Back to Journals » Clinical, Cosmetic and Investigational Dermatology » Volume 16
LncRNA FENDRR Suppresses Melanoma Growth via Influencing c-Myc mRNA Level
Received 22 February 2023
Accepted for publication 30 June 2023
Published 9 August 2023 Volume 2023:16 Pages 2119—2128
DOI https://doi.org/10.2147/CCID.S409622
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Jeffrey Weinberg
Changying Niu,1 Shenxing Tan2
1Dermatological Department, Affiliated Hospital of Weifang Medical University, Weifang, People’s Republic of China; 2Plastic Surgery, Affiliated Hospital of Weifang Medical University, Weifang, People’s Republic of China
Correspondence: Shenxing Tan, Tel +86 18754411279, Email [email protected]
Background: Long non-coding RNAs (lncRNAs) play an important role in the occurrence of melanoma. However, the specific molecular mechanisms that regulate its biological function are still poorly understood. Therefore, the main purpose of this study is to elucidate the internal mechanism of lncRNA-FENDRR as a biological marker for the occurrence of SKCM and its influence on its proliferation.
Results: FENDRR is low expressed in skin cutaneous melanoma (SKCM) tissues and appears to be at an even lower level as the tumor progresses. However, the high expression of FENDRR can affect the proliferation of SKCM cell line A375. The results of flow cytometry showed that after overexpression of FENDRR, the cell cycle was arrested in the G1/G0 phase. Bioinformatics analysis and RIP results showed that FENDRR could be combined with YTHDF1. Together, these complexes regulate c-Myc mRNA level and determine cell proliferation.
Conclusion: We found that overexpression of FENDRR can effectively inhibit SKCM, which provides a new theoretical basis for new therapeutic approaches and targeted RNA drugs.
Keywords: long non-coding RNAs, FENDRR, melanoma, cell proliferation
Introduction
Cutaneous melanoma is a kind of highly aggressive and metastatic cancer.1 It only occurs in 5% of all skin cancers but causes more than 75% of deaths.2,3 Melanoma can be cured by surgical removal at an early stage.4 However, when the disease progresses to an advanced stage (stage IV), the 5-year survival rate of patients decreases significantly, even to less than 20%.2 There have been a variety of methods in clinical treatment for advanced melanoma, such as radiotherapy, chemotherapy, immunotherapy or targeted therapy.5 Recent studies on the combination of immunotherapy and radiotherapy on melanoma found that this is a safe and promising way on melanoma treatment.6 However, the survival rate of patients has not been effectively improved. Therefore, more efforts are still needed on the exploration of the pathogenesis of melanoma.
Long non-coding RNAs (lncRNAs) were regarded as transcriptional noise at the initial stage of discovery. With in-depth studies, more and more lncRNAs have been proven to have specific functions.7,8 Especially, it is closely related to the occurrence of many cancers.9–13 For example, it has been found that lncRNA can affect histone H3 methylation by binding with EZH2, thus affecting p21 expression to regulate the proliferation of fine lung cancer.14 In addition, recent studies have found that lncRNA can also regulate gene expression by affecting mRNA stability, thus affecting the occurrence of diseases. LncRNA-MEG3 can bind to PTBP1 and thus affect the degradation of SHP mRNA, leading to liver injury.15 In general, lncRNAs are involved in the pathogenesis of a variety of diseases, especially cancer.
In this study, we found that lncRNA-FENDRR had a low expression level in SKCM tissues. However, low expression of FENDRR can lead to tumor deterioration and promote tumor cell proliferation. The internal mechanism of this phenomenon is discussed and deeply studied. This paper provides a potential diagnostic target for the diagnosis of SKCM.
