Back to Journals » OncoTargets and Therapy » Volume 13
LncRNA NEAT1 Regulates 5-Fu Sensitivity, Apoptosis and Invasion in Colorectal Cancer Through the MiR-150-5p/CPSF4 Axis
Authors Wang X, Jiang G, Ren W, Wang B, Yang C, Li M
Received 20 November 2019
Accepted for publication 4 June 2020
Published 1 July 2020 Volume 2020:13 Pages 6373—6383
DOI https://doi.org/10.2147/OTT.S239432
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Takuya Aoki
Xuesong Wang, Guosheng Jiang, Weidan Ren, Bo Wang, Chuanwei Yang, Meishuang Li
Department of Colorectal & Anal Surgery, Central Hospital of Cangzhou, Cangzhou 061000, Hebei, People’s Republic of China
Correspondence: Xuesong Wang Tel +86-18031792097
Email [email protected]
Background: Colorectal cancer (CRC) is one of the most prevalent malignancies in the world. Long non-coding RNA (lncRNA) nuclear enriched abundant transcript 1 (NEAT1) is involved in the development of many cancers. However, its role and mechanism in CRC progression still need further exploration.
Methods: The expression levels of lnc-NEAT1, microRNA-150-5p (miR-150-5p) and cleavage and polyadenylation specific factor 4 (CPSF4) were determined by quantitative real-time PCR (qRT-PCR). The sensitivity of cells to 5-fluorouracil (5-Fu) was measured by 3-(4,5-dimethyl-2 thiazolyl)-2,5-diphenyl-2-H-tetrazolium bromide (MTT) assay. Cell apoptosis and invasion were evaluated by flow cytometry and transwell assays, respectively. Western blot (WB) analysis was used to assess the levels of resistance-related proteins and CPSF4 protein. Besides, dual-luciferase reporter assay was used to verify the interactions among lnc-NEAT1, miR-150-5p and CPSF4. Also, mice xenograft models were used to determine the effect of lnc-NEAT1 on CRC tumor growth in vivo.
Results: In CRC, the expression of lnc-NEAT1 was upregulated and miR-150-5p was downregulated, and the expression of both was negatively correlated. Silencing of lnc-NEAT1 promoted the 5-Fu sensitivity, enhanced the apoptosis and suppressed the invasion of CRC cells. MiR-150-5p could be sponged by lnc-NEAT1, and its inhibitors could partially reverse the effect of lnc-NEAT1 silencing on CRC progression. Besides, CPSF4 could be targeted by miR-150-5p, and its overexpression also could invert the effect of lnc-NEAT1 knockdown on CRC progression. Further, CPSF4 expression was regulated by lnc-NEAT1 and miR-150-5p. In addition, interference of lnc-NEAT1 reduced tumor volume and improved the sensitivity of CRC to 5-Fu in vivo.
Conclusion: Lnc-NEAT1 acted as an oncogene in CRC through regulating CPSF4 expression by sponging miR-150-5p. The discovery of lnc-NEAT1/miR-150-5p/CPSF4 axis provided a novel approach for CRC genomic therapy strategy.
Keywords: CRC, lnc-NEAT1, CPSF4, miR-150-5p
Introduction
Colorectal cancer (CRC) is the third most common cause of cancer-related mortality in the world, with different incidence rates by sex and age.1,2 Currently, the incidence and mortality of CRC in China are on the rise in both males and females, and it is becoming the fastest growing cancer in our society.3,4 CRC patients have no obvious symptoms in the early stage, and the late stage is usually accompanied by abdominal pain, fever, wasting and other systemic symptoms.5 Hence, the exploration of effective screening targets is helpful for the early diagnosis of CRC.
In the human genome, 98% of the genes are not translated, and only about 20,000 genes can encode proteins. These RNAs that do not encode proteins are called non-coding RNAs (ncRNAs), among which those with more than 200 nucleotides (nts) in length are called long non-coding RNAs (lncRNAs).6 Many lncRNAs have been proven to be involved in the regulation of CRC progression, such as DILC, TCF7 and RUNX1-IT1.7–9 LncRNA nuclear enriched abundant transcript 1 (lnc-NEAT1) is a nuclear-restricted lncRNA found in recent years. Previous studies had suggested that lnc-NEAT1 was overexpressed in different types of solid tumors,10 and it could act as an oncogene in CRC.11,12 Therefore, exploring the mechanism of lnc-NEAT1 could provide more evidence for its role in CRC.
