature miR-130a was upregulated in IA. SAMMSON is localized in both the nucleus and cytoplasm, and direct interaction between SAMMSON and miR-130a was observed. SAMMSON overexpression suppressed miR-130a maturation in VSMCs and reduced the enhancing effects of miR-130a on VSMC cell proliferation.
Conclusion: SAMMSON is overexpressed in IA and suppresses VSMC proliferation via inhibiting miR-130a maturation.
Keywords: intracranial aneurysm, SAMMSON, miR-130a, maturation
Introduction
Intracranial aneurysm (IA), also widely known as brain aneurysm, is a severe cerebrovascular disorder caused by the cerebral vein or artery wall weakness.1,2 IA is commonly treated with ballooning or dilation of local blood vessels. IA may rupture or leak, and the ruptured IA could lead to bleeding in brain tissues, causing hemorrhagic stroke.3,4 It is estimated that more than 25% of IA patients will die after IA rupture, and IA complications cause deaths in another 25% of patients within 6 months after rupture.4–6 The ruptured IA is usually treated with surgical clipping or less invasive endovascular coiling.7 However, these two treatments might reduce blood flow to the brain and bleeding in the brain.7 At present, the prevention of IA rupture is still critical for IA treatment.
Previous studies have elucidated the correlation between IA development and many risk factors, such as alcoholism, tobacco consumption, hypertension, obesity, head trauma, and infections.2,8,9 Besides, alterations in the expression of some molecular factors may also contribute to IA.10 For instance, lncRNAs and miRNAs affect protein production and regulate other noncoding RNAs to regulate IA growth and rupture.11,12 Although lacking protein-coding capacity, lncRNAs and miRNAs participate in human diseases, including IA, mainly by affecting the expression of protein-coding genes. Therefore, regulating the expression of certain miRNAs or lncRNAs with critical functions in IA may provide potentials for IA treatment.11,12
MiR-130a enhances the proliferation of vascular smooth muscle cells (VSMCs),13 which participate in IA.14 It is still unknown whether miR-130a participates in IA. Our preliminary sequencing analysis revealed a close correlation between miR-130a and lncRNA SAMMSON, a well-studied cancer-related lncRNA,15 across IA samples. SAMMSON plays a critical role in cancer biology by interacting with critical signal pathways involved in cancer development, such as PI3K/Akt Pathway.15 However, the involvement of SAMMSON in IA is unknown. This study analyzed the potential interaction between SAMMSON and miR-130a in IA.
Materials and Methods
Patients and Blood Extraction
Aneurismal tissues were collected from 30 patients with ruptured IAs at West China Hospital, Sichuan University during microsurgical clipping. In addition, superficial temporal arteries (STAs) were collected from other 30 patients who underwent lateral frontal craniotomies and pterional craniotomies due to injuries at the same hospital. All tissue samples were stored at −80°C prior to subsequent RNA isolations. The study was approved by the Ethics Committee of West China Hospital, Sichuan University. All IAs and controls signed informed consent. Table 1 shows the clinical data of the two groups of participants.
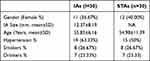 |
Table 1 Clinical Data of Two Groups of Participants |
VSMCs and Electric Transfection
Human VSMCs were purchased from Clonetics (San Diego, USA) and cultured in medium 231 containing smooth muscle growth supplements at 37°C in an incubator with 5% CO2. A total of 107 VSMCs were transfected with either 10 μg pcDNA3.1-SAMMSON expression vector, 50 nM miR-130a mimic, or their corresponding negative controls using Neon Electroporation Transfection system (Thermo Fisher Scientific). In addition, untreated cells were used as the control (C) cells. The subsequent analyses were performed at 48 h of post-transfection.
RNA Isolation and Analysis
Total RNAs were extracted from cells of each transfection group and tissue samples using the Direct-zol RNA Kit (ZYMO RESEARCH) and treated with DNase I to remove DNA. RNA quality was analyzed by Agilent 2100 Bioanalyzer (Agilent Technologies, USA). Only RNA samples with high quality were used for subsequent experiments. Otherwise, RNA isolations were repeated.
RT-qPCR
A total of 1000 ng total RNA were reverse transcribed into cDNA, followed by qPCR to determine the expression of SAMMSON and premature miR-130a using 18S rRNA as the endogenous control. Mature miR-130a expression was analyzed with All-in-One™ miRNA qRT-PCR Reagent Kit (Genecopoeia) by qPCR with U6 as the endogenous control. The method of 2−ΔΔCt was applied to normalize the expression of each gene to the corresponding controls.
