Back to Journals » Cancer Management and Research » Volume 12
LncRNA MAGI2-AS3 Affects Cell Invasion and Migration of Cervical Squamous Cell Carcinoma (CSCC) via Sponging miRNA-233/EPB41L3 Axis
Authors Hou A, Zhang Y, Fan Y, Zheng Y, Zhou X, Liu H
Received 22 July 2019
Accepted for publication 17 May 2020
Published 3 June 2020 Volume 2020:12 Pages 4209—4216
DOI https://doi.org/10.2147/CMAR.S224067
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Eileen O'Reilly
Anli Hou,1 Yali Zhang,2 Yujuan Fan,1 Yi Zheng,3 Xiuying Zhou,1 Huilan Liu1
1Department of Gynaecology, University of Chinese Academy of Sciences Shenzhen Hospital, Shenzhen City, Guangdong Province 518106, People’s Republic of China; 2Department of Gynaecology, Affiliated Hospital of Chengde Medical University, Chengde, Hebei 067000, People’s Republic of China; 3Department of Central Lab, University of Chinese Academy of Sciences Shenzhen Hospital, Shenzhen City, Guangdong Province 518106, People’s Republic of China
Correspondence: Anli Hou
Department of Gynaecology, University of Chinese Academy of Sciences Shenzhen Hospital, No. 4253 Songbai Road, Matian Street, Guangming District, Shenzhen City, Guangdong Province 518106, People’s Republic of China
Tel +86-755-27548704
Email [email protected]
Introduction: The incidence of cervical squamous cell carcinoma (CSCC) has expanded in recent years. However, the function of long non-coding RNA (lncRNA) MAGI2-AS3 in the occurrence and progression of CSCC remains unclear. Therefore, the role of lncRNA MAGI2-AS3 in cervical squamous cell carcinoma (CSCC) was investigated in our study.
Methods: We used qRT-PCR analysis to identify the level of MAGI2-AS3 mRNA expression in CSCC clinical samples and cell lines. We investigated cell migration and invasion of CSCC cells transfected with MAGI2-AS3, miR-233 mimic, or EPB41L3 with transwell assays. Bioinformatics analysis and a luciferase reporter assay were employed to predict the interaction between MAGI2-AS3 and miR-233.
Results: We found that MAGI2-AS3 and EPB41L3 were both downregulated in CSCC and the expression of this two was positively correlated. Bioinformatics analysis showed that MAGI2-AS3 might bind to miR-233, which could directly target EPB41L3. In CSCC cells, overexpression of MAGI2-AS3 led to upregulated, while overexpression of miRNA-233 led to downregulated expression of EPB41L3. However, MAGI2-AS3 and miR-233 did not affect the expression of each other. In addition, overexpression of MAGI2-AS3 and EPB41L3 led to inhibited cancer cell invasion and migration, while overexpression of miR-233 played an opposite role and attenuated the effects of overexpressing MAGI2-AS3.
Conclusion: MAGI2-AS3 may sponge miR-233 to upregulate EPB41L3, thereby inhibiting CSCC cell invasion and migration.
Keywords: lncRNA MAGI2-AS3, cervical squamous cell carcinoma, EPB41L3, miR-223
Introduction
As the most common type of cervical cancer, cervical squamous cell carcinoma (CSCC) causes high mortality rate in females.1,2 Even worse, young females can be affected by CSCC as well.1,2 Infections caused by human papillomavirus (HPV) are the main causes of CSCC.3 In effect, high prevalence of HPV infections is always accompanied by high incidence of CSCC.3 CSCCs are prone to develop tumor metastasis, such as lymph node metastasis and even long-distance metastasis.4 As a consequence, most CSCC patients are diagnosed at advanced stages and the survival is poor in most cases.5 A better understanding of the pathogenesis of CSCC is critical for the development of therapeutic approaches to treat CSCC.
Erythrocyte membrane protein band 4.1 like 3 (EPB41L3) has tumor suppressive functions in several types of cancer.6,7 For example, EPB41L3 suppresses tumor progression by regulating cancer cell behaviors, such as inhibiting tumor cell migration and promoting cell apoptosis.6,7 However, the mechanism remains poorly understood. EPB41L3 in cancer biology is frequently regulated by non-coding RNAs, such as miRNAs8 and long (> 200 nt) non-coding RNAs (lncRNAs).9 A recent study reported that miRNA-223 could target EPB41L3 to promote the invasion of gastric cancer cells.8 LncRNA MAGI2-AS3 has recently been characterized as a tumor suppressor in several types of cancer.10,11 Our preliminary bioinformatics analysis showed that MAGI2-AS3 may interact with miR-233. This study aimed to investigate the interaction between MAGI2-AS3 and miR-233 in CSCC.