LncRNAs have a variety of biological functions, mainly act as a sponge of microRNAs (miRNAs) to participate in the regulation of miRNAs on downstream target genes.13,14 As a gene regulator, miRNAs have been proved to be involved in the regulation of cancer progression.15 MiR-150-5p has been shown to be widely under-expressed in cancers and might function as a tumor suppressor in CRC.16,17 Therefore, the study on miR-150-5p could perfect its mechanism in CRC.
Cleavage and polyadenylation specific factor 4 (CPSF4) is a member of the CPSF complex and related to mRNA maturation.18,19 CPSF4 function as a target gene has been shown to participate in the regulation of many cancers, including lung cancer, breast cancer and thyroid cancer.20–22 Nevertheless, the exact role of CPSF4 in CRC progression are not well understood.
Based on the above reports, we explored the role and mechanism of lnc-NEAT1 in CRC progression. All these findings proved that lnc-NEAT1 acted as a pro-tumorigenic factor in the development of CRC, which might also provide a new perspective for the therapy of CRC.
Materials and Methods
Patient Tissues Collection
Tumor tissues and adjacent normal tissues of 30 CRC patients were obtained from the Central Hospital of Cangzhou. All patients not received any treatment and signed informed consent. This study was authorized by the Ethics Committee of Central Hospital of Cangzhou.
Cell Culture
CRC cell lines (SW480 and HCT116) were obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA), and normal colorectal epithelial cells (NCM460) were purchased from the Shanghai Enzyme Research Biotechnology Co., Ltd. (Shanghai, China). These cells were cultured in RPMI-1640 medium (Gibco, Waltham, MA, USA) containing 10% fetal bovine serum (FBS; Gibco) and 1% penicillin (100 U/mL)/streptomycin (0.1 mg/mL) (Invitrogen, Carlsbad, CA, USA) at 37°C in 5% CO2 atmosphere.
Quantitative Real-Time PCR (qRT-PCR)
Total RNAs were extracted using TRIzol reagent (Invitrogen). The first strand DNA synthesis was conducted using One Step miRNA cDNA Synthesis Kit (Takara, Dalian, China) for miR-150-5p and Universal RT-PCR Kit (Solarbio, Beijing, China) for genes, respectively. QRT-PCR was carried out using SYBR Green (Solarbio) on a Real-time PCR system (ABI, Foster City, CA, USA). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and U6 were used as internal controls. All primers were listed as below: lnc-NEAT1: F, 5ʹ-CTTCCTCCCTTTAACTTATCCATTCAC-3ʹ, R, 5ʹ-CTCTTCCTCCACCATTACCAACAATAC-3ʹ; CPSF4: F, 5ʹ-CTTCCTGCACATCGACCC-3ʹ, R, 5ʹ-AGATGACTCTCCGCGTGTG-3ʹ; GAPDH: F, 5ʹ-GGTGGTCTCCTCTGACTTCAA-3ʹ, R, 5ʹ-GTTGCTGTAGCCAAATTCGTTGT-3ʹ; miR-150-5p: 5ʹ-TCGGCGTCTCCCAACCCTTGTAC-3ʹ, R, 5ʹ-GTCGTATCCAGTGCAGGGTCCGAGGT-3ʹ; U6: F, 5ʹ-TGCGGGTGCTCGCTTCGGCAGC-3ʹ, R, 5ʹ-GGGTCCGAGGTGCACTGGATACGACAAAATATGG-3ʹ. Relative expression values were calculated using the 2−∆∆Ct method.
Cell Transfection
Small interference RNA targeting lnc-NEAT1 and CPSF4 (si-lnc-NEAT1 and si-CPSF4) or their negative control (si-NC), miR-150-5p mimics and inhibitors (anti-miR-150-5p) or their negative controls (miR-NC and anti-miR-NC), lentivirus-mediated short hairpin RNA targeting lnc-NEAT1 (sh-lnc-NEAT1) and its negative control (sh-NC) were synthesized by Genechem (Shanghai, China). Lnc-NEAT1 and CPSF4 overexpression plasmids or their negative control (pcDNA) were obtained from GeneCreate (Wuhan, China). SW480 and HCT116 cells transfection were executed using Lipofectamine 3000 (Invitrogen). Non-transfected cells were recorded as Control.
