Back to Journals » OncoTargets and Therapy » Volume 12
LncRNA HOTAIRM1/HOXA1 Axis Promotes Cell Proliferation, Migration And Invasion In Endometrial Cancer
Authors Li X, Pang L , Yang Z, Liu J, Li W, Wang D
Received 8 July 2019
Accepted for publication 12 October 2019
Published 13 December 2019 Volume 2019:12 Pages 10997—11015
DOI https://doi.org/10.2147/OTT.S222334
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Sanjeev K. Srivastava
Xianli Li,1 Li Pang,1 Zhuo Yang,1 Jing Liu,2 Weishan Li,2 Danbo Wang2
1Department of Obstetrics and Gynecology, Shengjing Hospital of China Medical University, Shenyang, Liaoning, People’s Republic of China; 2Department of Gynecology, Cancer Hospital of China Medical University, Liaoning Cancer Hospital and Institute, Shenyang, Liaoning, People’s Republic of China
Correspondence: Danbo Wang
Department of Gynecology, Cancer Hospital of China Medical University, Liaoning Cancer Hospital and Institute, 44. Xiaoheyan Road, Dadong District, Shenyang City, Liaoning Province 110042, People’s Republic of China
Tel +86-24-31916623
Email [email protected]
Background: Long non-coding RNA (lncRNA) microarray screening previously identified that HOXA transcript antisense RNA myeloid-specific 1 (HOTAIRM1) was significantly upregulated in type I endometrial cancer (EC). The present study aimed to determine the potential role of HOTAIRM1 and its sense transcript HOXA1 in the development and progression of type I EC.
Methods: We detected the expression levels of HOTAIRM1 and HOXA1 in type I EC tissues by quantitative real-time polymerase chain reaction (qRT-PCR) and Western blotting and analyzed associated clinical data. Gain- or loss-of-function experiments were used to investigate the biological function of HOTAIRM1 and HOXA1 in type I EC, both in vitro and in vivo.
Results: The expression levels of HOTAIRM1 and HOXA1 were significantly upregulated in type I EC tissues. Furthermore, the expression of HOTAIRM1 and HOXA1 were both significantly correlated with International Federation of Gynecology and Obstetrics (FIGO) stage and lymph node metastasis. The expression of HOTAIRM1 was significantly correlated with that of HOXA1. Knockdown of HOTAIRM1 significantly inhibited cell proliferation, migration, invasion and epithelial–mesenchymal transition (EMT) in vitro, while the over-expression of HOTAIRM1 led to the opposite effects. Moreover, we identified that HOTAIRM1 acts as a regulator for the expression of the HOXA1 gene in type I EC cells. As an oncogene, HOXA1 silencing also caused suppressive effects on tumors by inhibiting cell proliferation, migration and invasion. In addition, we also confirmed the role of HOTAIRM1 and HOXA1 in promoting tumor growth in vivo.
Conclusion: Our findings are the first to identify that HOTAIRM1 functions as an oncogene to promote cell proliferation, migration and invasion by regulating HOXA1 in type I EC. Therefore, the HOTAIRM1/HOXA1 axis is a novel potential prognostic biomarker and new potential therapeutic target for type I EC.
Keywords: long non-coding RNA, HOTAIRM1, HOXA1, type I endometrial cancer
Introduction
Endometrial cancer (EC) is one of the three most common malignancies of the female reproductive system. Over recent years, the incidence of EC has increased due to the increasing incidence of obesity and metabolic diseases, and the age of onset shows a trend for younger patients.1 In Western countries, EC has the highest incidence of female reproductive system malignancies.1 In 1983, in accordance with pathogenesis and biological behavior characteristics, Bokhman divided EC into two types: type I (estrogen dependent) and type II (non-estrogen dependent).2 Over recent years, it becomes more valuable to study the molecular pathogenesis of EC with more and more attention paid to its molecular classification. Although the incidence of type I EC accounts for approximately 80% of EC, its pathogenesis remains unclear. The identification of specific molecular markers that provide new ideas for the diagnosis and treatment of type I EC is highly desired.
Long non-coding RNA (lncRNA) is another widely concerned molecular marker after microRNA (miRNA) over recent years. However, our relative understanding of the role of lncRNA is far less extensive than for miRNA. Studies have shown that lncRNA can regulate gene expression in cells at epigenetic, transcriptional and posttranscriptional levels, and participate in a range of important regulatory processes such as X chromosome silencing, genomic imprinting, chromatin modification, transcriptional activation, posttranscriptional interference, the regulation of protein activity, and the intracellular transport of nucleic acids.3–6 Studies have also found that the differential expression or abnormal function of lncRNA is closely related to the occurrence and development of tumors. Some lncRNAs have been proven to regulate mRNA, miRNA, corresponding target genes and proteins via specific signal transduction pathways, and to play roles in the tumorigenesis and progression, cell differentiation, the cell cycle and apoptosis and other regulatory processes. Collectively, accumulating evidence suggests that lncRNA may represent a new molecular marker for the diagnosis, prognosis, metastasis of tumors, and therefore provide a new target for tumor therapy.7,8 Some research has indicated the role of lncRNAs in EC. For example, lncRNA DLEU1 combines with mTOR and then increases the expression of PI3K/AKT/mTOR pathway to promote EC tumorigenesis and progression.9 The high expression of lncRNA H19 in EC may regulate the expression level of its target gene HOXA10 by targeting miR-612, thus promoting cell proliferation to play a role in the development of EC.10 LncRNA ABHD11-AS1 can function as an oncogene to promote cell proliferation and invasion in EC by positively targeting cyclin D1.11
Our research group previously used the Arraystar Human LncRNA V3.0 microarray to screen the lncRNA expression profile of type I EC tissue and normal endometrial tissue as a control. We found that HOXA transcription antisense RNA myeloid-specific 1 (HOTAIRM1) is a significantly upregulated lncRNA in type I EC tissue. HOTAIRM1 was first discovered in the myeloid cell system by Zhang et al and is located on human chromosome 7p15.2.12 HOTAIRM1 regulates cell cycle progression during myeloid maturation in NB4 human promyelocytic leukemia cells and is significantly upregulated in acute myeloid leukemia.13,14 More recent studies have reported that the expression of HOTAIRM1 is significantly increased in pancreatic ductal adenocarcinoma, breast cancer and glioma, and also plays a role in promoting cancer.15–19 However, HOTAIRM1 is expressed at low levels in colorectal cancer, head and neck tumors, hepatocellular carcinoma and gastric cancer, and therefore plays the role of a tumor suppressor gene in such cases.20–23 This dual effect may be due to the differential roles of HOTAIRM1 in the pathogenesis of different types of malignancies.