Patients and Methods
Patients and Specimens
A total of 60 CSCC patients (females, age range from 34 to 66 years old, and the mean age of 50.2 ± 5.6 years old) were selected from 121 CSCC patients admitted at the University Of Chinese Academy of Sciences Shenzhen Hospital from August 2016 to April 2019. Inclusion criteria: 1) confirmed by histopathological tests; 2) new diagnosed CSCC cases; 3) willing to donate tissue biopsy; 4) no therapies were initiated. Exclusion criteria: 1) clinical disorders besides CSCC; 2) recurrent cases; 3) history of cancer. All the 60 CSCC were subjected to HPV infection detection and the results showed that all patients were infected by different variants of HPV (HPV positive). The patients were staged according to the standard proposed by AJCC, and 10, 10, 23 and 17 cases were classified to stage I–IV, respectively. All patients received biopsy to collect non-tumor (5 cm around tumor) cervical and CSCC tissues (0.019 to 0.025 g). All tissue specimens were confirmed by histopathological examinations. The whole study was performed in accordance with the Declaration of Helsinki and was approved by the Ethics Committee of aforementioned hospital. All patients understood the experimental principle and signed the written informed consent.
LncRNA–miRNA Interaction Prediction
The interaction between MAGI2-AS3 and miR-233 was predicted using an online program IntaRNA (http://rna.informatik.uni-freiburg.de/IntaRNA/Input.jsp). During prediction, miR-233 sequence was used as the “short sequence” and MAGI2-AS3 sequence was used as the “long sequence”.
Cell Culture
In vitro cell experiments were performed using CSCC cell lines SiHa and HeLa (ATCC, USA). SiHa and HeLa are both HPV-positive cell lines, which are consistent with the status of patients included in this study (all the patients were HPV-positive). Eagle’s Minimum Essential Medium (10% FBS) was used as the cell culture medium. Cells were cultured in a 5% CO2 incubator at 37 °C.
Lipofectamine 2000-Mediated Transient Transfections
Negative control miRNA and miR-233 mimic were obtained from Sangon (Shanghai, China). MAGI2-AS3 and EPB41L3 expression vectors were constructed using pcDNA3 vector by GenePharma (Shanghai, China). Cells (106) were transfected with 35 nM negative control miRNA (negative control group, NC), or 35 nM miR-233 mimic, or 10 nM empty pcDNA3 vector (NC), or MAGI2-AS3/EPB41L3 expression vector using lipofectamine 2000 (Sangon). Cells were collected at 24 h post-transfections to perform the following experiments.
RNA Extractions
Total RNAs were extracted from tissues (0.015 g grounded in liquid nitrogen) or 106 cells with 1 mL Trizol (Invitrogen, USA) to perform. All operations were performed following the protocol from Invitrogen, except that miRNAs were harvested using 80% ethanol instead of 75% ethanol to precipitate and wash RNA samples.
qPCR
All RNA samples were subjected to DNase I digestion to remove genomic DNAs. The iScript cDNA Synthesis Kit (Bio-Rad, USA) was used to perform reverse transcriptions. All qPCR reactions were prepared using the BlazeTaq™ SYBR Green qPCR Mix (Genecopoeia, Guangzhou, China). The expression levels of MAGI2-AS3 and EPB41L3 mRNA were detected with GAPDH as endogenous control. miRNA reverse transcriptions were performed using qScript microRNA cDNA Synthesis Kit (Quantabio, USA). The miScript SYBR Green PCR Kit (QIAGEN) was used to prepare all qPCR mixtures. The expression level of miR-223 was determined with U6 as endogenous control. Three replicate reactions were set for each experiment. The expression levels of MAGI2-AS3, EPB41L3 mRNA and miR-233 were normalized to endogenous controls using 2−ΔΔCT method.
