Back to Journals » OncoTargets and Therapy » Volume 13
LncRNA PART1 Exerts Tumor-Suppressive Functions in Tongue Squamous Cell Carcinoma via miR-503-5p
Authors Zhao X, Hong Y, Cheng Q, Guo L
Received 28 May 2020
Accepted for publication 25 August 2020
Published 7 October 2020 Volume 2020:13 Pages 9977—9989
DOI https://doi.org/10.2147/OTT.S264410
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Arseniy Yuzhalin
Xiqun Zhao1 ,* Yanqing Hong2 ,* Qingyuan Cheng,3 Lixin Guo4
1Department of Pediatric Dentistry, Jinan Stomatological Hospital, Jinan, Shandong 250001, People’s Republic of China; 2Prosthodontic Lab, Jinan Stomatological Hospital, Jinan, Shandong 250001, People’s Republic of China; 3Department of Stomatology, Jinan LiCheng District Hospital of Traditional Chinese Medicine, Jinan, Shandong 250001, People’s Republic of China; 4Department of Scientific Education, Jinan Stomatological Hospital, Jinan, Shandong 250001, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Lixin Guo
Department of Scientific Education, Jinan Stomatological Hospital, No. 101, Jing Liu Road, Shizhong District, Jinan, Shandong 250001, People’s Republic of China
Tel +86-15589970752
Email [email protected]
Background: Tongue squamous cell carcinoma (TSCC) accounts for one-third of oral cancers. Previous studies had reported that lncRNA/miRNA regulated the biological behaviors of different cancer cells. However, the mechanisms of PART1 in regulating tumorigenesis and TSCC development via targeting miR-503-5p had not been studied.
Methods: The expressions of PART1 and miR-503-5p in tissues and cultured cell lines were detected by qRT-PCR. StarBase 3.0 was used to predict the binding sites of PART1, then dual-luciferase assay and RNA pull-down assay were executed to confirm whether miR-503-5p was a target of PART1. TSCC cells were co-transfected with PART1-overexpressed plasmid or miR-503-5p mimics in vitro, and the transfection efficiency was evaluated through qRT-PCR. Western blot was performed to assess the expressions of EMT-related proteins. CCK-8 and clone formation assays were conducted to detect cell proliferation, TUNEL assay was used to detect apoptosis, and transwell assay was executed to test migration and invasion.
Results: The low PART1 expression and high miR-503-5p expression were found in TSCC tissues and cell lines (CAL-27 and SCC9). PART1 expression was positively correlated with patients’ prognosis. The targeting and binding relationship between PART1 and miR-503-5p was confirmed, and overexpressed PART1 diminished the expression of miR-503-5p as well. Moreover, PART1 facilitated apoptosis, inhibited proliferation, invasion and migration of TSCC cells, and these influences were impeded by miR-503-5p overexpression.
Conclusion: LncRNA PART1 played a cancer-suppressing role in TSCC by targeting miR-503-5p, which provided a potential target for TSCC treatment.
Keywords: tongue squamous cell carcinoma, LncRNA PART1, miR-503-5p, proliferation, invasion, migration
Introduction
Tongue squamous cell carcinoma (TSCC) is a common cancer that occurs in the oral cavity.1 TSCC always presents with local invasion, fast growth and obvious pain.2 The prognosis is still poor mainly because of the regional recurrence and lymph node metastasis.3 Lymph node ratio (LNR) and extranodal extension (ENE) are expected as promising prognostic tools to identify patients with worse disease-free survival (DFS) and overall survival (OS) in oral squamous cell carcinoma (OSCC).4 Despite the advanced therapies, the metastasis and recurrence of tumor cells remain the most aggressive factors of prognosis. Therefore, investigating the molecular mechanism involved in TSCC development and finding the effective molecular therapeutic targets are urgently needed.