Antisense lncRNA is transcribed from the opposite chains of protein or non-protein coding genes, and is closely related to the tumorigenesis and progression.24 LncRNA HOTAIRM1, located at the 5ʹ end of homeobox A (HOXA) gene cluster, is transcribed antisense to the HOXA genes and originates from the same CpG island that embeds the start site of HOXA1.12 Previous studies found that HOTAIRM1 could upregulate the expression of HOXA1 in glioblastoma multiforme and in myeloid-derived suppressor cells of lung cancer.19,25 HOXA1 is a member of the family of homeobox genes (HOX). The HOX genes are divided into four clusters: HOXA, HOXB, HOXC and HOXD. HOXA is located at 7p15.2 and the abnormal expression of HOXA will lead to the formation of abnormal morphological structure, thus leading to malignant transformation of cells into tumors. HOXA1 has been reported to be highly upregulated in a variety of tumors, such as breast cancer, oral squamous cell carcinoma, gastric cancer, and hepatocellular carcinoma, and is associated with poor clinical prognosis.26–29 HOXA1 is a key factor in the process of tumorigenesis and progression; this factor can regulate cell cycle, promote epithelial–mesenchymal transition (EMT), and can also improve the capacity for tumor cell proliferation, migration and invasion.28–30 However, as yet, the role of HOTAIRM1 and HOXA1 in EC and their mutual regulatory relationship have not been reported.
Therefore, based on our earlier microarray screening results, the present study aimed to investigate the expression and correlation of lncRNA HOTAIRM1 and HOXA1 gene in type I EC tissues, and study the effects of lncRNA HOTAIRM1 and its sense gene HOXA1 on the proliferation, migration and invasion of type I EC in vitro and in vivo.
Materials And Methods
Tissue Sample Collection
Tissues were acquired from 50 patients with type I EC undergoing surgery (the experimental group). During the same period, normal endometrial tissue was obtained from 50 patients undergoing hysterectomy for myoma of the uterus to form a control group. Mean patient age in the two groups was 50.62 ± 9.996 years and 47.60 ± 4.611 years; there was no statistically significant difference in this respect (P > 0.05). The patients in the experimental group did not receive chemotherapy, radiotherapy or biological therapy before surgery, and no other surgical, endocrine, immune and metabolic diseases were identified in the two groups. No hormone medication was given within the 3 months prior to surgery.
Fresh tissue samples, obtained under sterile conditions, were immediately placed in liquid nitrogen and frozen at −80ºC for long-term preservation. Meanwhile, we collected the clinical and pathological data of all recruited patients, including patient age, FIGO stage, tumor pathological grade, myometrial invasion and lymph node metastasis. All cases were confirmed by postoperative pathological diagnosis. All patients signed informed consent and compliance with the Declaration of Helsinki. The use of these specimens for research was approved by the approving ethics committee of Shengjing Hospital of China Medical University (Reference number: 2015PS103K).
Cell Lines And Cell Cultures
Two human endometrioid adenocarcinoma cell lines, Ishikawa (highly differentiated) and HEC-1A (medium differentiated), were purchased from the Shanghai Institute of Biochemistry and Cell Biology of the Chinese Academy of Sciences (Shanghai, China). The Ishikawa cell line and HEC-1A cell line were cultured in RPMI-1640 medium (Hyclone, Logan, UT, USA) and DMEM high glucose medium (Hyclone) respectively, supplemented with 10% fetal bovine serum (FBS, Bioind, Israel), 100 IU/mL penicillin and 100 μg/mL streptomycin (Gibco, Carlsbad, CA, USA). All cells were cultured in a sterile incubator at 37°C with 5% CO2.
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
Total RNA was extracted from tissues and cells using Trizol reagent (9108, Takara Bio Inc, Japan). Total RNA was then reversed transcribed into cDNA according to the instructions of the PrimeScript RT reagent kit (RR047A, Takara). qRT-PCR reactions were performed on a LightCycler 480 (Roche) in accordance with the SYBR Premix Ex Taq kit instructions (RR420A, Takara) and GAPDH was used as an internal reference gene. All operations were carried out under RNase-free conditions, and each sample was assayed in triplicate. The relative expression levels of lncRNA HOTAIRM1 and HOXA1 mRNA were calculated using the 2−ΔΔCt method. The primer sequences are shown in Table 1.
 |
Table 1 Primer Sequences For qRT-PCR |
Short Hairpin RNA (shRNA) Transfection And The Establishment Of Stable Cell Lines
The sequences of HOTAIRM1 shRNA, HOXA1 shRNA and negative control shRNA (GenePharma, Shanghai, China) are shown in Table 2. Ishikawa and HEC-1A cells were harvested from the logarithmic growth phase and seeded into six-well plates (Corning Life Sciences, Corning, NY, USA) until they were 50–70% confluent. We then transfected these cells using Opti-MEM I (Gibco) and Lipofectamine 2000 reagent (Life Technologies Corporation, Carlsbad, CA, USA). After 6 hrs, the complete medium was replaced. Stably transfected cells were then selected by G418 screening. Then, the silencing efficiency was detected by qRT-PCR.
