Back to Journals » OncoTargets and Therapy » Volume 13
lncRNA HAND2-AS1 Regulates Prostate Cancer Cell Growth Through Targeting the miR-106a-5p/RBM24 Axis
Authors Wei P, Yang J, Zhang D, Cui M, Li L
Received 16 January 2020
Accepted for publication 7 April 2020
Published 21 May 2020 Volume 2020:13 Pages 4523—4531
DOI https://doi.org/10.2147/OTT.S246274
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Carlos E Vigil
This paper has been retracted.
Pengtao Wei,1 Jing Yang,2 Dandan Zhang,3 Meng Cui,4 Lianjun Li5,6
1Department of Urology, Luoyang Central Hospital Affiliated to Zhengzhou University, Luoyang 471000, People’s Republic of China; 2Central Sterile Supply Department, Shandong Provincial Hospital Affiliated to Shandong University, Jinan, People’s Republic of China; 3Urinary Surgery, YiDu Central Hospital in Weifang City, Qingzhou, People’s Republic of China; 4Department of Gynecology, Shandong Provincial Maternity and Childcare Hospital, Jinan, People’s Republic of China; 5Department of Urology, Shandong Provincial Hospital Affiliated to Shandong University, Jinan 250014, People’s Republic of China; 6Department of Urology, Shandong Provincial Hospital Affiliated to Shandong First Medical University, Jinan 250014, People’s Republic of China
Correspondence: Lianjun Li
Department of Urology, Shandong Provincial Hospital Affiliated to Shandong University, Jinan 250014, P. R. China
Department of Urology, Shandong Provincial Hospital Affiliated to Shandong First Medical University, Jinan 250014, People’s Republic of China
Email [email protected]
Introduction: Increasing evidence has shown that abnormally expressed long non-coding RNA (lncRNA) plays crucial roles in prostate cancer (PCa) progression.
Materials and Methods: Here, we analyzed the expression level of lncRNA HAND2 antisense RNA 1 (HAND2-AS1) in PCa cells and tissues. Function assays were performed to investigate the biological roles of HAND2-AS1 in PCa cell behaviors. Bioinformatics methods, luciferase activity reporter assay, and RNA pull-down assay were performed to validate the connection of microRNA-106a-5p (miR-106a-5p) with HAND2-AS1. Also, the target of miR-106a-5p was explored using the same methods.
Results: Our results revealed HAND2-AS1 expression was decreased in both PCa cells and tissues. In vitro functional assays showed HAND2-AS1 could inhibit PCa cell proliferation and colony formation through promoting cell apoptosis. Dual-luciferase activity assays showed miR-106a-5p could directly bind with HAND2-AS1 and RNA binding motif protein 24 (RBM24). Mechanistically, we showed that HAND2-AS1 regulates PCa cell behaviors via targeting miR-106a-5p/RBM24 axis.
Conclusion: In summary, our results illustrated that HAND2-AS1 functions as miR-106a-5p sponge to regulate RBM24 expression, and to influence PCa progression.
Keywords: HAND2-AS1, miR-106a-5p, RBM24, prostate cancer
Introduction
Surgery or androgen deprivation therapy can be used for prostate cancer (PCa) treatment for patients at early stages.1 However, PCa remains a deadly malignancy for males in many countries and is even worse in developing countries.2,3 Therefore, it is essential to explore mechanisms underlying PCa progression to help better control PCa.
In recent years, many studies have indicated that long non-coding RNAs (lncRNAs) influence carcinogenesis.4 The improvements in high-throughput sequencing technology and bioinformatic analysis methods have identified numerous abnormally expressed lncRNAs in cancers.5,6
HAND2 antisense RNA 1 (HAND2-AS1) is a lncRNA reported to function as tumor suppressive lncRNA in numerous cancers. For instance, HAND2-AS1 could serve as a sponge for microRNA-340-5p to regulate BCL2L11 expression in ovarian cancer.7 In esophageal squamous cell carcinoma, HAND2-AS1 overexpression could result in cancer cell proliferation, migration, and invasion inhibition by regulating miR-21.8 In chronic myeloid leukemia, HAND2-AS1 was shown to inhibit cancer cell proliferation and promote apoptosis via regulating miR-1275.9 It is unclear whether HAND2-AS1 has a role in PCa.