Methods
Tissue and Cell Culture
Melanoma tissue and para-cancer tissue were collected from the Affiliated Hospital of Weifang Medical University between 2016 and 2022. Melanoma patients were recruited from the Affiliated Hospital of Weifang medical university between 2016 and 2022. This study was approved by the Affiliated Hospital of Weifang Medical University Ethics Committee, and the written informed consent of all participants was obtained before recruitment. Melanoma is confirmed by pathological examination. Clinical data have been collected and collated for clinicopathological analysis. All the procedures conform with the Declaration of Helsinki.16
A375 cells were purchased from Procell Life Science & Technology (Wuhan, China) and were cultured in basic medium with 100mL MEM (Gibco, 12492013, USA), 10mL Fetal Bovine Serum, FBS (Gibco, 10270-106, USA), 0.3mL gentamicin solution and 3mL 7.5% sodium carbonate solution. During cell passage, the cells were digested with 0.25% trypsin solution, until the cells were completely shed. Add cell culture medium to terminate digestion and pack according to the required proportion of passage.
During transfection, 5μg plasmid was premixed with 20μL LIPO2000 (Invitrogen, 11668027, USA) in Opti-MEM (Gibco, 11058021, USA). Let them stand at room temperature for 15 minutes. Then, slowly drop them into the medium. Mix gently and let culture for 24 hours before changing the medium. The cells were collected 72 hours later for follow-up experiments.
RNA Extraction and RT-qPCR
Cell precipitates or SKCM tissues were blown and lysed with 1mL Trizol (Invitrogen, 15596026, USA). Then, add 200 μL chloroform, mix vigorously and centrifuge to get supernatant. After adding isopropanol in equal volumes, the RNA was separated in an ice bath for 30 minutes. After centrifugation, the precipitation was washed with 75% ethanol, and the RNA was diluted to 300ng/ μL with ddH2O.
500ng total RNA was taken and reverse transcribed with a reverse transcription kit (Vazyme, R211-01, China). Q-PCR was then performed with SYBR Green Mix (Vazyme, R211-01, China) according to the instructions. The primers used in the experiment are as follows (5’ -3’):
GAPDH FW:CTGGGCTACACTGAGCACC
GAPDH RV:AAGTGGTCGTTGAGGGCAATG
FENDRR FW:TAAAATTGCAGATCCTCCG
FENDRR RV:AACGTTCGCATTTAGC
CDK4 FW:CTGGTGTTTGAGCATGTAGACC
CDK4 RV:GATCCTTGATCGTTTCGGCTG
CDK6 FW:TCTTCATTCACACCGAGTAGTGC
CDK6 RV:TGAGGTTAGAGCCATCTGGAAA
Cyclin D1 FW:TGGAGCCCGTGAAAAAGAGC
Cyclin D1 RV:TCTCCTTCATCTTAGAGGCCAC
Cyclin E1 FW:ACTCAACGTGCAAGCCTCG
Cyclin E1 RV:GCTCAAGAAAGTGCTGATCCC
c-Myc FW: GGTAGTGGAAAACCAGCAGCC
c-Myc RV: TAGAAATACGGCTGCACCGAG
Western Blot
RIPA (Abcam, ab156034, England) is used to lyse cells with a protease inhibitor (Transgen, DI111-01, China) in the ice bath for 30 minutes. Then, the Lysis products were centrifuged at 12000g and the supernatant was transferred to a new tube for protein quantification by a nanodrop (Thermo Scientific, ND-2000, USA). 20μg protein was added to the polyacrylamide gel. After electrophoresis, the protein was transferred to the PVDF membrane overnight at 4°C. The PVDF membrane (Beyotime, FFP19, China) was sealed with 5% skim milk powder for 1 hour and incubated overnight with a primary antibody. Incubate with secondary antibody for 1 h at room temperature the next day. The luminescent solution is then added for development. The list of primary antibodies is as follows: GAPDH (Abcam, ab181602, England), CDK4 (Abcam, ab199728, England), CDK6 (Abcam, ab124821, England), Cyclin D1 (Abcam, ab199728, England), Cyclin E1 (Abcam, ab133266, England). The secondary antibody used an HRP-conjugated goat anti-rabbit antibody (Beyotime, A0208, China).