Long non-coding RNAs (lncRNAs) are defined as a type of endogenous non-protein-coding RNAs that consists of over 200 nucleotides in length. LncRNAs regulate gene expression through various mechanisms and play a role in many physiological and pathological processes.5–9 LncRNAs medicate miRNAs expression through silencing at transcript level through chromatin modification or acting as endogenous sponges.10–12 MicroRNAs, as evolutionary conserved non-coding RNAs, which block protein production by base-pairing with target mRNA of functional proteins, thus leading to the pathological changes of diseases.13 In recent years, lncRNAs have become a hot spot of research and they contribute to tumorigenesis and metastasis.9,14 In TSCC, lncRNA has been reported to work as an oncogene or tumor suppressor gene and it always affects patient prognosis.15,16 For instance, the silencing of AFAP1-AS1 suppressed cell proliferation, migration and invasion in TSCC.17 Additionally, overexpression of lncRNA MEG3 reduced cell proliferation and enhanced apoptosis.18 LncRNA prostate androgen-regulated transcript 1 (PART1), located on chromosome 5q12, was discovered in a region harbor tumor suppressor genes.19 PART1 has been reported to strengthen the proliferation and suppress apoptosis in the prostate and NSCLC.20,21 In contrast, Li et al and Song et al have revealed that the PART1 level is positively correlated with the overall survival rate of TSCC and carried a good prognosis.15,22 Moreover, Zhang et al summarized that PART1 as a potential target for TSCC treatment could inhibit the metastasis and EMT of tumor cells by targeting miR-301b.16 Also, miR-503 was significantly up-regulated in laryngeal squamous cell carcinoma (LSCC), and it has been shown that it could cause tumorigenesis by silencing or disappearing tumor suppressor genes by negatively regulating the expression of programmed cell death 4 (PDCD4).23 Recently, more and more studies reported that miR-503-5p was significantly up-regulated in OSCC tissues and cells, which acted as an oncogene to promote the malignant behaviors (including proliferation, invasion and migration) of OSCC cells.24–26 Importantly, Song et al have simultaneously found the down-regulation of PART1 and up-regulation of miR-503 in TSCC,15 which confirmed that both lncRNA PART1 and miR-503 were associated with the occurrence and development of TSCC. However, the key mechanisms of how PART1 is involved in the regulation of tumorigenesis have not been reported.
In the current study, the levels of lncRNA PART1 and miR-503 in TSCC tissues and TSCC cell lines were detected. Furthermore, we examined the influences of them on the proliferation, invasion, migration and apoptosis of TSCC cells and explored the key molecular mechanisms in mediation of the development of TSCC, as well.
Materials and Methods
Clinical TSCC Tissue Collection
Forty cases pathologically confirmed TSCC and corresponding adjacent (>2 cm away from the tumor sites and has been confirmed by pathologist) tissue specimens were collected during surgery from 2013~2019 in Jinan stomatological hospital. Inclusion criteria: 1) samples were newly diagnosed primary TSCC cases; 2) patients did not receive chemoradiotherapy before surgery; 3) patients were willing to participate in a 5-year follow-up. Exclusion criteria: 1) patients had received any treatment before admission; 2) any histories of previous tumor; 3) patients had other clinical diseases. The follow-up study was conducted for 5 years until the patient died. Follow-up is done every 1–2 months through the visit of outpatient at clinic or phone call. The causes of patients’ deaths were confirmed by medical records or information provided by relatives; therefore, patients who died due to other reasons were excluded from this study. All tissue specimens were frozen and preserved in liquid nitrogen. The clinical-pathological data of TSCC patients were collected after pathological diagnosis for statistical analysis. This study passed the supervision and the review of the Medical Ethics Committee of the hospital and all patients provided informed consent, which complied with the requirements of Helsinki’s ethical principles, as well.
Cell Culture
Normal human oral keratinocyte cell line (NHOK), TSCC cell lines (CAL-27, SCC9 and SCC25) were bought from Shanghai cell bank, Chinese academy of sciences. Roswell Park Memorial Institute 1640 medium (RPIM-1640, Gibco, USA) with 10% fetal bovine serum (FBS, Hyclone, USA) was used for the incubation of NHOK cells, the 1:1 mixture of Dulbecco’s modified Eagle’s medium and Ham’s F12 (DMEM/F-12, Gibco, USA) supplied with 400 ng/mL hydrocortisone and 10% FBS was used for the incubation of TSCC cells. The cells were routinely cultivated in an incubator at 37°C with 5% CO2. When the confluence reached 80–90% (about 2–3 d), the passage was performed with a ratio of 1:3.
Cell Transfection
The pcDNA3.1-PART1 overexpression plasmid was constructed by GeneChem (Shanghai, China). Lipofectamine 2000 kit (Invitrogen, USA) was used to transfect CAL-27 and SCC9. In brief, TSCC cells were maintained overnight in 96-well plates with an antibiotic-free medium for transfection preparation. Then, 2μg pcDNA3.1-PART1 or pcDNA3.1-NC were transfected into 1×106 TSCC cells with Lipofectamine 2000 and gene expression was detected after 48 h of culture. The cells were divided into 3 groups: control, pcDNA3.1-NC, and pcDNA3.1-PART1. Similarly, miR-503-5p mimics (Sigma-Aldrich, USA) or mimics-NC were transfected into logarithmic TSCC cells and divided into 3 groups: control, mimics-NC and miR-503-5p mimics. Finally, the co-transfection of miR-503-5p mimics and pcDNA3.1-PART1 into TSCC was performed, and the cells were divided into 5 groups: control, mimics-NC+pcDNA3.1-NC, mimics-NC+pcDNA3.1-PART1, miR-503-5p mimics+pcDNA3.1-NC, and miR-503-5p mimics +pcDNA3.1-PART1.