 |
Table 2 shRNA Sequences |
Lentivirus Infection
Lentivirus containing the cDNA sequence of HOTAIRM1 (GV367-HOTAIRM1) and negative control virus (GV367-NC) were purchased from Genechem (Shanghai, China). We ensured that the negative control virus did not contain sequences that could affect the expression of any gene. Transfection was performed according to the manufacturer’s instructions. When the degree of cell fusion reached 60% in the six-well plate (Corning), the culture medium containing 10% fetal bovine serum was replaced, and polybrene (Genechem) was added at the final concentration of 5 μg/mL. The multiplicity of infection (MOI) for Ishikawa cell line infection was 50 while that for HEC-1A cell line infection was 1. After 8 hrs of transfection, the fresh complete medium was replaced. Green fluorescent protein was observed by fluorescence microscope for 72 hrs after transfection to evaluate transfection efficiency. The overexpression efficiency was determined by qRT-PCR.
To determine the overexpression and silencing effects of HOTAIRM1 on EC, we divided cells into five groups: Control group (without any shRNA or virus transfection, named “mock control”); GV367-NC group (transfection with negative control virus); GV367-HOTAIRM1 group; shRNA-NC group (transfection with negative control shRNA) and sh-HOTAIRM1 group. To determine the silencing effect of HOXA1 on EC, cells were divided into three groups: Control group; shRNA-NC group and sh-HOXA1 group.
Western Blot Assay
Total protein was extracted from tissues and cells using radioimmunoprecipitation assay buffer (RIPA buffer, Beyotime, China) with protease inhibitors (Beyotime). Protein concentration was then measured using a BCA Protein Assay Kit (Beyotime). Total protein was separated by 10% SDS-PAGE (Beyotime) and transferred to a PVDF membrane (Millipore, Billerica, MA, USA) by wet transfer. After blocking for 2 hrs with TBST solution containing 5% skimmed milk, the membranes were incubated with specific primary antibodies overnight at 4°C. The following morning, the membranes were incubated with a corresponding secondary antibody for 2 hrs at room temperature. Finally, the intensity of the protein bands was measured with C300 imaging system (Azure Biosystem, USA) by enhanced chemiluminescence (ECL, Thermo Scientific, USA). The relative-integrated density was analyzed using ImageJ software based on GAPDH as an internal control. The primary antibodies included HOXA1 (1:1000, Abcam, Eugene, USA), E-cadherin (1:1000, Cell Signaling Technology, CA, USA), N-cadherin (1:1000, Cell Signaling Technology), CyclinD1 (1:1000, Cell Signaling Technology) and GAPDH (1:10,000, Proteintech, Chicago, IL, USA). We also used an HRP-conjugated secondary antibody (1:2000, ZSGB-Bio Origene, Beijing, China). All assays were performed in triplicate.
Cell Proliferation Assay
The Cell Counting Kit-8 (CCK-8, Dojindo, Japan) was used to determine cell proliferation. For each group, cells were seeded into 96-well plates (Corning). Cell-free medium was used as a blank control group. After 24, 48, 72 and 96 hrs of culture, we added 10 μL CCK-8 reagent to each well followed by incubation for 3 hrs at 37°C. The absorbance of each well was then detected at a wavelength of 450 nm with an ELISA plate reader (Bio-Rad). The cell proliferation rate was then calculated as the ratio of the OD values. After calculating the relative proliferation rate of each groups at the four time points, we plotted a cell growth curve. All assays were performed in triplicate.
Transwell Assays
Transwell Migration Assay
Transwell plates (24-well chambers with 8 μm pore size; Corning) were used. For each group, cells (5 × 104) were resuspended in 200 μL of serum-free medium and seeded into the upper chamber of transwell plates. Then, 600 μL of 20% FBS medium was added to the lower chamber as a chemoattractant. After incubation for 18 hrs at 37ºC with 5% CO2, cells could be seen in the lower chamber. The upper chambers were removed and the cells on the top side of the chamber were wiped with cotton swabs. Migrated cells were then fixed with 4% paraformaldehyde for 20 mins, stained with 0.1% crystal violet for 30 mins and excess dye solution was washed off. Five randomly fields were then counted under an inverted light microscope and photographs were acquired (magnification, ×200).
Transwell Invasion Assay
Matrigel (BD, USA) was dissolved overnight in a 4°C refrigerator, diluted with precooled serum-free medium at a ratio of 1:6, coated on the bottom of an upper chamber in transwell plates (8 μm pores; Corning) for 4 hrs at 37°C with 5% CO2, and the transwell migration assay steps (described above) were performed after the Matrigel was fully solidified. Cells could be seen in the lower chamber after incubation for 26 hrs.
Cell Cycle Assay
For each group, cells were collected by routine digestion, and adjusted to a concentration of 1×106/mL. Then, 1 mL of single-cell suspension was fixed overnight with precooled 70% absolute ethyl alcohol at 4°C. The fixed cells were then mixed with 100 μL of RNase A reagent (Keygen Biotech, Nanjing, China) and incubated for 30 mins in a water bath at 37°C. The cells were then stained in 400 μL of propidium iodide reagent (Keygen Biotech) for another 30 mins in the dark. DNA content was then assayed at 488 nm using a FACSCalibur flow cytometer (Becton Dickinson). Finally, we analyzed the relative proportion of cells in G0/G1, S and G2/M phases using ModFit LT software (Verity Software House). All assays were performed in triplicate.