Luciferase Activity Assay
Before transfection, cells were well cultivated in a 48-well plate, and then were co-transfected with MAGI2-AS3 luciferase vector plasmid and miR-233 mimics or control mimics. The Firefly and Renilla luciferase activity was detected by Luc-Pair™ Duo-Luciferase Assay Kit 2.0 (GeneCopoeia, USA) at 48 h after transfection. The proportion of Firefly luciferase to the Renilla luciferase was counted as the relative luciferase activity.
Western Blot
RIPA solution (Sangon) was mixed with CSCC cells (1 mL per 106 cells) to extract total proteins. A BCA kit (Sangon) was used to measure the concentrations of protein samples. Protein samples were then incubated in boiled water for 5 min to denature proteins. To separate proteins, 12% SDS-PAGE gel was used to carry out electrophoresis. After electrophoresis, proteins were transferred to PVDF membranes, followed by blocking at 23°C for 1 h in PBS containing 5% non-fat milk. The first blotting was performed using GAPDH (1:1300, ab37168, Abcam) and EPB41L3 (1:1300, ab154071, Abcam) rabbit primary antibodies (4 °C for 12 h). The second blotting was performed using HRP goat anti-rabbit (IgG) antibody (1:1000; ab6721; Abcam, 23 °C for 2 h). RapidStep™ ECL detection reagent (EMD Millipore) was dropped onto the membranes to develop signals. All gray values were normalized using Image J v1.48 software.
In vitro Cell Migration and Invasion Assay
Eagle’s Minimum Essential Medium (1% FBS) was mixed with cells with a ratio of 1 mL per 4×104 cells. The upper Transwell chamber was added with 0.1 mL mixture containing 4×103 cells, and the lower chamber was filled with mixture of 80% Eagle’s Minimum Essential Medium and 20% FBS. Matrigel (Millipore) was used to coat the membranes before invasion assay. Cells were cultivated at 37 °C for 12 h. After that, 0.5% crystal violet (Sigma-Aldrich) was used to stain the membranes and cells were counted using a light microscope.
Data Analysis
Mean values of the 3 biological replicates of each experiment were calculated and used for all data analyses. Paired t test and ANOVA (one-way) in combination with Tukey’s test were used to explore differences between two types of tissues or among different cell transfection groups. Correlations were analyzed by linear regression. P < 0.05 was considered as statistically significant.
Results
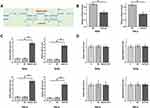
MAGI2-AS3 and EPB41L3 Were Positively Correlated in CSCC
The expression levels of MAGI2-AS3 and EPB41L3 in two types of tissue were measured by qPCR. Data were compared by paired t test. It was observed that the expression of MAGI2-AS3 (Figure 1A) and EPB41L3 (Figure 1B) were both downregulated in CSCC tissues compared to that in non-tumor tissues (p < 0.05). Correlations between MAGI2-AS3 and EPB41L3 were analyzed by linear regression. It was observed that the expression of MAGI2-AS3 and EPB41L3 were significantly and positively correlated in CSCC tissues (Figure 1C), but not in non-tumor tissues (Figure 1D). To further investigate the clinicopathological role of MAGI2-AS3 expression in CSCC patients, we divided 60 patients into low MAGI2-AS3 expression group (n = 35) and high MAGI2-AS3 expression group (n = 25). As presented in Table 1, the expression levels of MAGI2-AS3 in CSCC tissues were positively correlated with tumor diameter, HPV types and stages. These results indicated that the abnormal expression of MAGI2-AS3 played a crucial role in the progression of CSCC.
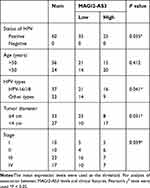 |
Table 1 Association with MAGI2-AS3 and the Clinical Pathological Characteristics of CSCC Patients |
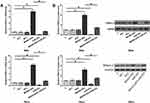
MiR-233 May Bind with MAGI2-AS3 but Did Not Regulate Its Expression
MiR-233 can directly target EPB41L3. We predicted the interaction between miR-233 and MAGI2-AS3 by IntaRNA. It can be observed that miR-233 may bind with MAGI2-AS3 at position 603 to 626 (Figure 2A). We constructed luciferase MAGI2-AS3 luciferase vector plasmid and miR-233 mimics or control mimics in both SiHa and HeLa cells. As shown in Figure 2B, the luciferase activity in these cells transfected with MAGI2-AS3 was reduced by miR-233 mimic treatment, compared with that in the control groups (Figure 2B). To further detect their interactions, miR-233 mimic and EPB41L3 expression vector were transfection into SiHa and HeLa cells. Compared to the C and NC group, expression levels of miR-233 and EPB41L3 were both significantly increased at 24 h post-transfections (Figure 2C, p < 0.05). However, overexpression of miR-233 did not affect the expression of MAGI2-AS3. Moreover, overexpression of MAGI2-AS3 also did not affect the expression of miR-233 (Figure 2D).