qRT-PCR
TRIZOL reagent (Invitrogen, USA) was applied for RNA extraction. Then, total RNA was synthesized to cDNA using PrimeScript RT reagent Kit (TaKaRa, Japan) according to the manufacturer’s specifications. Subsequently, the cDNA and SYBR Premix Ex Taq kit (TaKaRa, Japan) were used in qRT-PCR procedure and performed on the LightCycler 96 PCR instrument (Roche, USA). The amplification condition was described as follows: 95°C pre-denaturation for 10 min (94°C denaturation for 10 s, 62°C annealing for 15 s, 72°C extension for 35 s) × 40 cycles. GAPDH and U6 were used as internal references, and the primer sequences (Table 1) were synthesized by Shanghai Biotech Engineering Co., Ltd. The 2−ΔΔCt method was used for result analysis.
 |
Table 1 The Primer Sequences of qRT-PCR |
Western Blot
Lysed samples were analyzed with the BCA kit (ThermoFisher, USA) for protein concentration determination. The proteins were separated by 12% SDS-PAGE and transferred onto the PVDF membrane. Then, the PVDF membrane was blocked using 5% skim milk for 2 h and washed in TBS-T. The primary rabbit anti-human antibodies were added for immunoblotting as followed: β-actin (42 kDa, 1:1000, ab16769, Abcam, UK), N-cadherin (100 kDa, 1:1000, ab76057, Abcam, UK), Vimentin (67 kDa, 1:1000, ab8978, Abcam, UK) and E-cadherin (125 kDa, 1:1000, ab15148, Abcam, UK). After that, the samples were incubated at 4°C overnight and washed with TBS-T for 3 times and each time for 10 min. Horseradish peroxidase-labeled goat anti-rabbit IgG (1:10,000, Sigma, USA) was added and the membrane was incubated for 1 h and washed 3 times with TBS-T. Finally, chemiluminescence coloration, exposure, development, fixing, and data analysis were performed. β-actin was applied as an internal reference.
Cell Counting Kit-8 (CCK-8) Assay
The cell viability was determined using the CCK-8 kit (Abcam, UK). The transfected cells were made into single-cell suspension and seeded in a 96-well plate with 6×103 cells in each well. After different treatments, the plate was incubated at 37°C with 5% CO2 for different periods (0, 24, 48, and 72 h) and 10 μL CCK-8 solution was added into each well. The absorbance was measured by enzymatic reader (Biotek, USA) at 460 nm.
Colony Formation Assay
CAL-27 (800 cells/well) and SCC9 (2000 cells/well) cells were separately plated in 6-well plates after different transfections and incubated in DMEM/F-12 medium with 10% FBS. The cells were cultivated for 14 days and the medium was renewed every 3 days, the colonies were fixed by methanol for 20 min and stained using crystal violet. The inverted phase-contrast microscope (Olympus ckx53, Japan) was used to obtain the colony formation photographs and the number of colonies was automatically counted by ImageJ (1.48v) software.
Terminal Deoxynucleotidyl Transferase dUTP Nick End Labeling (TUNEL) Assay
The TUNEL kit (ab66108, Abcam, UK) was selected for staining DNA breakage. The transfected cells were incubated for 24 h and fixed with 4% formaldehyde for 30 min and washed with PBS. TdT enzyme reaction solution and FITC dUTP (labeling the DNA end of apoptosis cells) were added in turn and incubated in dark for 60 min. Finally, the DAPI staining solution was added to re-dye the nucleus and the cells were incubated in dark for 10 min. Then, the cells were rinsed 3 times with PBS and sealed. The apoptosis in each group was detected by microscopy (Olympus Ckx53, Japan) at 495 nm and 519 nm. The apoptosis rate was calculated through analyzing the relative fluorescence intensity automatically by ImageJ software (1.48V).
Transwell Invasion and Migration Assay
One day before the cells were inoculated in the transwell chamber, the surface of the upper chamber was spread with matrigel (without matrigel in migration experiments). In detail, 10% FBS DMEM/F-12 medium was added into the lower invasion-chamber and cell suspension was transferred into the upper invasion-chamber. The cells were maintained in the upper chamber with a density of 1×105 cells/well and incubated for 24 h. The matrigel and the cells in the upper chamber were removed with a cotton swab. The cells in lower chamber cells were fixed with 4% paraformaldehyde and stained by 0.1% crystal violet. Photographs were captured using microscopy (Olympus Ckx53, Japan).