Cell Viability Assay
This experiment was performed using 3-(4, 5-dimethyl-2 thiazolyl)-2, 5-diphenyl-2-H-tetrazolium bromide (MTT) Kit (Beyotime, Shanghai, China). After transfection, SW480 and HCT116 cells were treated with 5-fluorouracil (5-Fu) at different concentrations for 48 h. Then, MTT solution was added for further incubation for 4 h, followed by dimethylsulfoxide (DMSO) for dissolution. The absorbance was measured at 560 nm and half-maximal inhibitory concentration (IC50) was calculated to evaluate the 5-Fu sensitivity of cells.
Flow Cytometry
After transfection, SW480 and HCT116 cells were digested with trypsin and collected into the centrifuge tube. After centrifugation, cells were re-suspended with binding buffer and stained using Annexin V-FITC/PI Apoptosis Detection Kit (Yeasen, Shanghai, China). FITC fluorescence was monitored by Flow cytometer (Beckman Coulter, San Jose, CA, USA) and the apoptosis rate was calculated.
Cell Invasion Assay
Cell invasion was performed using Transwell chambers (Corning Inc., Corning, NY, USA). SW480 and HCT116 cells were seeded into the upper chambers coated with Matrigel (BD Biosciences, San Jose, CA, USA) and contained the serum-free medium. The lower chambers were added with RPMI-1640 medium containing 10% FBS. After 48 h, the lower chambers cells were fixed with methanol, stained with crystal violet, and then counted the number of invaded cells under a microscope (Shoif, Shanghai, China).
Western Blot (WB) Analysis
Total proteins were extracted using RIPA buffer (Beyotime). Equal amounts of proteins were isolated with 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel and transferred onto polyvinylidene fluoride (PVDF) membranes (Millipore, Billerica, MA, USA). Afterward, the membranes were closed with 5% non-fat milk and cultured with the primary antibodies against P-gp (1:500, Abcam, Cambridge, MA, USA), GST-π (1:1000, Abcam), CPSF4 (1:300, Bioss, Beijing, China) or GAPDH (1:300, Bioss) at 4°C overnight. After incubated with the secondary antibody (1:1000, Bioss) for 1 h, the protein signals were visualized using the ECL system (Beyotime).
Dual-Luciferase Reporter Assay
The sequences of lnc-NEAT1 or CPSF4 3ʹUTR containing the miR-150-5p binding sites or mutant binding sites were inserted into the pmirGLO reporter vector (Youbio, Changsha, China) to generate wild-type (lnc-NEAT1-WT and CPSF4 3ʹUTR-WT) or mutant-type (lnc-NEAT1-MUT and CPSF4 3ʹUTR-MUT) reporter vectors. SW480 and HCT116 cells were seeded into 24-well plates. After 24 h, the above report vectors were co-transfected with miR-150-5p mimics or miR-NC into SW480 and HCT116 cells. After transfection 48 h, the luciferase activities were tested using the Dual-luciferase Reporter Assay Kit (Genomeditech, Shanghai, China).
Mice Xenograft Models
One-month-old male BALB/c-nude mice (Laboratory animal center of southern medical university, Guangzhou, China) were used in this study. All experiments were authorized by the Animal Care Committee of Central Hospital of Cangzhou. Mice were subcutaneously injected with HCT116 cells transfected with sh-lnc-NEAT1 or sh-NC. After 7 d, 5-Fu (50 mg/kg) or PBS was intraperitoneally injected into mice every 4 d, and the tumor volume was calculated using the formula: volume = length × width2/2. After 27 d, the tumor was removed for weight measurement and further experiments. Animal studies were performed in compliance with the ARRIVE guidelines and the Basel Declaration. All animals received humane care according to the National Institutes of Health (USA) guidelines.