miR-106a-5p is an miRNA reported to have decreased expression in renal cell carcinoma tissues and cells.10 It was found that the inhibition of miR-106a-5p expression could promote cancer cell migration and invasion through interacting with PAK5.10 On the other hand, elevated miR-106a-5p expression was identified in glioblastoma, and the miR-106a-5p overexpression could facilitate cancer invasion via regulating adenomatosis polyposis coli.11 These data appear to be contradictory as miR-106a-5p acted as oncogene in some cancers and tumor suppressor in others, which might be explained by the imperfect binding between miRNAs and targets.10,11 It has been revealed that RNA binding motif protein 24 (RBM24), which is an RNA binding protein, could directly interact with p21 and enhance the mRNA stability.12 Moreover, RBM24 was revealed to have decreased expression in nasopharyngeal carcinoma and functioned as a direct target for miR-25 to inhibit cancer progression.13
In this study, we found decreased expression of HAND2-AS1 in PCa tissues and cells. Moreover, we showed HAND2-AS1 could regulate PCa cell proliferation, colony formation, and cell apoptosis in vitro. Moreover, the interaction of HAND2-AS1, miR-106a-5p, and RBM24 and their influences on PCa cell behaviors were further investigated to explore the mechanisms of HAND2-AS1 in PCa.
Materials and Methods
Cell Lines
PCa cells LNCaP, PC3 and DU145 and normal prostate epithelial cell P69 were acquired from Cell Bank of Chinese Academy of Science (Shanghai, China). Dulbecco’s Modified Eagle’s Medium (DMEM; Invitrogen, Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS; Invitrogen) was used to culture cells at a 37°C in a moist incubator containing 5% CO2.
Cell Transfection
The full length of HAND2-AS1 or RBM24 was cloned into pcDNA3.1 by GenScript (Nanjing, Jiangsu, China). miR-106a-5p mimic, miR-106a-5p inhibitor and negative control (NC-mimic, or NC-inhibitor) were purchased from RiboBio (Guangzhou, Guangdong, China). Small interfering RNA against HAND2-AS1 (si-HAND2-AS1) and negative control (NC-siR) were also obtained from RiboBio. Cell transfection was performed using Lipofectamine 2000 (Invitrogen). After 48 h of transfection, cells were collected for functional analyses.
Detection of HAND2-AS1, miR-106a-5p, and RBM24 Expression in Tumor Tissues and Normal Tissues
Expression levels of HAND2-AS1, miR-106a-5p, and RBM24 in PCa tissues and normal tissues were analyzed at ENCORI (http://starbase.sysu.edu.cn/).
Cell Proliferation Assay
Cells were plated in 96‐well plates to analyze cell proliferation rate using cell counting kit-8 (CCK‐8) method (Beyotime, Haimen, Jiangsu, China). CCK-8 reagent was added to the medium after seeding for 0, 1, 2, and 3 days. Then, optical density at the wavelength of 450 nm was measured using a microplate reader (Thermo Fisher Scientific, Inc.).
Colony Formation Assay
Cells were seeded in 6-well plates and incubated for 14 days to form colonies. Then, methanol was used to fix colonies, while crystal violet was used to stain these colonies. Finally, colony numbers were counted using a microscope.
Flow Cytometry Assay
Cells were collected, treated with trypsin, and stained with Annexin V-FITC and PI (Beyotime) in the dark following the supplier’s instructions. Cell apoptosis rate was analyzed using flow cytometry (BD Biosciences, San Jose, CA, USA).