Dual-Luciferase Reporter Assay
The StarBase 3.0 database was used to predict the binding sites of lncRNA PART1, and the mutant and wild sequences of PART1 were supplied by Shanghai Biotech Engineering Co., Ltd. The PART1 mutation and the wild-type fragment were combined with the pmirGLO luciferase reporter Vector (Promega, USA). The pmirGLO-PART1 WT or pmirGLO-PART1 MUT and miR-503-5p mimics or mimics-NC were co-transfected into SCC9 cells. Two days after transfection, the fluorescence activities of each group were detected by the Dual-Luciferase Kit (Promega, USA).
RNA Pull-Down
TSCC cells were transfected with miR-503-5p mimics and incubated for 48 h. Then, the cells were harvested and lysed. The biotin-labeled PART1 (Bio-PART1, probe sequence was shown in Table S1) was conjugated to streptavidin agarose beads (Invitrogen, USA). Cell lysate was mixed with Bio-PART1 or Bio-NC for 2 h, and then streptavidin agarose beads (Invitrogen, USA) were added and incubated for 1 h. The expression level of miR-503-5p in coprecipitated RNAs (Table S2) was determined by qRT-PCR as mentioned before.
Statistical Analysis
Each experiment was performed at least three times. Data were analyzed by SPSS v17.0 and GraphPad Prism 6, and the results were expressed as mean ± standard deviation (Mean ± SD). The t-test was used for the comparison between two groups, and One-Way ANOVA analysis of variance was used for the comparison among multiple groups. Tukey’s multiple comparisons test was conducted for the pairwise comparisons after ANOVA analysis. P < 0.05 was considered as statistically significant.
Results
PART1 Was Low Expressed in TSCC Tissues and Related to Patients’ Prognosis
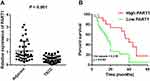
The detection of PART1 expression in TSCC and tumor-adjacent normal tissues was achieved via qRT-PCR. The results in Figure 1A suggested that PART1 expression was significantly lower (P < 0.001) in the TSCC group compared with the control adjacent group. To evaluate the relationship between clinical characteristics and PART1 expression, the correlation between PART1 expression level and the clinicopathological characteristics of TSCC patients was analyzed and the details were organized in Table 2. The results indicated a correlation between PART1 expression in TSCC tissues and tumor classification (P = 0.0024), clinical stage (P = 0.0057), lymph node metastasis (P = 0.000). In addition, the outcome of Kaplan–Meier survival analysis (Figure 1B) showed that low PART1 expression in TSCC tissue was correlated with a low overall survival rate (P = 0.0107).
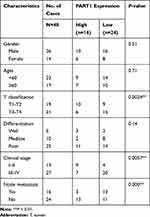 |
Table 2 Relationships Between the Expression of PART1 and Clinicopathologic Factors of TSCC Patients |
PART1 Inhibited the Proliferation and Promoted Apoptosis of TSCC Cells
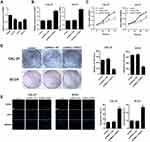
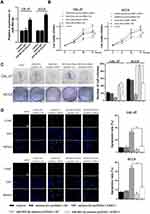
To examine the effect of PART1 on the proliferation of TSCC cells, qRT-PCR was applied to evaluate the expression of PART1 in NHOK and TSCC cell lines (including CAL-27, SCC9 and SCC25). The results in Figure 2A suggested that the expressions of PART1 in TSCC cell lines were significantly lower than NHOK (all P < 0.05). The CAL-27 and SCC9 cells were selected for subsequent experiments due to their more significant differences compared with NHOK (P < 0.01). The pcDNA3.1-PART1 or pcDNA3.1-NC were transfected into CAL-27 and SCC9 cells. The transfected efficiency was checked by qRT-PCR and the results reported that PART1 expression in pcDNA3.1-PART1 group was much higher than the control group and pcDNA3.1-NC group (P < 0.01), which indicated that the transfection was successful (Figure 2B). Cell viability was examined using the CCK-8 assay (Figure 2C), and the results showed that the cell viability in pcDNA3.1-PART1 group was lower than the control group and pcDNA3.1-NC group (P < 0.01). The effect of PART1 on cell proliferation was tested by colony formation assay (Figure 2D), and significant suppression of overexpressed PART1 on cell proliferation was found in pcDNA3.1-PART1 group relative to the control group and pcDNA3.1-NC group (P < 0.01). Meanwhile, the outcomes of the TUNEL assay showed an extraordinary increase of apoptosis in the pcDNA3.1-PART1 group compared with the control group and pcDNA3.1-NC group (P < 0.01), which indicated that overexpression of PART1 significantly promoted apoptosis (Figure 2E). In summary, PART1 diminished TSCC cell proliferation and promoted apoptosis.