Statistical Analysis
Data were shown as mean ± standard deviation (SD). SPSS 22.0 software (SPSS Inc., Chicago, IL, USA) was used for statistical analysis. One-way ANOVA, two-way ANOVA and Student’s t-test were used to compare the difference of groups. Three independent experiments were carried out for each assay. The correlation between lnc-NEAT1 and miR-150-5p or CPSF4 was analyzed using Pearson correlation analysis. P < 0.05 was considered to be significant.
Results
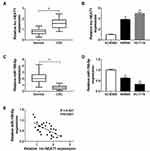
Lnc-NEAT1 Was Upregulated and MiR-150-5p Was Downregulated in CRC Tissues and Cells
To begin with, we detected the expression of lnc-NEAT1 in CRC, and the results showed that lnc-NEAT1 was significantly highly expressed in CRC tissues compared with adjacent normal tissues (Figure 1A). Besides, the expression of lnc-NEAT1 was markedly increased in two CRC cell lines (SW480 and HCT116) compared with NCM460 (Figure 1B). Meanwhile, we also found that miR-150-5p was obviously lower expressed in CRC tissues and cells (Figure 1C and D). Interestingly, correlation analysis revealed that the expression of lnc-NEAT1 was negatively correlated with miR-150-5p (Figure 1E). Given the opposite expression trend between this, we speculated that there might be an association between lnc-NEAT1 and miR-150-5p.
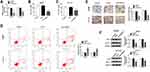
Knockdown of Lnc-NEAT1 Increased the CRC Cell Sensitivity to 5-Fu, Promoted Apoptosis and Inhibited Invasion in CRC Cells
To further explore the effect of lnc-NEAT1 on CRC, we transfected si-NC and si-lnc-NEAT1 into SW480 and HCT116 cells. The efficiency test results showed that si-lnc-NEAT1 had a good inhibitory effect on lnc-NEAT1 expression, which could be used for follow-up experiments (Figure 2A). Through detecting cell sensitivity, we found that the IC50 value of 5-Fu was significantly decreased after lnc-NEAT1 silencing, indicating that 5-Fu sensitivity of SW480 and HCT116 cells was markedly increased (Figure 2B and C). Flow cytometry results showed that lnc-NEAT1 knockdown promoted the apoptosis of SW480 and HCT116 cells (Figure 2D). Besides, we measured the invasion of CRC cells by transwell assay, as shown in Figure 2E, silenced-lnc-NEAT1 significantly blocked the number of invaded SW480 and HCT116 cells. Also, WB analysis results revealed that lnc-NEAT1 silencing markedly suppressed the protein levels of resistance-related proteins P-gp and GST-π in SW480 and HCT116 cells (Figure 2F). These results suggested that lnc-NEAT1 played an active role in the progression of CRC cells.
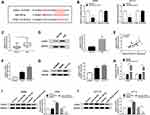
MiR-150-5p Could Be Sponged by Lnc-NEAT1 and Its Inhibitors Could Invert the Effects of Lnc-NEAT1 Knockdown on CRC Cell Progression
To confirm the relationship between lnc-NEAT1 and miR-150-5p, we performed the bioinformatics predictions. Surprisingly, the StarBase2.0 tool predicted that miR-150-5p had complementary binding sites with lnc-NEAT1 (Figure 3A). Besides, dual-luciferase reporter assay results revealed that overexpression of miR-150-5p markedly reduced the luciferase activity of lnc-NEAT1-WT in SW480 and HCT116 cells, but did not affect lnc-NEAT1-MUT (Figure 3B). Meanwhile, we also detected the effect of lnc-NEAT1 expression on miR-150-5p expression. The increased expression of lnc-NEAT1 indicated that the transfection efficiency of lnc-NEAT1 overexpression plasmid was excellent in SW480 and HCT116 cells (Figure 3C). As shown in Figure 3D, knockdown of lnc-NEAT1 upregulated the expression of miR-150-5p, while overexpressed lnc-NEAT1 inhibited miR-150-5p level. In addition, we co-transfected si-lnc-NEAT1 and anti-miR-150-5p into SW480 and HCT116 cells to investigate the role of miR-150-5p on CRC cell progression. The decreased expression of miR-150-5p in SW480 and HCT116 cells revealed that the transfection efficiency of anti-miR-150-5p was good (Figure 3E). The statistical results of IC50 values showed that inhibition of miR-150-5p partially recovered the promoting effect of lnc-NEAT1 knockdown on the 5-Fu sensitivity of SW480 and HCT116 cells (Figure 3F). Furthermore, flow cytometry and transwell assays results indicated that the promotion effect of silenced-lnc-NEAT1 on apoptosis and suppression effect of it on invasion could be inverted by miR-150-5p inhibitors (Figure 3G and H). Meanwhile, the increased expression of P-gp and GST-π after miR-150-5p knockdown also confirmed the recovery effect of miR-150-3p on the sensitivity of CRC cells to 5-Fu (Figure 3I and J). These results suggested that miR-150-5p played a vital role in the regulation of lnc-NEAT1 on CRC progression.