Tumor Xenograft Implantation In Nude Mice
Female BALB/c nude mice (4 weeks of age) were purchased from Beijing Huafukang Bioscience Co. Inc. (Beijing, China) and fed in specific pathogen-free (SPF) conditions. To explore the role of HOTAIRM1, the mice were randomly divided into five groups (five mice in each group): Control group, GV367-NC group, GV367-HOTAIRM1 group, shRNA-NC group and sh-HOTAIRM1 group. For subcutaneous implantation, 2 × 106 cells were injected subcutaneously into the right back of the mice in each group, and all groups were marked for identification purposes. To determine the effect of HOXA1, the mice were randomly divided into three groups (five mice in each group): Control group, shRNA-NC group and sh-HOXA1 group, which were all injected subcutaneously with 3 × 106 cells. The length and width of tumors in experimental mice were measured with calipers every 4 days after tumor formation. Tumor volume (V) was calculated using the following formula: V= (length × width2)/2. Forty days after implantation, the mice were sacrificed and the transplanted tumors were completely isolated, photographed and weighed. The research was approved by the approving ethics committee of Shengjing Hospital of China Medical University (Reference number: 2015PS103K). All experiments were performed following Shengjing Hospital’s and national guidelines and regulations.
Statistical Analysis
Data were analyzed by SPSS version 20.0 statistical software (SPSS Inc. Chicago, USA). All data are expressed as mean ± standard deviation (SD). The Student’s t-test was used to compare differences between the two groups. One-way analysis of variance (ANOVA) was used for multiple comparisons. The chi-square test was used to compare HOTAIRM1/HOXA1 expression and clinicopathological features. Pearson’s correlation analysis was used for correlation analysis. Differences were considered to be significant if P < 0.05 (two-tailed).
Results
Expression Levels Of lncRNA HOTAIRM1 And HOXA1 Gene In Type I EC Tissues And Correlation Analysis
Expression Levels Of HOTAIRM1 And HOXA1 In Type I EC Tissues
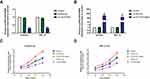
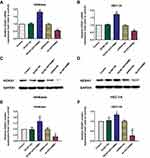
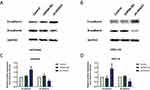
In type I EC tissues, qRT-PCR showed that the expression of HOTAIRM1 (14.69 ± 9.95) was significantly higher than that in normal endometrium tissues (2.36 ± 2.78, P < 0.01; Figure 1A), and that the expression of HOXA1 mRNA (9.00 ± 7.02) was also significantly higher than that in normal endometrium tissues (2.04 ± 2.23, P < 0.01; Figure 1B). Western blotting further revealed that the expression of HOXA1 protein in type I EC tissues (1.08 ± 0.52) was significantly higher than that in normal endometrium tissues (0.61 ± 0.35, P < 0.01; Figure 1C and D).
Relationship Between The Expression Of HOTAIRM1 And HOXA1 And Clinicopathological Characteristics In Type I EC
In accordance with the literature method,31 we used the median HOTAIRM1 expression level (Y = 12.474) and divided 50 type I EC tissues into a high-HOTAIRM1 expression group (n = 25) and a low-HOTAIRM1 expression group (n = 25, Figure 1E). Using the same method, 50 type I EC tissues were divided into high- and low-HOXA1 expression groups (25 cases per group) using the median HOXA1 mRNA expression level (Y = 7.299, Figure 1F). The HOTAIRM1 and HOXA1 mRNA expression levels were both significantly correlated with International Federation of Gynecology and Obstetrics (FIGO) stage and lymph node metastasis (P < 0.05), but were not correlated with patient age, differentiation and myometrial invasion (P > 0.05; Table 3).
 |
Table 3 Relationship Between The Expression Of HOTAIRM1/HOXA1 And The Clinicopathological Variables Of Patients |
Correlation Between HOTAIRM1 And Its Sense Transcript HOXA1
Statistical analysis further showed that there was a significant positive correlation between HOTAIRM1 and HOXA1 mRNA expression in type I EC tissues (r = 0.746, P < 0.01; Figure 1G). There was also significant positive correlation between HOTAIRM1 and HOXA1 protein expression (r = 0.530, P < 0.01; Figure 1H).
The Influence Of HOTAIRM1 On The Biological Behavior Of EC Cells Lines In Vitro
Knockdown And Overexpression Efficiency Of HOTAIRM1 In Type I EC Cells
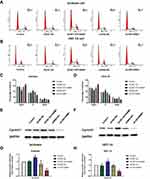
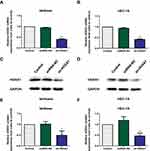
To investigate the role of HOTAIRM1 in the progression of EC, we first established stable transfected cell lines for the knockdown and overexpression of HOTAIRM1 in Ishikawa and HEC-1A cells. The transfection efficiency was detected by qRT-PCR. Compared with the shRNA-NC group and the Control group, the expression of HOTAIRM1 in the sh-HOTAIRM1 group was significantly decreased, while the expression of HOTAIRM1 in the GV367-HOTAIRM1 group was significantly increased compared with the GV367-NC group and the Control group (P < 0.01; Figure 2A and B).
Influence Of HOTAIRM1 On The Proliferation Of EC Cell Lines
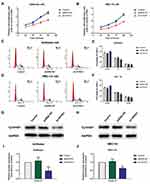
The CCK8 assay was used to verify the effect of HOTAIRM1 on the proliferation of Ishikawa and HEC-1A cells. The proliferation rate was significantly higher in the GV367-HOTAIRM1 group than that in the GV367-NC and Control groups (P < 0.05), and was significantly lower in the sh-HOTAIRM1 group than that in the shRNA-NC and Control groups (P < 0.05; Figure 2C and D).