PART1 Inhibited Invasion and Migration of TSCC Cells
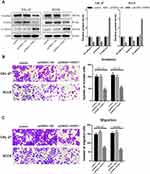
To investigate the mechanism of PART1 on metastasis of TSCC cells, western-blot was executed to test the expression of EMT-related proteins (Figure 3A). EMT induces the invasion and migration of cancer cells through the weakened or lost intercellular adhesion.27,28 The results showed that the expression of N-cadherin and Vimentin was reduced (P < 0.01) while E-cadherin expression was up-regulated (P < 0.01) in pcDNA3.1-PART1 group compared with the control group and pcDNA3.1-NC group, which proved that overexpression of PART1 inhibited the EMT process in CAL-27 and SCC9 cells. Furthermore, transwell assay was used to detect the effects of PART1 on invasion and migration of CAL-27 and SCC9 cells (Figure 3B and 3C), the results showed that compared with the control group and pcDNA3.1-NC group, the number of invaded and migrated cells in pcDNA3.1-PART1 group was decreased (P < 0.01). To sum up, PART1 inhibited the EMT process, thereby inhibiting the invasion and migration of TSCC cells.
miR-503-5p Was a Target of PART1
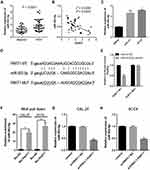
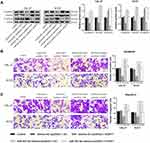
The expression of miR-503-5p in TSCC tissue and cell lines was evaluated by qRT-PCR. The result suggested that the expression of miR-503-5p in TSCC tissue was significantly higher than the control group (Figure 4A, P < 0.0001), and a significant negative correlation was found between PART1 and miR-503-5p (Figure 4B, r2 = 0.2245, P = 0.0003). Meanwhile, the expression of miR-503-5p in TSCC cell lines (CAL-27 and SCC9) was higher than NHOK cell line (Figure 4C, P < 0.01). The binding sites of PART1 on miR-503-5p were predicted by StarBase3.0 (Figure 4D), and the outcomes of dual-luciferase Reporter Assay verified that overexpression of miR-503-5p decreased the luciferase activity of PART1-WT reporter gene in SCC9 cells (Figure 4E, P < 0.01), while it did not change the luciferase activity of PART1-MUT reporter gene (P < 0.01). The enrichment of miR-503-5p by PART1 was detected through RNA pull-down assay (Figure 4F) and the results showed that biotin-labeled PART1 could significantly enrich the expression of miR-503-5p in SCC9 cells (P < 0.01). Besides, qRT-PCR was applied to detect the effect of PART1 overexpression on miR-503-5p expression and the results indicated that compared with the control group, the expression of miR-503-5p in pcDNA3.1-PART1 group was decreased in SCC9 cells (Figure 4H, P < 0.01), the similar tendency was also found in CAL-27 cells (Figure 4G, P < 0.01). Therefore, all findings above indicated that miR-503-5p was a target of PART1 and the expression of miR-503-5p was impeded by PART1.
Overexpression of miR-503-5p Impeded the Effects of PART1 on TSCC Cells
Firstly, we transfected CAL-27 and SCC9 cells with miR-503-5p mimics, and the transfection efficiency was verified by qRT-PCR (Figure 5A). The outcomes reported that the expression of miR-503-5p in miR-503-5p mimics group was higher than the control group and mimics-NC group (P < 0.01). Subsequently, we also explored whether PART1 played a key role in TSCC by mediating miR-503-5p. To this end, CAL-27 and SCC9 cells were co-transfected with miR-503-5p mimics and pcDNA3.1-PART1, and the biological behaviors (including cell proliferation, apoptosis, invasion and migration) were checked. As shown in Figure 5B, the cell viability was decreased in mimics-NC+pcDNA3.1-PART1 group relative to that in the control group, mimics-NC+pcDNA3.1-NC group and miR-503-5p mimics+pcDNA3.1-PART1 group (P < 0.01), which suggested that miR-503-5p alleviated the inhibitory effect of PART1 on cell viability. Moreover, the proliferation (Figure 5C) and apoptosis (Figure 5D) of TSCC cells were detected separately by colony formation and TUNEL assays. The results reported that the ability of cell proliferation of TSCC cells was decreased in mimics-NC+pcDNA3.1-PART1 group relative to that in the control group, mimics-NC+pcDNA3.1-NC group and miR-503-5p mimics+pcDNA3.1-PART1 group (P < 0.01). However, a converse tendency of cell apoptosis was found among these groups (P < 0.01). Subsequently, the EMT-related proteins (Figure 6A) and the abilities of invasion (Figure 6B) and migration (Figure 6C) were detected separately by Western blot and transwell assays. The results reported that the levels of N-cadherin and Vimentin were decreased (P < 0.05) while E-cadherin expression was increased (P < 0.01) in mimics-NC+pcDNA3.1-PART1 group relative to the control group and miR-503-5p mimics+pcDNA3.1-PART1 group (P < 0.01). Meanwhile, Figure 6B and 6C showed similar tendencies of invasion and migration which were consistent with the expressions of N-cadherin and Vimentin. These findings suggested that the combined overexpression of PART1 and miR-503-5p relieved the inhibitory effects of single overexpression of PART1 on the biological behaviors of TSCC cells. To sum up, the overexpression of miR-503-5p impeded the effects of over-expressed PART1 on TSCC cells.