RNA Extraction and Quantitative Real-Time PCR (qRT-PCR)
RNA was extracted from cells using Trizol (Invitrogen) following the manufacturer’s instructions. Using the PrimerScript kit, RNA was reverse transcribed into complementary DNA. qRT-PCR was performed with ABI 7500 (Applied Biosystems, Foster City, CA, USA) using SYBR Green (Takara, Dalian, Liaoning, China). Primers used in this work were as follows: HAND2-AS1: 5ʹ-GGGTGTTTACGTAGACCAGAACC-3ʹ (forward) and 5ʹ-CTTCCAAAAGCCTTCTGCCTTAG-3ʹ (reverse); RBM24: 5ʹ-GGCCAACGTGAACCTGGCATACTT-3ʹ (forward) and 5ʹ-GGCAGGTATCCCGAAAGGTCTTTGT-3ʹ (reverse); GAPDH: 5ʹ-ATCACTGCCACCCAGAAGAC-3ʹ (forward) and 5ʹ-TTTCTAGACGGCAGGTCAGG-3ʹ (reverse); miR-106a-5p: 5ʹ-GATGCTCAAAAAGTGCTTACAGTGCA-3ʹ (forward) and 5ʹ-TATGGTTGTTCTGCTCTCTGTCTC-3ʹ (reverse); U6 snRNA: 5ʹ-AGAGCCTGTGGTGTCCG-3ʹ (forward) and 5ʹ-CATCTTCAAAGCACTTCCCT-3ʹ (reverse). U6 snRNA was used as internal control for miR-106a-5p, while GAPDH was used as control for the rest genes.
Dual-Luciferase Reporter Assay
miR-106a-5p ranks top for all the targets for HAND2-AS1, while RBM24 ranks top among targets for miR-106a-4p. Sequence of HAND2-AS1 or RBM24 containing predicted miR-106a-5p binding sequence was inserted into Dual‐luciferase Expression Vector (Promega, Madison, WI, USA) to generate HAND2-AS1 wild‐type (HAND2-AS1‐wt) or RBM24 wild-type (RBM24-wt). The mutant luciferase activity vector was built with/site-direct mutagenesis kit (Takara) and then these vectors were named HAND2-AS1-mt or RBM24-mt. After 48 h transfection, relative luciferase activities were measured using Dual‐luciferase reporter assay system (Promega).
RNA Pull-Down Assay
The biotin labeled miR-106a-5p was synthesized by RiboBio. The biotin-miR-106a-5p was incubated with cell lysate, and then incubated with streptavidin magnetic agarose beads for 1 h. After washing the beads, RNA was isolated with Trizol and then subjected to qRT-PCR.
Statistical Analysis
Data were analyzed with GraphPad Prism 6.0 and then presented as mean ± SD. Differences in groups were analyzed using Student’s t-test or one‐way analysis of variance (ANOVA) and Tukey post hoc test. P value less than 0.05 indicated significant difference.
Results
HAND2-AS1 Expression Was Decreased in PCa Tissues and Cells
Expression level of HAND2-AS1 in PCa tissues and normal tissues was analyzed with ENCORI and we found HAND2-AS1 expression was decreased in PCa tissues compared with normal tissues (Figure 1A). Meanwhile, qRT-PCR results indicated that HAND2-AS1 expression was lower in PCa cells than in normal cell line (Figure 1B).
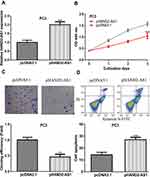
HAND2-AS1 Overexpression Inhibited PCa Cell Growth
HAND2-AS1 expression level was elevated by pHAND2-AS1 transfection (Figure 2A). CCK-8 assay indicated HAND2-AS1 overexpression inhibited cell proliferation (Figure 2B) and the colony formation assay suggested that HAND2-AS1 overexpression inhibited colony formation (Figure 2C). Moreover, flow cytometry analysis revealed that pHAND2-AS1 transfection promoted cell apoptosis (Figure 2D).
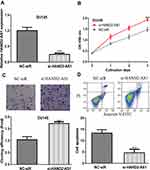
HAND2-AS1 Knockdown Promoted PCa Cell Growth
qRT-PCR showed that introduction of si-HAND2-AS1 decreased the levels of HAND2-AS1 in PCa cells (Figure 3A). CCK-8 assay, colony formation assay, and flow cytometry assay indicated that knockdown of HAND2-AS1 promoted cell growth (Figure 3B-D).