MiR-150-5p Directly Targeted CPSF4
Meanwhile, TargetScan Human 7.2 prediction analysis showed that CPSF4 3ʹUTR had complementary sites with miR-150-5p (Figure 4A). Dual-luciferase reporter assay verification showed that miR-150-5p mimics remarkably suppressed the luciferase activity of CPSF4 3ʹUTR-WT in SW480 and HCT116 cells, while had no effect on CPSF4 3ʹUTR-MUT (Figure 4B). Through detecting the expression of CPSF4, we found that the mRNA and protein expression levels of CPSF4 were remarkably elevated in CRC tissues (Figure 4C and D). Besides, correlation analysis indicated that CPSF4 expression was positively correlated with lnc-NEAT1 (Figure 4E). We also measured the expression of CPSF4 in CRC cells and NCM460 cells and discovered that CPSF4 mRNA and protein expression levels were increased in CRC cells (Figure 4F and G). Furthermore, we detected the effect of miR-150-5p expression on CPSF4 expression. The increased level of miR-150-5p in SW480 and HCT116 cells indicated that the transfection efficiency of miR-150-5p mimics was excellent (Figure 4H). WB results showed that CPSF4 protein level was promoted by miR-150-5p inhibition, while suppressed by miR-150-5p overexpression in SW480 and HCT116 cells (Figure 4I and J). Hence, all data revealed that CPSF4 was a target of miR-150-5p.
CPSF4 Overexpression Reversed the Effects of Silenced-Lnc-NEAT1 on 5-Fu Sensitivity, Apoptosis and Invasion in CRC Cells
To further confirm the role of CPSF4 in the regulation of lnc-NEAT1 on CRC progression, we co-transfected si-lnc-NEAT1 and CPSF4 overexpression plasmid into SW480 and HCT116 cells. The mRNA and protein levels of CPSF4 in SW480 and HCT116 cells were increased by CPSF4 overexpression plasmid, suggesting that its transfection efficiency was excellent (Figure 5A and B). The detection of IC50 values in SW480 and HCT116 cells revealed that overexpressed-CPSF4 could reverse the promoting effect of lnc-NEAT1 knockdown on the sensitivity of CRC cells to 5-Fu (Figure 5C). Besides, flow cytometry and transwell assays results indicated that CPSF4 overexpression restored the promotion effect of lnc-NEAT1 silencing on apoptosis, as well as the suppression effect of it on invasion in SW480 and HCT116 cells (Figure 5D and E). Further, the inhibition effect of silenced-lnc-NEAT1 on the protein levels of P-gp and GST-π in SW480 and HCT116 cells also could be inverted by CPSF4 overexpression (Figure 5F and G). These results showed that CPSF4 participated in the regulation of lnc-NEAT on CRC progression.
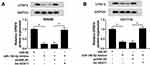
CPSF4 Expression Was Regulated by MiR-150-5p and Lnc-NEAT1
To verify the regulation of lnc-NEAT1 on CPSF4, we co-transfected miR-150-5p mimics and lnc-NEAT1 overexpression plasmid into SW480 and HCT116 cells. Through detecting the CPSF4 protein level in SW480 and HCT116 cells, we found that the suppression effect of miR-150-5p overexpression on CPSF4 expression could be reversed by lnc-NEAT1 overexpression (Figure 6A and B). These results confirmed that CPSF4 was indeed the downstream target gene of the lnc-NEAT1/miR-150-5p axis.