Nuclear Fractionation Assay
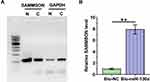
Nuclear/Cytosol Fractionation Kit (BioVision, # K266) was applied to prepare nuclear and cytosolic samples from VSMCs. Briefly, after cell lysis, the homogenate was centrifuged for 10 min at 2500 g (4°C). The supernatant was used as the cytosolic fraction, and the nuclear pellet was collected to serve as the nuclear fraction. After that, RNA isolation from these two fractions was performed, followed by RTs and PCR to determine the expression of SAMMSON with GAPDH as the endogenous control. Electrophoresis was performed with 1% agarose gel to separate PCR products. After EB staining, images were taken with MyECL Imager (Bio-Rad).
RNA Pull-Down Assay
Vectors with T7 promoter expressing premature miR-130a or NC miRNA were used to prepare transcripts of both premature miR-130a or NC miRNA through in vitro transcription using the T7 RNA polymerase MEGAscript™ T7 Transcription Kit (Cat # AMB13345, Thermo Fisher Scientific). Both transcripts were labeled with biotin using the Pierce™ Biotin 3ʹ End DNA Labeling Kit (Cat # PI89818, VWR) and named Bio-miR-130a and Bio-NC, respectively. The two miRNAs were then transfected into VSMCs as described above. After the preparation of cell lysis at 48h of post-transfection, biotin-ligated miRNAs were pulled down using streptavidin agarose magnetic beads (Life Technologies). The pull-down samples were subjected to RNA isolations and RT-qPCR to determine the expression of SAMMSON.
BrdU Incorporation Assay
Cell proliferation after transfections was analyzed by BrdU incorporation. In brief, transfected cells were seeded onto a 96-well plate to a density of 6000 cells per well. After cultured for 48 h, BrdU was added into each well to a final concentration of 10 µM. After cultured for another 24 h, cells were fixed for 30 min and then incubated with peroxidase-coupled anti-BrdU-antibody (Sigma–Aldrich) for one hour. After washed with PBS three times, cells were further incubated with peroxidase substrate for 1 h. Finally, OD values at 450 nm were measured.
Statistical Analysis
All in vitro experiments were performed in three biological replicates. Two independent groups were compared by unpaired t test. Multiple independent groups were compared by ANOVA Tukey’s test. A P<0.05 was statistically significant.
Results
Changed SAMMSON and miR-130a Levels in IA
SAMMSON and miR-130a levels in IAs and STAs were analyzed by RT-qPCR. The results illustrated that the levels of SAMMSON (Figure 1A, p<0.01) and premature miR-130a (Figure 1B, p<0.01) were lower while mature miR-130a was higher in IA than in STA (Figure 1C, p<0.01). Therefore, SAMMSON downregulation and increased miR-130a maturation are likely involved in IA. Correlation analysis revealed that SAMMSON was positively correlated with premature miR-130a (Figure 1D) but inversely correlated with mature miR-130a (Figure 1E).
Subcellular Location of SAMMSON and Its Direct Interaction with Premature miR-130a
Nuclear fractionation assay was performed to analyze the subcellular localization of SAMMSON in VSMCs. With GAPDH as the endogenous control, our data clearly showed that SAMMSON was detected in both nuclear (N) and cytosolic (C) fractions. Therefore, SAMMSON might traffic between the nucleus and cytoplasm (Figure 2A). RNA pull-down assay was conducted with Bio-miR-130a to examine the direct interaction between premature miR-130a and SAMMSON. Compared to Bio-NC pull-down samples, a significantly higher SAMMSON level was detected in Bio-miR-130a pull-down samples (Figure 2B, p<0.01). As premature miRNAs are localized in the nucleus, we concluded that SAMMSON in the nucleus might directly interact with premature miR-130a.
SAMMSON Suppresses miR-130a Maturation in VSMCs
SAMMSON or mature miR-130a was overexpressed in VSMCs. The follow-up RT-qPCR analyses illustrated the increased expression of both SAMMSON and miR-130a in VSMCs from 24 h to 72h (Figure 3A, p<0.05). In addition, SAMMSON overexpression in VSMCs significantly increased premature miR-130a level (Figure 3B, p<0.05) but significantly decreased mature miR-130a level (Figure 3C, p<0.05) in VSMCs. Therefore, SAMMSON might suppress miR-130a maturation in VSMCs.