Cell Proliferation Assay
Cell Counting Kit-8 (CCK8) (Beyotime, C0038, China) was used to examine the cell proliferation ability of A375 (Procell Life Science & Technology, Wuhan, China). A375 cells were cultured in 96-well plates (3000 cells/well). After different incubation times, 100 μL CCK-8 solution was added and incubated at 37°C for 2 hours. Then, the absorbance at 450 nm was measured with a microplate analyzer.
5-Ethynyl-2′-Deoxyuridine Staining
A375 cells were plated in Petri dishes, and 5-ethynyl-2’ -deoxyuridine (EDU) (Beyotime, ST067-50mg, China) was added after the cell turn rate reached 50%, and then cultured for 8 hours. Subsequently, a DNA Proliferation In Vitro Detection Kit (Guangzhou Ribobio, C10310-1, China) was used for Detection. A fluorescence microscope was used to observe and count.
Flow Cytometry
A375 cells were digested with 0.25% trypsin, and the digestion was terminated with medium and precipitated by centrifugation at 500g 3 min for subsequent detection. To assess cell apoptosis, cell precipitates were resuspended with binding buffer (Invitrogen, V13246, USA) and mixed with 5μL fluorescein isothiocyanate labelled Annexin V and 5μL propidium iodide.
To detect the cell cycle, we resuspended cell precipitation with 75% ethanol treatment. After 24 hours, it was mixed with cell cycle fluid and detected by flow cytometry.
RNA-Binding Protein Immunoprecipitation (RIP)
When A375 cells grew to 90% confluences, they were collected by centrifugation after trypsin digestion. The cells were resuspended with an equal volume of lysate and then lysed in an ice bath for 5 min. Seventy-five-microliter Protein-A magnetic beads (Beyotime, P2108-5mL, China) were washed twice with NT-2 Buffer (Tris-HCL, 50mM; NaCl, 150mM; MgCl2, 1 mM; Nonidet P-40, 0.05%) and were resuspended with 100 μL NT-2 Buffer. Five-microgram YTHDF1 antibody (Abcam, ab220162, England) was added and mixed at room temperature for 1 h. Wash with NT-2 6 times and resuspend with 900 μL. Add 100 μL pyrolysis product and mix at 4 °C for 3 hours. Wash the magnetic beads 5 times. Add 150 μL protease K Buffer and incubate at 5 °C for 30 min. Add 400μL phenol, chloroform and isoamyl alcohol (125:24:1), vibrate vigorously for 15S, and centrifuge at 20,000 × g at room temperature for 10 min. The supernatant was added with an equal volume of chloroform and centrifuged for 15 s at 20,000 × g room temperature for 10 min. Add 50μL of 5M ammonium acetate, 15μL of 7.5M LiCl, 5 μL of 5mg/mL glycogen, and 850μL of anhydrous ethanol. The samples were placed overnight at −80 °C and centrifuged at 20,000 × g at 4 °C for 30 min. The precipitation was washed with 500 μL 80% ethanol. The sample was dissolved in 10~20 μL RNASE-FREE water for Western blot examination.
Bioinformatics Analysis and Statistics
Gene Expression Profiling Interactive Analysis (GEPIA, http://gepia.cancer-pku.cn/)17 was used to analyze the SKCM disease-free survival difference in FENDRR low group and FENDRR high group. FENDRR is normalized by ACTB. Group Cutoff was set median (Cutoff-High 50%, Cutoff-Low 50%). SKCM was chosen as dataset. The data were collected from the Cancer Genome Atlas Database (TCGA).
The FENDRR expression profile in different SKCM stages was plotted by Stage plot functions in GEPIA database.
FENDRR protein interaction was predicted by ENCORI18 (http://starbase.sysu.edu.cn). For the high stringency of the predicted results, we set CLIP Data as 3.