Influence Of HOTAIRM1 On The Cell Cycle Of EC Cell Lines
Flow cytometry was used to analyze the effects of HOTAIRM1 on the cell cycle distribution of Ishikawa and HEC-1A cells. Data showed that compared with the shRNA-NC group and the Control group, the proportion of cells in G0/G1 phase in the sh-HOTAIRM1 group was significantly increased, while the proportion of cells in S phase was significantly decreased (P < 0.05). Compared with the GV367-NC group and the Control group, the proportion of cells in G0/G1 phase in the GV367-HOTAIRM1 group was significantly decreased, while the proportion of cells in S phase was significantly increased (P < 0.05). No significant difference was observed in G2/M phase (P > 0.05; Figure 3A–D).
CyclinD1 is an important regulatory factor of the cell cycle. The increased expression of CyclinD1 can promote the evolution from G1 phase to S phase and promote cell proliferation; the abnormal expression of CyclinD1 is closely related to the occurrence and development of various tumors.32 Western blotting further showed that compared with the shRNA-NC group and the Control group, the expression levels of CyclinD1 in the sh-HOTAIRM1 group were significantly reduced in Ishikawa and HEC-1A cells and that the expression levels of CyclinD1 in the GV367-HOTAIRM1 group were significantly increased compared with the GV367-NC group and the Control group (P < 0.05; Figure 3E–H).
Influence Of HOTAIRM1 On Cell Migration And Invasion In EC Cell Lines
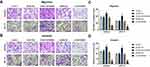
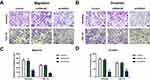
Transwell assays were used to determine the effect of HOTAIRM1 on the migration and invasion ability of Ishikawa and HEC-1A cells. Data showed that compared with the GV367-NC group and the Control group, the numbers of migrating and invading transmembrane cells in the GV367-HOTAIRM1 group were both significantly increased (P < 0.05). Compared with the shRNA-NC group and the Control group, the numbers of migrating and invading transmembrane cells in the sh-HOTAIRM1 group were significantly decreased (P < 0.05; Figure 4).
Influence Of HOTAIRM1 On EMT In EC Cell Lines In Vitro
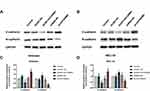
Epithelium-mesenchymal transition (EMT) is closely related to tumor invasion and metastasis. Western blotting was used to analyze the expression of E-cadherin and N-cadherin, which are both molecular markers of EMT, with the expression of HOTAIRM1 changed in Ishikawa and HEC-1A cells. Data showed that the expression level of N-cadherin was significantly decreased and the expression level of E-cadherin was significantly increased in the sh-HOTAIRM1 group compared with the shRNA-NC group and the Control group (P < 0.05). Furthermore, the expression level of N-cadherin was significantly increased, while E-cadherin expression level was significantly decreased in the GV367-HOTAIRM1 group compared with the GV367-NC group and the Control group (P < 0.05; Figure 5).
LncRNA HOTAIRM1 Regulates The Expression Of HOXA1 In EC Cell Lines
The effects of HOTAIRM1 knockdown and overexpression on HOXA1 gene expression were analyzed in Ishikawa and HEC-1A cell lines. qRT-PCR showed that the expression level of HOXA1 mRNA in the GV367-HOTAIRM1 group was significantly increased compared with the GV367-NC group and the Control group, while the expression level of HOXA1 mRNA in the sh-HOTAIRM1 group was significantly decreased compared with the shRNA-NC group and the Control group (P < 0.05; Figure 6A and B). Western blotting showed that the expression level of HOXA1 protein in the GV367-HOTAIRM1 group was significantly increased, while the expression level of HOXA1 protein in the sh-HOTAIRM1 group was significantly decreased (P < 0.05; Figure 6C–F).
HOXA1 Partly Mediates The Effect Of HOTAIRM1 On Type I EC Biology
Knockdown Efficiency Of HOXA1 In Type I EC Cells
To determine whether HOTAIRM1 promotes type I EC progression by regulating HOXA1, we next silenced the expression of HOXA1 using sh-RNA and established stable-transfected cell lines in Ishikawa and HEC-1A cells. qRT-PCR and Western blotting were then used to detect the transfection efficiency. The expression of HOXA1 mRNA and protein in the sh-HOXA1 group were both significantly decreased compared with the shRNA-NC group and the Control group. (P < 0.05; Figure 7).
Influence Of HOXA1 On The Proliferation Of EC Cell Lines
The CCK8 assay showed that the proliferation rate was significantly lower in the sh-HOXA1 group than that in the shRNA-NC and Control groups (P < 0.05; Figure 8A and B).
Influence Of HOXA1 On The Cell Cycle Of EC Cell Lines
Flow cytometry showed that compared with the shRNA-NC group and the Control group, the proportion of cells in G0/G1 phase in the sh-HOXA1 group was significantly increased in Ishikawa and HEC-1A cells, while the proportion of cells in S phase was significantly decreased (P < 0.05). No significant difference was observed in G2/M phase (P > 0.05; Figure 8C–F). Western blotting results showed that the expression levels of Cyclin D1 in the sh-HOXA1 group were significantly decreased (P < 0.05; Figure 8G–J).
Influence Of HOXA1 On Cell Migration And Invasion In EC Cell Lines
Transwell assays showed that compared with the shRNA-NC group and the Control group, the numbers of migrating and invading transmembrane cells in the sh-HOXA1 group were both significantly decreased (P < 0.05; Figure 9).
Influence Of HOXA1 On EMT In EC Cell Lines In Vitro
Western blotting showed that compared with the shRNA-NC group and the Control group, the expression level of N-cadherin was significantly decreased and the expression level of E-cadherin was significantly increased in the sh-HOXA1 group of Ishikawa and HEC-1A cells (P < 0.05; Figure 10).