Roles of SAMMSON and miR-130a in VSMC Proliferation
BrdU incorporation assay was utilized to analyze VSMC proliferation after overexpression of SAMMSON and/or miR-130a. The results clearly demonstrated an inhibitory effect of SAMMSON on cell proliferation and enhanced effects of miR-130a on cell proliferation (p<0.05). Moreover, the co-transfection assay illustrated that SAMMSON suppressed the enhancing effects of miR-130a on VSMC proliferation (Figure 4, p<0.05).
 |
Figure 4 Effects of SAMMSON and miR-130a in VSMC proliferation. BrdU incorporation assay was performed to analyze VSMC proliferation after overexpression of SAMMSON and/or miR-130a. *p<0.05. |
Discussion
Our study first reported the interaction between SAMMSON and miR-130a in IA. We observed an altered expression of SAMMSON and miR-130a (both mature and premature form) in IA. Remarkably, miR-130a maturation in VSMCs is regulated by SAMMSON, suggesting the potential application of SAMMSON and miR-130a in the treatment of IA.
A recent study showed that miR-30a is upregulated in a rat spontaneous hypertension model, and miR-30a overexpression targets GAX to promote VSMC proliferation.13 Hypertension is a major risk factor for IA.2,8,9 Therefore, miR-130a might also participate in IA. This study first reported upregulation of mature miR-130a and decreased premature miR-130a expression in IA. Therefore, the increased maturation of miR-130a but not the increased transcription of miR-130a may participate in IA. The study also confirmed the enhancing effects of miR-130a on VSMC proliferation. The altered VMSC proliferation plays different roles at different stages of IAs.16 The increased VMSC proliferation rate promotes the growth of IA at early stages and suppresses IA rupture at late stages. Therefore, accurate regulation of miR-130a expression might suppress IA growth and rupture.
At present, the upstream regulation of miR-130a in VSMC proliferation is unknown. This study showed that SAMMSON is downregulated in IA, and SAMMSON overexpression in VSMCs suppresses miR-130a maturation, suggesting that SAMMSON is a novel molecular inhibitor of miR-130a maturation. Furthermore, SAMMSON could be detected in both the nucleus and cytoplasm. It has been well established that premature miRNAs are mainly localized in the nucleus while mature miRNAs are mainly localized in the cytoplasm.17 Therefore, we hypothesized that SAMMSON might sponge premature miR-130a in the nucleus to suppress its maturation. This hypothesis is supported by the observation of the direct interaction between SAMMSON and miR-130a.
The study is limited by the relatively small size of patients and lack of in vivo functional analyses of SAMMSON and miR-130a. Therefore, our conclusions remain to be further validated.
Conclusion
Overall, our data illustrated that SAMMSON overexpression might suppress miR-130a maturation in VSMCs to participate in IAs. Therefore, regulating the expression of SAMMSON and miR-130a may improve the treatment of IA.
Availability of Supporting Data
The data that support the findings of this study are available on request from the corresponding author. The data are not publicly available due to their containing information that could compromise the privacy of research participants.
Ethical Approval and Consent to Participate
All patients signed the written informed consent. All experiments were approved by the Ethics Committee of West China Hospital, Sichuan University. Procedures operated in this research completely complied with the standards set out in the Announcement of Helsinki and Laboratory Guidelines of Research in China.
Author Contributions
Wen Pan, Chao You: study concepts, literature research, clinical studies, data analysis, experimental studies, manuscript writing and review; Yuan Gao: study design, literature research, experimental studies and manuscript editing; Weifeng Wan: definition of intellectual content, clinical studies, data acquisition and statistical analysis; Wenfeng Xiao: data acquisition, manuscript preparation and data analysis.
All authors have read and approved the submission of the manuscript. All authors contributed to data analysis, drafting or revising the article, have agreed on the journal to which the article will be submitted, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Funding
There is no funding to report.
Disclosure
All authors have no conflicts of interest. We declare that we do not have any commercial or associative interest that represents a conflict of interest connected with the work submitted.