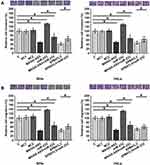
MAGI2-AS3 Upregulated EPB41L3 Through miR-233
The effects of overexpression of MAGI2-AS3 and miR-233 on the expression of EPB41L3 were assessed by Western blot and qPCR. Comparing to C and NC (NC1, empty pcDNA3 transfection; NC2, negative control miRNA transfection) groups, overexpression of MAGI2-AS3 upregulated, while overexpression of miR-233 downregulated expression of EPB41L3 at both mRNA (Figure 3A) and protein (Figure 3B) levels (p < 0.05). In addition, overexpression of miR-233 reduced the effects of overexpressing MAGI2-AS3 (p < 0.05).
MAGI2-AS3 Inhibited SiHa and HeLa Cell Invasion and Migration Through EPB41L3 and miR-233
After transfections, cell invasion and migration were evaluated by transwell assays. Comparing to C and NC (NC1, empty pcDNA3 transfection; NC2, negative control miRNA transfection) groups, overexpression of MAGI2-AS3 and EPB41L3 led to inhibited cancer cell invasion (Figure 4A) and migration (Figure 4B), while overexpression of miR-233 attenuated the effects of overexpressing MAGI2-AS3 or EPB41L3 (p < 0.05).
Discussion
Cervical squamous cell carcinoma is the most prevalent gynecologic tumor caused by HPV infection. Even with extensive studies, the mechanisms underlying the tumor initiation and progression remain largely unknown. Patients are confronted with limited clinical application including surgical operations and chemotherapies, which are usually lagged behind, as well as effective biomarkers. This study was the first to show the role of MAGI2-AS3 in CSCC as a bona fide tumor suppressor and also a biomarker for clinical diagnosis. We found that MAGI2-AS3 may sponge miR-233 to upregulate EPB41L3, thereby inhibiting the invasion and migration of CSCC cells. The tumor suppressive function of MAGI2-AS3 has been studied in several types of cancer.10–12 In breast cancer, MAGI2-AS3 targets the Fas/FasL signaling pathway to suppress the proliferation of cancer cells.10 In bladder cancer, MAGI2-AS3 inhibits cancer progression by regulating the expression of CCDC19.11 In hepatocellular carcinoma, MAGI2-AS3 inhibits both cell migration and proliferation by interacting with miR-374b-5p and SMG1.12 To the best of our knowledge, this study is the first to report the downregulation of MAGI2-AS3 in CSCC. Our results also showed inhibited CSCC cell invasion and migration after the overexpression of MAGI2-AS3. Our data suggest the tumor suppressive role of MAGI2-AS3 in CSCC.
MiRNAs target gene expression by direct cleavage or translation inhibition.13,14 For the regulation of lncRNA expression, cleavage might be the only mechanism.15 Our study showed that miR-233 can bind with MAGI2-AS3. However, overexpression experiments showed that overexpression of MAGI2-AS3 has no significant effects on the expression of miR-233. Recent studies have revealed that lncRNAs can sponge miRNAs to inhibit their functions.16,17 It has been reported that MAGI2-AS3 can sponge miR-15b-5p to participate in bladder cancer.11 In the present study, we observed that MAGI2-AS3 may sponge miR-233 to upregulate EPB41L3. It was reported that miR-233 can directly target tumor suppressive EPB41L3 in gastric cancer.8 Our results showed that miR-233 may also target EPB41L3 in CSCC. Therefore, our study identified a novel MAGI2-AS3/miR-233/EPB41L3 pathway in CSCC. However, miR-233 can also target other genes, such as Irak1.18 Our future studies will investigate the involvement of other targets of miR-233 in CSCC.