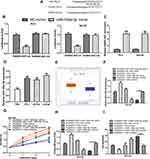
HAND2-AS1 Functioned as a Sponge for miR-106a-5p
The binding module of HAND2-AS1 and miR-106a-5p was presented in Figure 4A. Luciferase activity reporter assay indicated that miR-106a-5p overexpression inhibited luciferase activity in cells with HAND2-AS1-wt transfection (Figure 4B). RNA pull-down assay indicated the direct interaction of HADN2-AS1 and miR-106a-5p (Figure 4C). Moreover, we showed that miR-106a-5p expression level was increased in PCa tissues and cells (Figure 4D and 4E). qRT-PCR showed that pHAND2-AS1 transfection decreased miR-106a-5p expression, while miR-106a-5p mimic transfection increased miR-106a-5p expression (Figure 4F). Functional assays showed groups with miR-106a-5p mimic transfection exhibited higher cell growth ability compared with the other groups (Figure 4G-I).
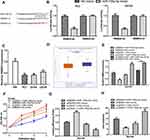
HAND2-AS1 Served as a Sponge of miR‐106a‐5p and Regulated RBM24
Bioinformatic tool showed RBM24 was a possible target for miR-106-5p, as shown in Figure 5A. Luciferase intensity in PCa cells was significantly reduced after co-transfection of RBM24-wt and miR-106a-5p mimic compared with that of RBM24-wt and NC-mimic (Figure 5B). Exploration of RBM24 expression level in PCa showed RBM24 levels were significantly decreased in PCa tissues and cells (Figure 5C and D). Results of qRT-PCR showed RBM24 expression level was stimulated by pRBM24 but inhibited by miR-106a-5p mimic (Figure 5E). Functional assays showed cell growth ability was highest in groups with miR-106a-5p mimic transfection but lowest in those with pRBM24 transfection (Figure 5F-H).
Discussion
Numerous lncRNAs have been identified as aberrantly expressed in PCa and participating in PCa progression.14 For example, Jiang et al revealed lncRNA NEAT1 was expressed at a high level in docetaxel-resistant PCa cells and exerted an oncogenic effect by regulating ACSL4 expression through sponging both miR-34a-5p and miR-204-5p.15 Moreover, lncRNA CASC15 was also found to be expressed at a high level in PCa tissues and cells.16 Moreover, the knockdown of CASC15 was found to suppress PCa cell migration and invasion through functioning as a sponge for miR-200a-3p.16
In this work, we first validated the downregulated status of HAND2-AS1 in both PCa tissues and cells. Gain-of and loss-of experiments indicated that HADN2-AS1 overexpression inhibits PCa cell proliferation, colony formation, and promotes apoptosis, whereas knockdown of HADN2-AS1 exerted opposite effects on PCa cell behaviors. These results suggested a tumor suppressive role of HAND2-AS1 in PCa, which is consistent with its role in cancers including ovarian cancer, esophageal squamous cell carcinoma, and chronic myeloid leukemia.7–9 In recent years, the competing endogenous RNA (ceRNA) theory was used to understand the functions of lncRNA.17 The connection of HAND2-AS1 and miR-106a-5p was validated by luciferase activity reporter assay and RNA pull-down assay. Here, we showed miR-106a-5p overexpression could stimulate – while miR-106a-5p knockdown could inhibit – PCa cell growth. These results indicated that miR-106a-5p serves as an oncomiR in PCa, which is consistent with its role in glioblastoma.11 Moreover, rescue experiments showed that miR-106a-5p was a functional target of HAND2-AS1. In addition, we found RBM24 was a putative target for miR-106a-5p, and the overexpression of RBM24 decreases PCa cell growth, suggesting its tumor suppressive role in PCa. In this work, the functional analyses revealed that lncRNA HAND2-AS1 exerted a tumor suppressive effect in PCa by regulating RBM24 expression through serving as a sponge for miR-106a-5p.
Conclusion
To sum up, we identified that HAND2-AS1 was remarkably decreased in PCa tissues and cells. Besides, HAND2-AS1 could inhibit PCa cell proliferation and colony formation by promoting apoptosis through downregulating RBM24 via sponging miR-106a-5p. These results suggested that HAND2-AS1 functioned as a tumor suppressive lncRNA in PCa and may be a potential therapeutic target for PCa.
Author Contributions
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Zhu Y, Yang XQ, Han CT, et al. Pathological features of localized prostate cancer in China: a contemporary analysis of radical prostatectomy specimens. PLoS One. 2015;10(3):e121076.