References
1. Wang X, Zhu C, Leng Y, Degnan AJ, Lu J. Intracranial aneurysm wall enhancement associated with aneurysm rupture: a systematic review and meta-analysis. Acad Radiol. 2019;26(5):664–673. doi:10.1016/j.acra.2018.05.005
2. Jin D, Song C, Leng X, Han P. A systematic review and meta-analysis of risk factors for unruptured intracranial aneurysm growth. Int J Surg. 2019;69:68–76. doi:10.1016/j.ijsu.2019.07.023
3. Juvela S, Korja M. Intracranial aneurysm parameters for predicting a future subarachnoid hemorrhage: a Long-Term Follow-up Study. Neurosurgery. 2017;81(3):432–440. doi:10.1093/neuros/nyw049
4. de Souza MR, Fagundes CF, Rabelo NN, Teixeira MJ, Figueiredo EG. Association between intracranial aneurysm and meningiomas: an integrative survival Analysis with identification of prognostic factors. Clin Neurol Neurosurg. 2020;198:106128. doi:10.1016/j.clineuro.2020.106128
5. Niu X, Wang T, Li J, et al. An integrative survival analysis with identification of prognostic factors in the patients with coexisting glioma and intracranial aneurysm. World Neurosurg. 2018;111:e592–e600. doi:10.1016/j.wneu.2017.12.126
6. Ruiz-Sandoval JL, Cantú C, Chiquete E, et al. Aneurysmal subarachnoid hemorrhage in a Mexican multicenter registry of cerebrovascular disease: the RENAMEVASC study. J Stroke Cerebrovasc Dis. 2009;18(1):48–55. doi:10.1016/j.jstrokecerebrovasdis.2008.09.019
7. Darsaut TE, Findlay JM, Magro E, et al. Surgical clipping or endovascular coiling for unruptured intracranial aneurysms: a pragmatic randomised trial. J Neurol Neurosurg Psychiatry. 2017;88(8):663–668. doi:10.1136/jnnp-2016-315433
8. Zholdybayeva EV, Medetov YZ, Aitkulova AM, et al. Genetic risk factors for intracranial aneurysm in the Kazakh population. J Mol Neurosci. 2018;66(1):135–145. doi:10.1007/s12031-018-1134-y
9. Björkman J, Frösen J, Tähtinen O, et al. Aneurysm size is the strongest risk factor for intracranial aneurysm growth in the Eastern Finnish population. Neurosurgery. 2019;84(5):1098–1103. doi:10.1093/neuros/nyy161
10. Xu Z, Rui YN, Hagan JP, Kim DH. Intracranial aneurysms: pathology, genetics, and molecular mechanisms. Neuromolecular Med. 2019;21(4):325–343. doi:10.1007/s12017-019-08537-7
11. Li H, Wang W, Zhang L, et al. Identification of a long noncoding RNA-associated competing endogenous RNA network in intracranial aneurysm. World Neurosurg. 2017;97:684–692.e684. doi:10.1016/j.wneu.2016.10.016
12. Gao G, Zhang Y, Yu J, et al. Long non-coding RNA MALAT1/microRNA-143/VEGFA signal axis modulates vascular endothelial injury-induced intracranial aneurysm. Nanoscale Res Lett. 2020;15(1):139. doi:10.1186/s11671-020-03357-2
13. Wu WH, Hu CP, Chen XP, et al. MicroRNA-130a mediates proliferation of vascular smooth muscle cells in hypertension. Am J Hypertens. 2011;24(10):1087–1093. doi:10.1038/ajh.2011.116
14. Miyata T, Minami M, Kataoka H, et al. Osteoprotegerin prevents intracranial aneurysm progression by promoting collagen biosynthesis and vascular smooth muscle cell proliferation. J Am Heart Assoc. 2020;9(17):e015731. doi:10.1161/JAHA.119.015731
15. Ni H, Wang K, Xie P, Zuo J, Liu W, Liu C. LncRNA SAMMSON knockdown inhibits the malignancy of glioblastoma cells by inactivation of the PI3K/Akt pathway. Cell Mol Neurobiol. 2021;41(1):79–90. doi:10.1007/s10571-020-00833-2
16. Frösen J, Tulamo R, Paetau A, et al. Saccular intracranial aneurysm: pathology and mechanisms. Acta Neuropathol. 2012;123(6):773–786. doi:10.1007/s00401-011-0939-3
17. Li XG, Wang YB. SRPK1 gene silencing promotes vascular smooth muscle cell proliferation and vascular remodeling via inhibition of the PI3K/Akt signaling pathway in a rat model of intracranial aneurysms. CNS Neurosci Ther. 2019;25(2):233–244. doi:10.1111/cns.13043
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.