In conclusion, MAGI2-AS3 plays a tumor suppressive role in CSCC by sponging miR-233 to upregulate EPB41L3, thereby inhibiting the invasion and migration of CSCC cells.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Intaraphet S, Kasatpibal N, Siriaunkgul S, et al. Prognostic impact of histology in patients with cervical squamous cell carcinoma, adenocarcinoma and small cell neuroendocrine carcinoma. Asian Pac J Cancer Prev. 2013;14(9):5355–5360. doi:10.7314/APJCP.2013.14.9.5355
2. Wang SS, Sherman ME, Hildesheim A, Lacey JV
3. Castellsague X, Diaz M, de Sanjose S, et al. Worldwide human papillomavirus etiology of cervical adenocarcinoma and its cofactors: implications for screening and prevention. J Natl Cancer Inst. 2006;98(5):303–315. doi:10.1093/jnci/djj067
4. Trattner M, Graf AH, Lax S, et al. Prognostic factors in surgically treated stage ib-iib cervical carcinomas with special emphasis on the importance of tumor volume. Gynecol Oncol. 2001;82(1):11–16. doi:10.1006/gyno.2001.6252
5. Hosaka M, Watari H, Mitamura T, et al. Survival and prognosticators of node-positive cervical cancer patients treated with radical hysterectomy and systematic lymphadenectomy. Int J Clin Oncol. 2011;16(1):33–38. doi:10.1007/s10147-010-0123-0
6. Bernkopf DB, Williams ED. Potential role of EPB41L3 (protein 4.1B/Dal-1) as a target for treatment of advanced prostate cancer. Expert Opin Ther Targets. 2008;12(7):845–853. doi:10.1517/14728222.12.7.845
7. Dafou D, Grun B, Sinclair J, et al. Microcell-mediated chromosome transfer identifies EPB41L3 as a functional suppressor of epithelial ovarian cancers. Neoplasia. 2010;12(7):579–589. doi:10.1593/neo.10340
8. Li X, Zhang Y, Zhang H, et al. miRNA-223 promotes gastric cancer invasion and metastasis by targeting tumor suppressor EPB41L3. Mol Cancer Res. 2011;9(7):824–833. doi:10.1158/1541-7786.MCR-10-0529
9. Zhu L, Yang N, Chen J, et al. LINC00052 upregulates EPB41L3 to inhibit migration and invasion of hepatocellular carcinoma by binding miR-452-5p. Oncotarget. 2017;8(38):63724–63737. doi:10.18632/oncotarget.18892
10. Yang Y, Yang H, Xu M, et al. Long non-coding RNA (lncRNA) MAGI2-AS3 inhibits breast cancer cell growth by targeting the Fas/FasL signalling pathway. Hum Cell. 2018;31(3):232–241. doi:10.1007/s13577-018-0206-1
11. Wang XM, Liu Y, Fan YX, et al. LncRNA PTCSC3 affects drug resistance of anaplastic thyroid cancer through STAT3/INO80 pathway. Cancer Biol Ther. 2018;19(7):590–597. doi:10.1080/15384047.2018.1449610
12. Yin Z, Ma T, Yan J, et al. LncRNA MAGI2-AS3 inhibits hepatocellular carcinoma cell proliferation and migration by targeting the miR-374b-5p/SMG1 signaling pathway. J Cell Physiol. 2019;234(10):18825–18836. doi:10.1002/jcp.28521
13. Ambros V. The functions of animal microRNAs. Nature. 2004;431(7006):350–355. doi:10.1038/nature02871
14. Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi:10.1016/j.cell.2009.01.002
15. Jeggari A, Marks DS, Larsson E. miRcode: a map of putative microRNA target sites in the long non-coding transcriptome. Bioinformatics. 2012;28(15):2062–2063. doi:10.1093/bioinformatics/bts344
16. Liang WC, Fu WM, Wong CW, et al. The lncRNA H19 promotes epithelial to mesenchymal transition by functioning as miRNA sponges in colorectal cancer. Oncotarget. 2015;6(26):22513–22525. doi:10.18632/oncotarget.4154
17. Wu XS, Wang F, Li HF, et al. LncRNA-PAGBC acts as a microRNA sponge and promotes gallbladder tumorigenesis. EMBO Rep. 2017;18(10):1837–1853. doi:10.15252/embr.201744147
18. Wang H, Hao P, Zhang H, Xu C, Zhao J. MicroRNA-223 inhibits lipopolysaccharide-induced inflammatory response by directly targeting Irak1 in the nucleus pulposus cells of intervertebral disc. IUBMB Life. 2018;70(6):479–490. doi:10.1002/iub.1747
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.