HOTAIRM1 And HOXA1 Regulate Tumor Growth In Vivo
HOTAIRM1 Promotes Tumor Growth In Vivo
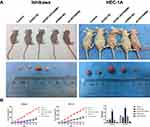
Following the knockdown and overexpression of HOTAIRM1, we then used Ishikawa and HEC-1A cells for subcutaneous implantation modeling to investigate the effect of HOTAIRM1 on the growth of transplanted tumors in nude mice. Data showed that the transplanted tumors formed early, grew fast and that the volume and weight of the transplanted tumors were significantly increased in the GV367-HOTAIRM1 group compared with the GV367-NC group and the Control group. In contrast, the transplanted tumors formed late, grew slowly and the volume and weight of the transplanted tumors were significantly decreased in the sh-HOTAIRM1 group compared with the shRNA-NC group and the Control group (P < 0.05; Figure 11).
HOXA1 Promotes Tumor Growth In Vivo
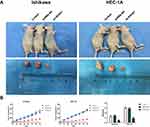
Following the knockdown of HOXA1, we used Ishikawa and HEC-1A cells for subcutaneous implantation modeling to investigate the effect of HOXA1 on the growth of transplanted tumors in nude mice. Data showed that the transplanted tumors formed late, grew slowly and that the volume and weight of the transplanted tumors were significantly decreased in the sh-HOXA1 group compared with the shRNA-NC group and the Control group (P < 0.05; Figure 12).
Discussion
In our previous study, using an Arraystar Human LncRNA V3.0 microarray, we found that lncRNA HOTAIRM1 was one of the significantly upregulated lncRNAs in type I EC. In this continuation study, we demonstrated, for the first time, that the antisense lncRNA HOTAIRM1, and its sense gene HOXA1, were significantly upregulated in type I EC tissues compared with normal endometrial tissues. The expression levels of HOTAIRM1 and HOXA1 both showed a significant and positive correlation with FIGO stage and lymph node metastasis. Statistical analysis showed that the expression level of HOTAIRM1 was significantly and positively correlated with the expression level of both HOXA1 mRNA and protein in type I EC tissues. Furthermore, we found that HOTAIRM1 and HOXA1 significantly promoted cell proliferation, migration, invasion and EMT in vitro. In addition, HOTAIRM1 regulated the expression of HOXA1 gene. In vivo studies further confirmed that HOTAIRM1 and HOXA1 promoted subcutaneous tumorigenesis in nude mice.
Over recent years, studies have confirmed that lncRNA is closely associated with occurrence and development of tumors. However, the role of lncRNA in the development of EC is still unclear. In the present study, we found that the expression of HOTAIRM1 in type I EC tissues was significantly higher than that in normal endometrium and was positively correlated with both FIGO stage and lymph node metastasis. Recent studies have found that the expression of HOTAIRM1 is significantly increased in pancreatic ductal adenocarcinoma, breast cancer, and glioma, and thus plays a role in the promotion of cancer.15–19 However, the role of HOTAIRM1 in type I EC is still unclear. In the present study, in vitro cell experiments revealed that following HOTAIRM1 knockdown or overexpression, the proliferation, migration and invasion ability of Ishikawa and HEC-1A cell lines showed obvious changes with the same trend. Knocking down the expression of HOTAIRM1 increased the proportion of cells in G0/G1 phase, decreased the proportion of cells in S phase, reduced the expression of Cyclin D1 and N-cadherin protein and increased the expression of E-cadherin, while these results were reversed after the overexpression of HOTAIRM1. These findings illustrated that high-expression level of HOTAIRM1 may promote the development of type I EC, and may be closely associated with the clinical outcomes of patients. The role of HOTAIRM1 in type I EC therefore deserves further attention.
LncRNA can be divided into 5 main types: sense lncRNA, antisense lncRNA, bidirectional lncRNA, intronic LncRNA and intergenic lncRNA.33 Antisense lncRNAs have been found to be associated with various malignant tumors. Antisense lncRNAs are long non-coding RNAs that originate from opposite DNA strands of the same genomic locus.24 Studies have demonstrated that antisense lncRNAs often regulate their sense genes via a positive or negative expression relationship. For example, lncRNA DNAJC3-AS1 promotes the progression of osteosarcoma by upregulating its sense-cognate gene DNAJC3.34 Furthermore, the increased expression of antisense lncRNA SPINT1-AS1 predicts a poor prognosis in colorectal cancer and is negatively correlated with its sense transcript SPINT1.35 In the present study, we investigated an antisense lncRNA, HOTAIRM1, which is an antisense transcript of HOXA1 gene that originates from the opposite strand of the HOXA1 locus. Li et al previously reported that HOTAIRM1 promoted tumor growth and invasion by upregulating the HOXA1 gene in glioblastoma multiforme.19 Tian et al further found that HOTAIRM1 could enhance the expression of HOXA1 in myeloid-derived suppressor cells in lung cancer.25 In our present study, we investigated whether HOTAIRM1 also modulated the development and function of type I EC by targeting its sense gene HOXA1. First, we confirmed the expression levels of both HOXA1 mRNA and protein in type I EC tissues were significantly higher than that in normal endometrium. Then, similar to HOTAIRM1, we found that the expression level of HOXA1 mRNA was also closely related to FIGO stage and lymph node metastasis. Importantly, statistical analysis found that there was a significant positive correlation between the expression of HOXA1 and the expression of HOTAIRM1. In addition, the upregulation of HOTAIRM1 caused an increase in the expression of HOXA1, while the downregulation of HOTAIRM1 caused a reduction in the expression of HOXA1. Further research showed that shRNA-mediated HOXA1-knockdown inhibited proliferation, migration, invasion, and EMT in type I EC cells; these findings were consistent with the functional changes that occurred after silencing the expression of HOTAIRM1 in type I EC cells. Further, in vivo tumorigenesis experiments in nude mice also proved that the expression of HOTAIRM1 and HOXA1 was positively correlated with the capacity of tumorigenesis. Collectively, these results prove that lncRNA HOTAIRM1 upregulates the expression of HOXA1 and may be one of the most important molecular mechanisms in the promotion of tumorigenesis and progression in type I EC.