Interference of Lnc-NEAT1 Inhibited CRC Tumor Growth in vivo
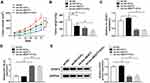
To further explore the effect of lnc-NEAT1 on CRC tumor growth, we constructed the mice xenograft model in vivo using HCT116 cells stably transfected with sh-lnc-NEAT1 or sh-NC. It was found that 5-Fu and lnc-NEAT1 knockdown could significantly inhibit the tumor volume of CRC, and the inhibitory effect was more obvious when the two co-existed (Figure 7A). Also, we measured the tumor weight and found that the tumor weight of the 5-Fu-treated group and sh-lnc-NEAT1 group was significantly reduced, and the tumor weight was lower when the two were treated together (Figure 7B). According to qRT-PCR analysis, we found that sh-lnc-NEAT1 could inhibit lnc-NEAT1 expression, while the treatment of 5-Fu did not affect the expression of lnc-NEAT1 (Figure 7C). In addition, we also detected the expression of miR-150-5p and CPSF4 in the tumors. As shown in Figure 7D and E, we discovered that compared to the negative groups, miR-150-5p expression was markedly enhanced and CPSF4 protein level was obviously restrained in the lnc-NEAT1 knockdown group with or without treatment with 5-Fu. Therefore, we speculated that lnc-NEAT1 silencing could enhance the inhibitory ability of 5-Fu on tumor growth, which might be because lnc-NEAT1 silencing promoted the sensitivity of CRC to 5-Fu.
Discussion
At present, many studies have confirmed that lncRNAs are related to the occurrence of CRC through multiple pathways, including regulation of cell metastasis, autophagy and proliferation.23–25 Studies have shown that lnc-NEAT1 is abnormally expressed in different types of cancer, and its dysregulation may be related to tumorigenesis.26–28 Herein, we selected lnc-NEAT1 to future study and discovered that lnc-NEAT1 expression was elevated in CRC. Silencing of lnc-NEAT1 enhanced 5-Fu sensitivity, promoted apoptosis and inhibited invasion in CRC cells in vitro, which was consistent with the previous studies in CRC.12,29 Besides, lnc-NEAT1 knockdown also reduced CRC tumor growth and enhanced the sensitivity of CRC to 5-Fu in vivo. The conclusion that the expression of lnc-NEAT1 could affect the sensitivity of CRC to 5-Fu also provided a new solution for the clinical reduction of the drug resistance in CRC. Of course, these results also need to be further studied in the scope of some big data to prove that lnc-NEAT1 expression can be used as an effective biomarker to evaluate the progression of CRC patients in clinical practice.
The role of miRNAs has been well elucidated since their function was discovered. In our study, we here found that miR-150-5p was decreased in CRC tissues and cells, which was consistent with the previous study.16 Besides, miR-150-5p could be absorbed by lnc-NEAT1, and its expression was regulated by lnc-NEAT1. MiR-150-5p is not only involved in the regulation of CRC progression but also has been proved to be related to the poor prognosis of CRC patients.30 Functional experiments confirmed that miR-150-5p inhibitors could reverse the effect of silenced-lnc-NEAT1 on CRC sensitivity, apoptosis and invasion. Meanwhile, further analysis revealed that CPSF4 was a downstream target of miR-150-5p. It had been proved that CPSF4 was overexpressed in many cancers, including CRC.31,32 Our data also showed that CPSF4 expression was markedly increased in CRC tissues and cells. The gain- and loss-of functional tests also confirmed the role of CPSF4, that was, its overexpression could reverse the regulatory effect of lnc-NEAT1 on CRC sensitivity, apoptosis and invasion.
Although the regulatory effect of lncRNAs on the progression of CRC has been confirmed, the molecular mechanism of lncRNAs still needs further study. Our study explored the effect of lnc-NEAT1 on the 5-Fu sensitivity, apoptosis and invasion of CRC and confirmed the positive influence of lnc-NEAT1 on the progression of CRC. However, this study has some limitations. The restoration of the function of miR-150-5p to lnc-NEAT1 is partial, and there may be other miRNAs involved in the regulation of lnc-NEAT1 on CRC progression, which needs further study. In addition, the molecular mechanism by which lnc-NEAT1 reduced 5-Fu sensitivity is still unclear, which can be further discussed the role and mechanism of lnc-NEAT1 on CRC drug-resistant cell lines in the future to enrich the functions of lnc-NEAT1.