Discussion
Increasing numbers of reports have proved that lncRNAs act as miRNA decoys or sponges in human carcinomas, their abnormal expressions are related to the development and progression of tumor.29–31 LncRNA PART1 has been reported to have controversial roles in cancers. In detail, PART1 acted as oncogenic factors in bladder cancer32 and hepatocellular carcinoma,33 while it acted as tumor suppressors in oral squamous cell carcinoma22 and glioblastoma.34,35 However, the biological effects and underlying mechanisms of PART1 on TSCC were unclear. In our study, we found that the expression levels of lncRNA PART1 and miR-503-5p were inversely correlated with each other in TSCC tissues, and overexpression of PART1 in TSCC cell lines inhibited miR-503 expression. In addition, we found that expression of PART1 was positively correlated with the prognosis of overall survival rates of TSCC patients. And miR-503-5p expression was diminished in pcDNA3.1-PART1 transfected cells. The overexpressed PART1 and miR-503-5p showed opposite effects on proliferation, apoptosis migration, and invasion of TSCC cells. These findings suggested that PART1 played a tumor-suppressive role in TSCC progression.
A growing number of studies have proved that lncRNAs and miRNAs coordinate the development of TSCC. For instance, Zhu et al found that lncRNA MALAT1 strengthened the proliferation, migration and invasion of TSCC cells by targeting miR-140-5p.31 Zhang et al reported that lncRNA KCNQ1OT1 potentiated the TSCC growth and chemo-resistance via regulating miR-211-5p.36 Wu et al demonstrated that lncRNA CACS15 accelerated the migration and invasion of TSCC cells by down-regulating miR-124.37 PART1 could mediate the secretion of cytokines and metastasis of tumor cells to affect cancer progression.15 The expression of PART1 was impeded in glioma tissues and exerted suppressive functions on malignant behavior by targeting miR-190a-3p in glioma cell lines.35 Similarly, PART1 was down-regulated in TSCC tissues of patients and a higher level of PART1 was positively correlated with better prognosis.15 Different miRNAs showed conversed effects in TSCC, as well. For instance, miR-219 expression was down-regulated in TSCC tissues and cell lines and up-regulated miR-219 restrained the growth and metastasis of TSCC through targeting protein kinase CI.38 miR-21 was overexpressed in TSCC cell line CAL-27 and silenced miR-21 inhibited the abilities of proliferation and invasion, arrested cell cycle and induced apoptosis.39 miR-503 acted as an oncogene in certain cancers, for instance, miR-503 induced EMT and the progression of breast cancer through its inhibitive effects on SMAD2 and E-cadherin.40 The metastasis of tumor cells is closely related to EMT. The E-cadherin expression in cells was down-regulated, the adhesion between cells was lost, and the abilities of migration and invasion were enhanced.41,42 In the present study, EMT-related proteins, E-cadherin was up-regulated, N-cadherin and Vimentin were down-regulated in PART1 overexpressed group, which confirmed the anticarcinogenic effect of PART1 on TSCC cells. Meanwhile, all these findings indicated that PART1 acted as a suppressive factor and miR-503 acted as an oncogene, and the targeting effect of PART1 on miR-503 regulated the progression of TSCC.
Some limitations existed in the current study. For instance, the downstream target genes of PART1/miR-503-5p axis remained unclear and they will be explored in the near future. Similarly, studies with more samples and the in vivo drug resistance tests on animal models were required to further explore whether PART1 is appropriate to be a therapeutic target for TSCC.
In conclusion, PART1 contributed to apoptosis, inhibited the proliferation, invasion and migration abilities of TSCC cells by targeting miR-503-5p, thus slowing down the development of TSCC.
Abbreviations
TSCC, tongue squamous cell carcinoma; LncRNAs, long non-coding RNAs; PART1, prostate androgen-regulated transcript 1; EMT, epithelial–mesenchymal transition; SDS-PAGE, sodium dodecyl sulfate-polyacrylamide gels; LSCC, laryngeal squamous cell carcinoma; PDCD4, programmed cell death 4; NHOK, normal human oral keratinocyte cell line; SDS-PAGE, sodium dodecyl sulfate-polyacrylamide gels; CCK-8, Cell Counting Kit-8; TUNEL, terminal deoxynucleotidyl transferase dUTP nick end labeling.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
There is no funding to report.