SPSS statistical data version, using PRISM7 statistical analysis. Data are expressed as mean ± standard deviation. t-test was used for significant difference analysis, and P < 0.05 indicates that the difference is statistically significant. The experiment was repeated three times, and the error bar represented the standard deviation.
Results
LncRNA-FENDRR Was Low Expressed in SKCM Tissues and Decreased the Overall Survival Rate
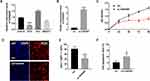
To explore the specific expression level of LncRNA-FENDRR in SKCM tissues, we selected SKCM patients from 2016 to 2022 for study. During that time, 98 patients underwent surgery. Among these samples, 78 samples with low expression of FENDRR, compared to normal tissue, accounted for 78.57% of the total samples. Notably, FENDRR expression levels were very low in 24 of the 25 metastatic melanomas (Table 1). We extracted RNA from the metastatic melanomas and adjacent tissue samples and detected the relative expression level of FENDRR. The results showed that FENDRR was low expressed in SKCM tissues (Figure 1A).
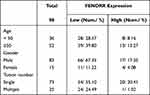 |
Table 1 The Expression Level of FENDRR in All 98 Clinical Samples |
To better illustrate the relationship between FENDRR and SKCM patients’ survival, Kaplan–Meier survival curves were drawn using data collected by GEPIA (Figure 1B), which indicated that FENDRR with low expression significantly reduced survival (low = 198, high = 221). These results suggested that the expression of FENDRR would influence SKCM occurrence and growth. The level of FENDRR in all stages of SKCM was analysed by GEPIA. The results indicated that, with the development of SKCM at different stages, FENDRR had been continuing low expressed, and the expression was slight lower median in Stage II and Stage IV (Figure 1C). In a more detailed SKCM development stage analysis, FENDRR would show a low-expression trend when SKCM was about to transition to the next stage (Figure 1D).
LncRNA-FENDRR was low expressed in SKCM tissue, and the lower the expression level was, the worse the prognosis was. Moreover, the higher the degree of tumor malignancy or the downward stage of tumor development, the lower the expression trend of FENDRR was.
Increased Expression of LncRNA-FENDRR Affects the Proliferation of SKCM Cells
We detected the expression of FENDRR in four SKCM cell lines (B16F10, A375, M14, WM3211). And then, we chose A375 for the future experimental study (Figure 2A, p < 0.001, n = 3). We enhanced the FENDRR level, by using FENDRR overexpression plasmid in A375 SKCM cell line. FENDRR level of the overexpression group was about 10.5 times that of ctrl (Figure 2B, p < 0.001, n = 3).
In the CCK-8 experiment, after overexpression of FENDRR at 24, 48, 72, 96 hours, the absorbance of A375 decreased significantly compared with ctrl (Figure 2C, n = 3). This showed overexpressed FENDRR decreased the proliferation ability of A375.
To further analyze the mechanism of FENDRR affecting A357 viability, we subsequently performed an EDU incorporation experiment and apoptosis detection. The number of EDU-stained cells was significantly decreased in the overexpression group compared to the control group (Figure 2D and E). We then examined the apoptosis of A375 using flow cytometry. The results showed a slight increase in apoptosis in the overexpression group (Figure 2F).
Increased Expression of LncRNA-FENDRR Influences the Normal Cell Cycle
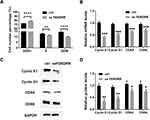
To better understand the mechanism of FENDRR inhibited cell proliferation, we examined the cell cycle of A375 by flow cytometry. In the overexpressing FENDRR group, the number of cells in G0/G1 phase increased significantly, while the number of cells in S phase decreased, compared with the control group (Figure 3A). These results suggested that a high FENDRR level may impede the transition of A375 from G1 to S phase.