2. Litwin MS, Tan HJ. The diagnosis and treatment of prostate cancer: a review. JAMA. 2017;317(24):2532–2542. doi:10.1001/jama.2017.7248
3. Guo T, Wang XX, Fu H, Tang YC, Meng BQ, Chen CH. Early diagnostic role of PSA combined miR155 detection in prostate cancer. Eur Rev Med Pharmacol Sci. 2018;22(6):1615–1621. doi:10.26355/eurrev_201803_14568
4. Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136(4):629–641. doi:10.1016/j.cell.2009.02.006
5. Zhao X, Tang DY, Zuo X, Zhang TD, Wang C. Identification of lncRNA-miRNA-mRNA regulatory network associated with epithelial ovarian cancer cisplatin-resistant. J Cell Physiol. 2019;234(11):19886–19894. doi:10.1002/jcp.28587
6. Wang B, Tang D, Zhang Z, Wang Z. Identification of aberrantly expressed lncRNA and the associated TF-mRNA network in hepatocellular carcinoma. J Cell Biochem. 2020;121(2):1491–1503. doi:10.1002/jcb.29384
7. Chen J, Lin Y, Jia Y, Xu T, Wu F, Jin Y. LncRNA HAND2-AS1 exerts anti-oncogenic effects on ovarian cancer via restoration of BCL2L11 as a sponge of microRNA-340-5p. J Cell Physiol. 2019;234(12):23421–23436. doi:10.1002/jcp.28911
8. Yan Y, Li S, Wang S, et al. Long noncoding RNA HAND2-AS1 inhibits cancer cell proliferation, migration, and invasion in esophagus squamous cell carcinoma by regulating microRNA-21. J Cell Biochem. 2019;120(6):9564–9571. doi:10.1002/jcb.28233
9. Yang JR, Shi MX, Zeng Y. LncRNA HAND2-AS1 inhibits proliferation and promotes apoptosis of chronic myeloid leukemia cells by sponging with micRNA-1275. Eur Rev Med Pharmacol Sci. 2019;23(5):2103–2111. doi:10.26355/eurrev_201903_17254
10. Pan Y-J, Wei -L-L, Wu X-J, Huo F-C, Mou J, Pei D-S. MiR-106a-5p inhibits the cell migration and invasion of renal cell carcinoma through targeting PAK5. Cell Death Dis. 2017;8(10):e3155. doi:10.1038/cddis.2017.561
11. Li D, Wang Z, Chen Z, et al. MicroRNA-106a-5p facilitates human glioblastoma cell proliferation and invasion by targeting adenomatosis polyposis coli protein. Biochem Biophys Res Commun. 2016;481(3–4):245–250. doi:10.1016/j.bbrc.2016.10.132
12. Jiang Y, Zhang M, Qian Y, Xu E, Zhang J, Chen X. Rbm24, an RNA-binding protein and a target of p53, regulates p21 expression via mRNA stability. J Biol Chem. 2014;289(6):3164–3175. doi:10.1074/jbc.M113.524413
13. Hua WF, Zhong Q, Xia TL, et al. RBM24 suppresses cancer progression by upregulating miR-25 to target MALAT1 in nasopharyngeal carcinoma. Cell Death Dis. 2016;7(9):e2352. doi:10.1038/cddis.2016.252
14. Hua JT, Chen S, He HH. Landscape of noncoding RNA in prostate cancer. Trends Genet. 2019;35(11):840–851. doi:10.1016/j.tig.2019.08.004
15. Jiang X, Guo S, Zhang Y, et al. LncRNA NEAT1 promotes docetaxel resistance in prostate cancer by regulating ACSL4 via sponging miR-34a-5p and miR-204-5p. Cell Signal. 2019;65:109422. doi:10.1016/j.cellsig.2019.109422
16. Zhang C, Wang GX, Fu B, Zhou XC, Li Y, Li YY. LncRNA CASC15 promotes migration and invasion in prostate cancer via targeting miR-200a-3p. Eur Rev Med Pharmacol Sci. 2019;23(19):8303–8309. doi:10.26355/eurrev_201910_19141
17. Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta stone of a hidden RNA language? Cell. 2011;146(3):353–358. doi:10.1016/j.cell.2011.07.014
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.