Disclosure
The authors declare no conflict of interest.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70(1):7–30.
2. Wu K, Wei J, Liu Z, et al. Can pattern and depth of invasion predict lymph node relapse and prognosis in tongue squamous cell carcinoma. BMC Cancer. 2019;19(1):714. doi:10.1186/s12885-019-5859-y
3. Sano D, Myers JN. Metastasis of squamous cell carcinoma of the oral tongue. Cancer Metastasis Rev. 2007;26(3–4):645–662.
4. Mascitti M, Rubini C, De Michele F, et al. American Joint Committee on Cancer staging system 7th edition versus 8th edition: any improvement for patients with squamous cell carcinoma of the tongue? Oral Surg Oral Med Oral Pathol Oral Radiol. 2018;126(5):415–423.
5. Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. 2016;17(1):47–62.
6. Shi X, Sun M, Liu H, Yao Y, Song Y. Long non-coding RNAs: a new frontier in the study of human diseases. Cancer Lett. 2013;339(2):159–166.
7. Moran VA, Perera RJ, Khalil AM. Emerging functional and mechanistic paradigms of mammalian long non-coding RNAs. Nucleic Acids Res. 2012;40(14):6391–6400.
8. Castro-Oropeza R, Melendez-Zajgla J, Maldonado V, Vazquez-Santillan K. The emerging role of lncRNAs in the regulation of cancer stem cells. Cell Oncol. 2018;41(6):585–603.
9. Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21(11):1253–1261.
10. Hao NB, He YF, Li XQ, Wang K, Wang RL. The role of miRNA and lncRNA in gastric cancer. Oncotarget. 2017;8(46):81572–81582. doi:10.18632/oncotarget.19197
11. Han P, Chang CP. Long non-coding RNA and chromatin remodeling. RNA Biol. 2015;12(10):1094–1098. doi:10.1080/15476286.2015.1063770
12. Ballantyne MD, McDonald RA, Baker AH. lncRNA/MicroRNA interactions in the vasculature. Clin Pharmacol Ther. 2016;99(5):494–501. doi:10.1002/cpt.355
13. Macfarlane LA, Murphy PR. MicroRNA: biogenesis, Function and Role in Cancer. Curr Genomics. 2010;Nov;11(7):537–561. doi:10.2174/138920210793175895
14. Chen L, Yao H, Wang K, Liu X. Long Non-Coding RNA MALAT1 Regulates ZEB1 Expression by Sponging miR-143-3p and Promotes Hepatocellular Carcinoma Progression. J Cell Biochem. 2017;118(12):4836–4843. doi:10.1002/jcb.26158
15. Song Y, Pan Y, Liu J. Functional analysis of lncRNAs based on competitive endogenous RNA in tongue squamous cell carcinoma. PeerJ. 2019;7:e6991. doi:10.7717/peerj.6991
16. Zhang S, Cao R, Li Q, Yao M, Chen Y, Zhou H. Comprehensive analysis of lncRNA-associated competing endogenous RNA network in tongue squamous cell carcinoma. PeerJ. 2019;7:e6397. doi:10.7717/peerj.6397
17. Wang ZY, Hu M, Dai MH, et al. Upregulation of the long non-coding RNA AFAP1-AS1 affects the proliferation, invasion and survival of tongue squamous cell carcinoma via the Wnt/beta-catenin signaling pathway. Mol Cancer. 2018;17(1):3. doi:10.1186/s12943-017-0752-2
18. Jia LF, Wei SB, Gan YH, et al. Expression, regulation and roles of miR-26a and MEG3 in tongue squamous cell carcinoma. Int j Cancer. 2014;135(10):2282–2293. doi:10.1002/ijc.28667
19. Lin B, White JT, Ferguson C, et al. PART-1: a novel human prostate-specific, androgen-regulated gene that maps to chromosome 5q12. Cancer Res. 2000;60(4):858–863.
20. Sun M, Geng D, Li S, Chen Z, Zhao W. LncRNA PART1 modulates toll-like receptor pathways to influence cell proliferation and apoptosis in prostate cancer cells. Biol Chem. 2018;399(4):387–395. doi:10.1515/hsz-2017-0255
21. Li M, Zhang W, Zhang S, Wang C, Lin Y. PART1 expression is associated with poor prognosis and tumor recurrence in stage I-III non-small cell lung cancer. J Cancer. 2017;8(10):1795–1800. doi:10.7150/jca.18848
22. Li S, Chen X, Liu X, et al. Complex integrated analysis of lncRNAs-miRNAs-mRNAs in oral squamous cell carcinoma. Oral Oncol. 2017;73:1–9. doi:10.1016/j.oraloncology.2017.07.026
23. Shuang Y, Zhou X, Li C, Huang Y, Zhang L. MicroRNA503 serves an oncogenic role in laryngeal squamous cell carcinoma via targeting programmed cell death protein 4. Mol Med Rep. 2017;16(4):5249–5256. doi:10.3892/mmr.2017.7278
24. Fei Y, Shan W, Chen X. MiR-503-5p functions as an oncogene in oral squamous cell carcinoma by targeting Smad7. Histol Histopathol. 2020;22:18220.