Q-PCR showed that high expression of FENDRR would reduce the expression levels of Cyclin E1 and Cyclin D1, and CDK4, and CDK6 (Figure 3B, P < 0.001, n = 3). Similarly, the above results were also confirmed by Western blot (Figure 3C and D). The low expression of these proteins is the direct cause of A375 stagnation in G1 phase.
FENDRR Influences A375 Proliferation via Reducing c-Myc
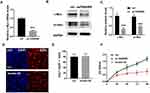
We next examined the expression of the proto-oncogene c-Myc. In the Q-PCR experiment, the mRNA level of c-Myc was significantly reduced after overexpression of FENDRR (Figure 4A, P < 0.001, n = 3). Western blot also showed that the low levels of c-Myc and phosphorylated c-Myc (pc-Myc) in the overexpressed FENDRR group (Figure 4B and C).
To explore whether high FENDRR-induced phenomenon could be rescued, we transfected plasmids that both expressed FENDRR and c-Myc in A375 cells. EDU experiments showed that the inhibited cell proliferation ability was significantly increased after c-Myc overexpression (Figure 4D and E). And CCK-8 assay showed that a higher level of c-Myc would restore its cell viability to the normal level at 48, 72, 96 hours after transfection compared to control group (Figure 4F).
FENDRR Regulates the mRNA Level of c-Myc by Binding to YTHDF1
ENCORI website predicted that FENDRR may interact with YTHDF1, CNBP and other RNA-binding proteins (RBPs) (Table 2). A previous study has found that YTHDF1 is involved in the stability regulation of c-Myc mRNA.19 We speculated that FENDRR may combine with YTHDF1 to regulate the stability of c-Myc mRNA.
 |
Table 2 Bioinformatics Predicts FENDRR-Binding Proteins |
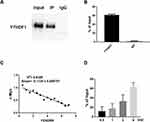
RIP experiment showed that YTHDF1 could be combined with FENDRR (Figure 5A and B). Next, we simultaneously overexpressed YTHDF1 with different doses of FENDRR. Q-PCR assay claimed a negative correlation between FENDRR level and c-Myc expression level (Figure 5C). Moreover, different transfection doses also led to a cascade difference in the amount of FENDRR bound by YTHDF1 (Figure 5D).
Discussion
Melanoma is an aggressive and highly lethal skin cancer. Advanced melanoma has an extremely high fatality rate and a poor 5-year prognosis. To diagnose melanoma earlier and treat melanoma earlier, researchers and medical workers have done a lot of practice and research work. However, this cancer is still poorly understood and there is no good treatment for it.
With the deepening of understanding of lncRNAs, a large number of studies have found that lncRNAs are involved in the occurrence and development of melanoma. Leucci et al found that lncRNA-sammson could combine with p32 to affect mitochondrial homeostasis and promote the occurrence of melanoma.20 However, more literatures have shown that lncRNAs can co-regulate the development of melanoma by regulating miRNAs.21–23 With the consent of the patient, we collected and analyzed the surgical tissues collected since 2016. We found that the expression level of lncRNA-FENDRR was very low in melanoma, compared to the paracancerous tissues. Combined with data collected by GEPIA, the lowest levels of FENDRR tended to be found in late-stage SKCM tissues. Notably, when SKCM is about to deteriorate and move to the next stage, FENDRR will appear at a lower degree of expression. This suggests that FENDRR may be involved in regulating the expression of proto-oncogenes or affecting the biological function of cells.
We selected SKCM cell line A375 with low FENDRR to further investigate the effect of FENDRR on SKCM. We found that the high expression of FENDRR can effectively affect the cell cycle of A375, thereby affecting cell proliferation. This finding is similar to the role of FENDRR in other types of cancer, such as breast cancer,24 cholangiocarcinoma,25 cervical cancer,26 etc. However, in the above studies, FENDRR acted more like a sponge to adsorb different miRNAs to play its function.