In summary, our results showed that lnc-NEAT1 enhanced CPSF4 expression via sponging miR-150-5p to accelerate CRC progression. The revelation of the lnc-NEAT1/miR-150-5p/CPSF4 axis enriched the regulatory network of lnc-NEAT1 and provided a complete basis for lnc-NEAT1 acted as a therapeutic target of CRC.
Disclosure
The authors declare that they have no conflicts of interest in this work.
References
1. Siegel RL, Miller KD, Fedewa SA, et al. Colorectal cancer statistics, 2017. CA Cancer J Clin. 2017;67(3):177–193. doi:10.3322/caac.21395
2. Brenner H, Kloor M, Pox CP. Colorectal cancer. Lancet. 2014;383(9927):1490–1502. doi:10.1016/S0140-6736(13)61649-9
3. Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. doi:10.3322/caac.21338
4. Wang X, Kuang YY, Hu XT. Advances in epigenetic biomarker research in colorectal cancer. World J Gastroenterol. 2014;20(15):4276–4287. doi:10.3748/wjg.v20.i15.4276
5. Compton CC. Colorectal carcinoma: diagnostic, prognostic, and molecular features. Mod Pathol. 2003;16(4):376–388. doi:10.1097/01.MP.0000062859.46942.93
6. Boon RA, Jae N, Holdt L, et al. Long noncoding RNAs: from clinical genetics to therapeutic targets? J Am Coll Cardiol. 2016;67(10):1214–1226. doi:10.1016/j.jacc.2015.12.051
7. Gu LQ, Xing XL, Cai H, et al. Long non-coding RNA DILC suppresses cell proliferation and metastasis in colorectal cancer. Gene. 2018;666:18–26. doi:10.1016/j.gene.2018.03.100
8. Jin FS, Wang HM, Song XY. Long non-coding RNA TCF7 predicts the progression and facilitates the growth and metastasis of colorectal cancer. Mol Med Rep. 2018;17(5):6902–6908. doi:10.3892/mmr.2018.8708
9. Gong J, Tian J, Lou J, et al. A functional polymorphism in lnc-LAMC2-1:1 confers risk of colorectal cancer by affecting miRNA binding. Carcinogenesis. 2016;37(5):443–451. doi:10.1093/carcin/bgw024
10. Yu X, Li Z, Zheng H, et al. NEAT1: a novel cancer-related long non-coding RNA. Cell Prolif. 2017;50(2). doi:10.1111/cpr.12368.
11. Wu Y, Yang L, Zhao J, et al. Nuclear-enriched abundant transcript 1 as a diagnostic and prognostic biomarker in colorectal cancer. Mol Cancer. 2015;14:191. doi:10.1186/s12943-015-0455-5
12. Yu HM, Wang C, Yuan Z, et al. LncRNA NEAT1 promotes the tumorigenesis of colorectal cancer by sponging miR-193a-3p. Cell Prolif. 2019;52(1):e12526. doi:10.1111/cpr.12526
13. Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505(7483):344–352. doi:10.1038/nature12986
14. Salmena L, Poliseno L, Tay Y, et al. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146(3):353–358. doi:10.1016/j.cell.2011.07.014
15. Reddy KB. MicroRNA (miRNA) in cancer. Cancer Cell Int. 2015;15:38. doi:10.1186/s12935-015-0185-1
16. Chen X, Xu X, Pan B, et al. miR-150-5p suppresses tumor progression by targeting VEGFA in colorectal cancer. Aging (Albany NY). 2018;10(11):3421–3437. doi:10.18632/aging.101656
17. Chen X, Zeng K, Xu M, et al. SP1-induced lncRNA-ZFAS1 contributes to colorectal cancer progression via the miR-150-5p/VEGFA axis. Cell Death Dis. 2018;9(10):982. doi:10.1038/s41419-018-0962-6
18. Kaufmann I, Martin G, Friedlein A, et al. Human Fip1 is a subunit of CPSF that binds to U-rich RNA elements and stimulates poly(A) polymerase. EMBO J. 2004;23(3):616–626. doi:10.1038/sj.emboj.7600070