25. Han L, Cheng J, Li A. hsa_circ_0072387 Suppresses Proliferation, Metastasis, and Glycolysis of Oral Squamous Cell Carcinoma Cells by Downregulating miR-503-5p. Cancer Biother Radiopharm. 2020;17.
26. Zheng X, Wu K, Liao S, et al. MicroRNA-transcription factor network analysis reveals miRNAs cooperatively suppress RORA in oral squamous cell carcinoma. Oncogenesis. 2018;7(10):79. doi:10.1038/s41389-018-0089-8
27. Ye X, Weinberg RA. Epithelial-Mesenchymal Plasticity: A Central Regulator of Cancer Progression. Trends Cell Biol. 2015;25(11):675–686. doi:10.1016/j.tcb.2015.07.012
28. Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15(3):178–196.
29. Liz J, Esteller M. lncRNAs and microRNAs with a role in cancer development. Biochim Biophys Acta. 2016;1859(1):169–176. doi:10.1016/j.bbagrm.2015.06.015
30. Chan JJ, Tay Y. Noncoding RNA:RNA Regulatory Networks in Cancer. Int J Mol Sci. 2018;19(5):5. doi:10.3390/ijms19051310
31. Zhu M, Zhang C, Chen D, Chen S, Zheng H. lncRNA MALAT1 potentiates the progression of tongue squamous cell carcinoma through regulating miR-140-5p-PAK1 pathway. Onco Targets Ther. 2019;12:1365–1377. doi:10.2147/OTT.S192069
32. Hu X, Feng H, Huang H, et al. Downregulated Long Noncoding RNA PART1 Inhibits Proliferation and Promotes Apoptosis in Bladder Cancer. Technol Cancer Res Treat. 2019;18:1533033819846638. doi:10.1177/1533033819846638
33. Ye J, Zhang J, Lv Y, et al. Integrated analysis of a competing endogenous RNA network reveals key long noncoding RNAs as potential prognostic biomarkers for hepatocellular carcinoma. J Cell Biochem. 2019;120(8):13810–13825. doi:10.1002/jcb.28655
34. Zhang X-Q, Sun S, Lam K-F, et al. A long non-coding RNA signature in glioblastoma multiforme predicts survival. Neurobiol Dis. 2013;58:123–131. doi:10.1016/j.nbd.2013.05.011
35. Jin Z, Piao L, Sun G, Lv C, Jing Y, Jin R. Long Non-Coding RNA PART1 Exerts Tumor Suppressive Functions in Glioma via Sponging miR-190a-3p and Inactivation of PTEN/AKT Pathway. Onco Targets Ther. 2020;13:1073–1086.
36. Zhang S, Ma H, Zhang D, et al. LncRNA KCNQ1OT1 regulates proliferation and cisplatin resistance in tongue cancer via miR-211-5p mediated Ezrin/Fak/Src signaling. Cell Death Dis. 2018;9(7):742. doi:10.1038/s41419-018-0793-5
37. Wu X, Ma J, Chen J, Huang H. LncRNA CACS15 regulates tongue squamous cell carcinoma cell behaviors and predicts survival. BMC Oral Health. 2019;19(1):231. doi:10.1186/s12903-019-0924-0
38. Song KB, Liu WJ, Jia SS. miR-219 inhibits the growth and metastasis of TSCC cells by targeting PRKCI. Int J Clin Exp Med. 2014;7(9):2957–2965.
39. Wang Y, Zhu Y, Lv P, Li L. The role of miR-21 in proliferation and invasion capacity of human tongue squamous cell carcinoma in vitro. Int J Clin Exp Pathol. 2015;8(5):4555–4563.
40. Zhao Z, Fan X, Jiang L, et al. miR-503-3p promotes epithelial-mesenchymal transition in breast cancer by directly targeting SMAD2 and E-cadherin. J Genetics Genomics. 2017;44(2):75–84.
41. Mathias RA, Simpson RJ. Towards understanding epithelial-mesenchymal transition: a proteomics perspective. Biochim Biophys Acta. 2009;1794(9):1325–1331.
42. Mittal V. Epithelial-Mesenchymal Transition in Tumor Metastasis. Annu Rev Pathol. 2018;24(13):395–412.
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.