In our study, the expression level of FENDRR showed a negative correlation with the expression level of c-Myc. Overexpression of the proto-oncogene c-Myc can effectively rescue the trend of reduced cell proliferation caused by high expression of FENDRR. Considering that FENDRR expression tended to decline when SKCM was about to develop to the next stage, we inferred that FENDRR might be involved in the post-transcriptional regulation of c-Myc. ENCORI predicted that YTHDF1 and CNBP, which regulate mRNA homeostasis, could bind to FENDRR. The RIP experiment verified that YTHDF1 and FENDRR were indeed combined. In previous studies, it was found that YTHDF1 affected the regulation of METTL3 on c-Myc mRNA stability by recognizing m6A.15 In a dose-dependent experiment, we proved that the increase of FENDRR dose showed an increasing trend of binding amount with YTHDF1, which also led to a gradual decrease of c-Myc mRNA expression. This may be because the combination of FENDRR and YTHDF1 hinders the recognition of m6A by YTHDF1, thus may affect the stability of mRNA. It may also affect the c-Myc translation process, resulting in excessive mRNA degradation. This needs to be followed by further research.
In conclusion, we found that FENDRR can be used as a potential marker for the early diagnosis of SKCM. Furthermore, FENDRR is involved in the regulation of c-Myc mRNA level, affecting cell cycle and proliferation, as well as the deterioration degree of SKCM. This suggested that FENDRR could be a potential target for future research on SKCM.
Data Sharing Statement
Data were deposited at Mendeley: https://data.mendeley.com/datasets/kcgpw3wm6f/draft?a=370e05f4-85d3-4d1c-8b81-f05ab1159cec.
Ethical Approval
This study was approved by the Affiliated hospital of Weifang Medical College Ethics Committee, and the written informed consent of all participants was obtained before recruitment.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This work was supported by Shandong Provincial Medical and Health Science and Technology Development Plan Project (No.202104101031).
Disclosure
All the authors declared that there is no competing interest.
References
1. Kremenovic M, Schenk M, Lee DJ. Clinical and molecular insights into BCG immunotherapy for melanoma. J Intern Med. 2020;288:625–640. doi:10.1111/joim.13037
2. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70(1):7–30. doi:10.3322/caac.21590
3. Freddie B, Jacques F, Isabelle S, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;2018:254.
4. Ross MI, Gershenwald JE. Evidence-based treatment of early-stage melanoma. J Surg Oncol. 2011;104(4):341–353. doi:10.1002/jso.21962
5. Beatriz D, José M, Lopes P, et al. Melanoma treatment in review. Immunotargets Ther. 2018;2018:178.
6. Tagliaferri L, Lancellotta V, Fionda B, et al. Immunotherapy and radiotherapy in melanoma: a multidisciplinary comprehensive review. Hum Vaccin Immunother. 2022;18(3):1903827. doi:10.1080/21645515.2021.1903827
7. Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. 2016;17:47–62. doi:10.1038/nrg.2015.10
8. Deveson IW, Hardwick SA, Mercer TR, Mattick JS. The dimensions, dynamics, and relevance of the mammalian noncoding transcriptome. Trends Genet Tig. 2017;33:464. doi:10.1016/j.tig.2017.04.004
9. Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: a new paradigm. Cancer Res. 2017;77(15):3965–3981.