In order to exert biological functions, antisense lncRNA can regulate the expression of sense genes at the transcriptional and post-transcriptional level via four major mechanisms: mechanisms related to transcription, RNA–DNA interactions, RNA–RNA interactions in the nucleus and RNA–RNA interactions in the cytoplasm.36 Wang et al previously showed that HOTAIRM1 interacted with Polycomb Repressive Complex 2 (PRC2) and histone demethylase UTX/MLL to regulate chromatin conformation and then influenced transcriptional activity of HOXA gene cluster.37 Li et al previously reported that HOTAIRM1 regulates the expression of the HOXA1 gene in glioblastoma multiforme. HOTAIRM1 has been shown to mediate the demethylation of histone H3K9 and H3K27 and reduced DNA methylation levels by sequestering epigenetic modifiers G9a and EZH2, which are H3K9me2 and H3K27me3 specific histone methyltransferases, and DNA methyltransferases away from the transcription start sites of HOXA1 gene.19 In the present study, we also confirmed that HOTAIRM1 positively regulates the expression of HOXA1 and promotes the occurrence and development of type I EC. Thus, we speculated that HOTAIRM1 may regulate the expression of its sense gene HOXA1 by acting on the HOXA1 promoter to change chromosome status and then affect the transcriptional activity of HOXA1 gene. The specific mechanism involved remains unknown and is currently being investigated.
Conclusion
Collectively, our results demonstrate that the HOTAIRM1/HOXA1 axis may be involved in promoting tumorigenesis and progression of type I EC and that HOTAIRM1 may exert its function by regulating HOXA1, at least in part. Our data indicate that lncRNA HOTAIRM1/HOXA1 axis represents a novel prognostic biomarker and new potential therapeutic target for type I EC. However, the exact mechanism involved, and the signaling pathways downstream remain unknown and need to be investigated further.
Acknowledgments
This study was supported by National Natural Science Foundation of China (81771556).
Disclosure
The author reports no conflicts of interest in this work.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7–34. doi:10.3322/caac.v69.1
2. Bokhman JV. Two pathogenetic types of endometrial carcinoma. Gynecol Oncol. 1983;15:10–17. doi:10.1016/0090-8258(83)90111-7
3. Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–159. doi:10.1038/nrg2521
4. Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi:10.1016/j.cell.2009.02.006
5. Clark MB, Mattick JS. Long noncoding RNAs in cell biology. Semin Cell Dev Biol. 2011;22:366–376. doi:10.1016/j.semcdb.2011.01.001
6. Sánchez Y, Huarte M. Long non-coding RNAs: challenges for diagnosis and therapies. Nucleic Acid Ther. 2013;23:15–20. doi:10.1089/nat.2012.0414
7. Li C, Chen J, Zhang K, Feng B, Wang R, Chen L. Progress and prospects of long noncoding RNAs (lncRNAs) in hepatocellular carcinoma. Cell Physiol Biochem. 2015;36:423–434. doi:10.1159/000430109
8. Liu Y, Sharma S, Watabe K. Roles of lncRNA in breast cancer. Front Biosci (Schol Ed). 2015;7:94–108. doi:10.2741/s427
9. Du Y, Wang L, Chen S, Liu Y, Zhao Y. LncRNA DLEU1 contributes to tumorigenesis and development of endometrial carcinoma by targeting mTOR. Mol Carcinog. 2018;57:1191–1200. doi:10.1002/mc.v57.9
10. Zhang L, Wang DL, Yu P. LncRNA H19 regulates the expression of its target gene HOXA10 in endometrial carcinoma through competing with miR-612. Eur Rev Med Pharmacol Sci. 2018;22:4820–4827. doi:10.26355/eurrev_201808_15617
11. Liu Y, Wang LL, Chen S, Zong ZH, Guan X, Zhao Y. LncRNA ABHD11-AS1 promotes the development of endometrial carcinoma by targeting cyclin D1. J Cell Mol Med. Epub 2018 May 25.
12. Zhang X, Lian Z, Padden C, et al. A myelopoiesis-associated regulatory intergenic noncoding RNA transcript within the human HOXA cluster. Blood. 2009;113:2526–2534. doi:10.1182/blood-2008-06-162164
13. Zhang X, Weissman SM, Newburger PE. Long intergenic non-coding RNA HOTAIRM1 regulates cell cycle progression during myeloid maturation in NB4 human promyelocytic leukemia cells. RNA Biol. 2014;11:777–787. doi:10.4161/rna.28828
14. Díaz-Beyá M, Brunet S, Nomdedéu J, et al. The lincRNA HOTAIRM1, located in the HOXA genomic region, is expressed in acute myeloid leukemia, impacts prognosis in patients in the intermediate-risk cytogenetic category, and is associated with a distinctive microRNA signature. Oncotarget. 2015;6:31613–31627. doi:10.18632/oncotarget.5148
15. Zhou Y, Gong B, Jiang ZL, et al. Microarray expression profile analysis of long non-coding RNAs in pancreatic ductal adenocarcinoma. Int J Oncol. 2016;48:670–680. doi:10.3892/ijo.2015.3292
16. Luo Y, He Y, Ye X, et al. High expression of long noncoding RNA HOTAIRM1 is associated with the proliferation and migration in pancreatic ductal adenocarcinoma. Pathol Oncol Res. Epub 2019 Jan 6.