19. Nemeroff ME, Barabino SM, Li Y, et al. Influenza virus NS1 protein interacts with the cellular 30 kDa subunit of CPSF and inhibits 3ʹend formation of cellular pre-mRNAs. Mol Cell. 1998;1(7):991–1000. doi:10.1016/S1097-2765(00)80099-4
20. Wu J, Miao J, Ding Y, et al. MiR-4458 inhibits breast cancer cell growth, migration, and invasion by targeting CPSF4. Biochem Cell Biol. 2019;97(6):722–730. doi:10.1139/bcb-2019-0008
21. Yi C, Wang Y, Zhang C, et al. Cleavage and polyadenylation specific factor 4 targets NF-kappaB/cyclooxygenase-2 signaling to promote lung cancer growth and progression. Cancer Lett. 2016;381(1):1–13. doi:10.1016/j.canlet.2016.07.016
22. Li Z, Xu X, Li Y, et al. Synergistic antitumor effect of BKM120 with prima-1Met via inhibiting PI3K/AKT/mTOR and CPSF4/hTERT signaling and reactivating mutant P53. Cell Physiol Biochem. 2018;45(5):1772–1786. doi:10.1159/000487786
23. Liu L, Wang HJ, Meng T, et al. lncRNA GAS5 inhibits cell migration and invasion and promotes autophagy by targeting miR-222-3p via the GAS5/PTEN-signaling pathway in CRC. Mol Ther Nucleic Acids. 2019;17:644–656. doi:10.1016/j.omtn.2019.06.009
24. Si Y, Yang Z, Ge Q, et al. Long non-coding RNA Malat1 activated autophagy, hence promoting cell proliferation and inhibiting apoptosis by sponging miR-101 in colorectal cancer. Cell Mol Biol Lett. 2019;24:50. doi:10.1186/s11658-019-0175-8
25. Liu Y, Zhou J, Wang S, et al. Long non-coding RNA SNHG12 promotes proliferation and invasion of colorectal cancer cells by acting as a molecular sponge of microRNA-16. Exp Ther Med. 2019;18(2):1212–1220. doi:10.3892/etm.2019.7650
26. Li X, Deng S, Pang X, et al. LncRNA NEAT1 silenced miR-133b promotes migration and invasion of breast cancer cells. Int J Mol Sci. 2019;20(15):3616.
27. Pang Y, Wu J, Li X, et al. NEAT1/miR124/STAT3 feedback loop promotes breast cancer progression. Int J Oncol. 2019. doi:10.3892/ijo.2019.4841
28. Dong P, Xiong Y, Yue J, et al. Long noncoding RNA NEAT1 drives aggressive endometrial cancer progression via miR-361-regulated networks involving STAT3 and tumor microenvironment-related genes. J Exp Clin Cancer Res. 2019;38(1):295. doi:10.1186/s13046-019-1306-9
29. Zhong F, Zhang W, Cao Y, et al. LncRNA NEAT1 promotes colorectal cancer cell proliferation and migration via regulating glial cell-derived neurotrophic factor by sponging miR-196a-5p. Acta Biochim Biophys Sin (Shanghai). 2018;50(12):1190–1199. doi:10.1093/abbs/gmy130
30. Zou SL, Chen YL, Ge ZZ, et al. Downregulation of serum exosomal miR-150-5p is associated with poor prognosis in patients with colorectal cancer. Cancer Biomark. 2019;26(1):69–77. doi:10.3233/CBM-190156
31. Chen W, Qin L, Wang S, et al. CPSF4 activates telomerase reverse transcriptase and predicts poor prognosis in human lung adenocarcinomas. Mol Oncol. 2014;8(3):704–716. doi:10.1016/j.molonc.2014.02.001
32. Yang Q, Fan W, Zheng Z, et al. Cleavage and polyadenylation specific factor 4 promotes colon cancer progression by transcriptionally activating hTERT. Biochim Biophys Acta Mol Cell Res. 2019;1866(10):1533–1543. doi:10.1016/j.bbamcr.2019.07.001
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.