10. Cao Q, Yang W, Ji X, Wang W. Long non-coding RNA ST8SIA6-AS1 promotes lung adenocarcinoma progression through sponging miR-125a-3p. Front Genet. 2020;11. doi:10.3389/fgene.2020.597795
11. Li M, Liang M, Lan T, Wu X, Peng B. Four immune-related long non-coding RNAs for prognosis prediction in patients with hepatocellular carcinoma. Front Mol Biosci. 2020;7. doi:10.3389/fmolb.2020.566491
12. Jia H, Wu D, Zhang Z, Li S. Regulatory effect of the MAFGAS1/miR1505p/MYB axis on the proliferation and migration of breast cancer cells. Int J Oncol. 2020;58(1):33–44. doi:10.3892/ijo.2020.5150
13. Wang S, Cheng M, Zheng X, Zheng L, Chen W. Interactions between lncRNA TUG1 and miR-9-5p modulate the resistance of breast cancer cells to doxorubicin by regulating eIF5A2. Onco Targets Ther. 2020;13:13159–13170. doi:10.2147/OTT.S255113
14. Dandan Y, Xiyi L, Jun S, et al. Long noncoding RNA AFAP1-AS1 predicts a poor prognosis and regulates non–small cell lung cancer cell proliferation by epigenetically repressing p21 expression. Mol Cancer. 2018;17(1):92. doi:10.1186/s12943-018-0836-7
15. Zhang L, Yang ZH, Trottier J, Barbier O, Wang L. Long noncoding RNA MEG3 induces cholestatic liver injury by interaction with PTBP1 to facilitate Shp mRNA decay. Hepatology. 2017;65(2):604–615. doi:10.1002/hep.28882
16. World Medical Association. World Medical Association Declaration of Helsinki: ethical principles for medical research involving human subjects. JAMA. 2013;310(20):2191–2194. doi:10.1001/jama.2013.281053
17. Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98–W102. doi:10.1093/nar/gkx247
18. Li J-H, Liu S, Zhou H, Qu L-H, Yang J-H. starBase v2. 0: decoding miRNA-ceRNA, miRNA-ncRNA and protein–RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42(D1):D92–D7. doi:10.1093/nar/gkt1248
19. Zhao W, Cui Y, Liu L, et al. METTL3 facilitates oral squamous cell carcinoma tumorigenesis by enhancing c-Myc stability via YTHDF1-mediated m(6)A modification. Mol Ther Nucleic Acids. 2020;20:1–12. doi:10.1016/j.omtn.2020.01.033
20. Leucci E, Vendramin R, Spinazzi M, et al. Melanoma addiction to the long non-coding RNA SAMMSON. Nature. 2016;531(7595):518–+. doi:10.1038/nature17161
21. Xia Y, Zhou Y, Han H, Li P, Wei W, Lin NX. lncRNA NEAT1 facilitates melanoma cell proliferation, migration, and invasion via regulating miR-495-3p and E2F3. J Cell Physiol. 2019;234(11):19592–19601. doi:10.1002/jcp.28559
22. Han CF, Tang FJ, Chen J, et al. Knockdown of lncRNA-UCA1 inhibits the proliferation and migration of melanoma cells through modulating the miR-28-5p/HOXB3 axis. Exp Ther Med. 2019;17(5):4294–4302. doi:10.3892/etm.2019.7421
23. Luan W, Ding Y, Ma S, Ruan H, Wang J, Lu F. Long noncoding RNA LINC00518 acts as a competing endogenous RNA to promote the metastasis of malignant melanoma via miR-204-5p/AP1S2 axis. Cell Death Dis. 2019;10. doi:10.1038/s41419-019-2090-3
24. Li Y, Zhang W, Liu P, et al. Long non-coding RNA FENDRR inhibits cell proliferation and is associated with good prognosis in breast cancer. Onco Targets Ther. 2018;11:1403–1412. doi:10.2147/OTT.S149511
25. Qin X, Lu M, Zhou Y, Li G, Liu Z. LncRNA FENDRR represses proliferation, migration and invasion through suppression of survivin in cholangiocarcinoma cells. Cell Cycle. 2019;18(8):889–897. doi:10.1080/15384101.2019.1598726
26. Zhu Y, Zhang X, Wang L, et al. FENDRR suppresses cervical cancer proliferation and invasion by targeting miR-15a/b-5p and regulating TUBA1A expression. Cancer Cell Int. 2020;20(1). doi:10.1186/s12935-020-01223-w
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.