17. Su X, Malouf GG, Chen Y, et al. Comprehensive analysis of long non-coding RNAs in human breast cancer clinical subtypes. Oncotarget. 2014;5:9864–9876. doi:10.18632/oncotarget.v5i20
18. Chen Y, Wu JJ, Lin XB, et al. Differential lncRNA expression profiles in recurrent gliomas compared with primary gliomas identified by microarray analysis. Int J Clin Exp Med. 2015;8:5033–5043.
19. Li Q, Dong C, Cui J, Wang Y, Hong X. Over-expressed lncRNA HOTAIRM1 promotes tumor growth and invasion through up-regulating HOXA1 and sequestering G9a/EZH2/Dnmts away from the HOXA1 gene in glioblastoma multiforme. J Exp Clin Cancer Res. 2018;37:265. doi:10.1186/s13046-018-0941-x
20. Wan L, Kong J, Tang J, et al. HOTAIRM1 as a potential biomarker for diagnosis of colorectal cancer functions the role in the tumor suppressor. J Cell Mol Med. 2016;20:2036–2044. doi:10.1111/jcmm.12892
21. Zheng M, Liu X, Zhou Q, Liu G. HOTAIRM1 competed endogenously with miR-148a to regulate DLGAP1 in head and neck tumor cells. Cancer Med. Epub 2018 Jun 14.
22. Zhang Y, Mi L, Xuan Y, et al. LncRNA HOTAIRM1 inhibits the progression of hepatocellular carcinoma by inhibiting the Wnt signaling pathway. Eur Rev Med Pharmacol Sci. 2018;22:4861–4868. doi:10.26355/eurrev_201808_15622
23. Lu R, Zhao G, Yang Y, et al. Long noncoding RNA HOTAIRM1 inhibits cell progression by regulating miR-17-5p/PTEN axis in gastric cancer. J Cell Biochem. 2019;120:4952–4965. doi:10.1002/jcb.v120.4
24. Rosikiewicz W, Makałowska I. Biological functions of natural antisense transcripts. Acta Biochim Pol. 2016;63:665–673. doi:10.18388/abp.2016_1350
25. Tian X, Ma J, Wang T, et al. Long non-coding RNA HOXA transcript antisense RNA myeloid-specific 1-HOXA1 axis downregulates the immunosuppressive activity of myeloid-derived suppressor cells in lung cancer. Front Immunol. 2018;9:473. doi:10.3389/fimmu.2018.00473
26. Liu J, Liu J, Lu X. HOXA1 upregulation is associated with poor prognosis and tumor progression in breast cancer. Exp Ther Med. 2019;17:1896–1902. doi:10.3892/etm.2018.7145
27. Bitu CC, Destro MF, Carrera M, et al. HOXA1 is overexpressed in oral squamous cell carcinomas and its expression is correlated with poor prognosis. BMC Cancer. 2012;12:146. doi:10.1186/1471-2407-12-146
28. Yuan C, Zhu X, Han Y, et al. Elevated HOXA1 expression correlates with accelerated tumor cell proliferation and poor prognosis in gastric cancer partly via cyclin D1. J Exp Clin Cancer Res. 2016;35:15. doi:10.1186/s13046-016-0294-2
29. Zha TZ, Hu BS, Yu HF, Tan YF, Zhang Y, Zhang K. Overexpression of HOXA1 correlates with poor prognosis in patients with hepatocellular carcinoma. Tumour Biol. 2012;33:2125–2134. doi:10.1007/s13277-012-0472-6
30. Wang H, Liu G, Shen D, et al. HOXA1 enhances the cell proliferation, invasion and metastasis of prostate cancer cells. Oncol Rep. 2015;34:1203–1210. doi:10.3892/or.2015.4085
31. Motoyama K, Inoue H, Nakamura Y, Uetake H, Sugihara K, Mori M. Clinical significance of high mobility group A2 in human gastric cancer and its relationship to let-7 microRNA family. Clin Cancer Res. 2008;14:2334–2340. doi:10.1158/1078-0432.CCR-07-4667
32. John RR, Malathi N, Ravindran C, Anandan S. Mini review: multifaceted role played by cyclin D1 in tumor behavior. Indian J Dent Res. 2017;28:187–192. doi:10.4103/ijdr.IJDR_697_16
33. Qi P, Du X. The long non-coding RNAs, a new cancer diagnostic and therapeutic gold mine. Mod Pathol. 2013;26:155–165. doi:10.1038/modpathol.2012.160
34. Liang R, Liu Z, Chen Z, et al. Long noncoding RNA DNAJC3-AS1 promotes osteosarcoma progression via its sense-cognate gene DNAJC3. Cancer Med. 2019;8:761–772. doi:10.1002/cam4.2019.8.issue-2
35. Li C, Li W, Zhang Y, et al. Increased expression of antisense lncRNA SPINT1-AS1 predicts a poor prognosis in colorectal cancer and is negatively correlated with its sense transcript. Onco Targets Ther. 2018;11:3969–3978. doi:10.2147/OTT.S163883
36. Faghihi MA, Wahlestedt C. Regulatory roles of natural antisense transcripts. Nat Rev Mol Cell Biol. 2009;10:637–643. doi:10.1038/nrm2738
37. Wang XQ, Dostie J. Reciprocal regulation of chromatin state and architecture by HOTAIRM1 contributes to temporal collinear HOXA gene activation. Nucleic Acids Res. 2017;45:1091–1104. doi:10.1093/nar/gkw966
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.