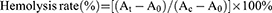Back to Journals » International Journal of Nanomedicine » Volume 18
Liver-Specific Ionizable Lipid Nanoparticles Mediated Efficient RNA Interference to Clear “Bad Cholesterol”
Authors Huang C, Zhang Y, Su J, Guan X, Chen S, Xu X, Deng X, Zhang L , Huang J
Received 10 September 2023
Accepted for publication 1 December 2023
Published 19 December 2023 Volume 2023:18 Pages 7785—7801
DOI https://doi.org/10.2147/IJN.S434908
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Yan Shen
Chuangjia Huang,1,2,* Yu Zhang,3,* Jianfen Su,1,2,* Xiaoling Guan,1,2 Sheng Chen,1,2 Xiaowei Xu,1,2 Xiaohua Deng,1,2 Lingmin Zhang,1,2 Jionghua Huang1
1Department of Cardiology, Guangdong Provincial Key Laboratory of Major Obstetric Diseases, Guangdong Provincial Clinical Research Center for Obstetrics and Gynecology, The Third Affiliated Hospital of Guangzhou Medical University, Guangzhou, 510150, People’s Republic of China; 2Guangzhou Municipal and Guangdong Provincial Key Laboratory of Molecular Target & Clinical Pharmacology, the NMPA and State Key Laboratory of Respiratory Disease, School of Pharmaceutical Sciences and the Fifth Affiliated Hospital, Guangzhou Medical University, Guangzhou, 511436, People’s Republic of China; 3Department of Pharmaceutics, School of Pharmacy, Shenyang Pharmaceutical University, Shenyang, 110016, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Jionghua Huang; Lingmin Zhang, Email [email protected]; [email protected]
Background: High-level low-density lipoprotein cholesterol (LDL-C) plays a vital role in the development of atherosclerotic cardiovascular disease. Low-density lipoprotein receptors (LDLRs) are scavengers that bind to LDL-C in the liver. LDLR proteins are regulated by proprotein convertase subtilisin/kexin type 9 (PCSK9), which mediates the degradation of LDLR and adjusts the level of the plasma LDL-C. The low expression of PCSK9 leads to the up-regulation of liver LDLRs and the reduction of plasma LDL-C. Hepatocytes are attractive targets for small interfering RNA (siRNA) delivery to silence Pcsk9 gene, due to their significant role in LDL-C regulation.
Methods: Here, a type of liver-specific ionizable lipid nanoparticles is developed for efficient siRNA delivery. This type of nanoparticles shows high stability, enabling efficient cargo delivery specifically to hepatocytes, and a membrane-active polymer that reversibly masks activity until an acidic environment is reached.
Results: Significantly, the siPcsk9 (siRNA targeting to Pcsk9)-loaded nanoparticles (GLP) could silence 90% of the Pcsk9 mRNA in vitro. In vivo study showed that the improved accumulation of GLP in the liver increased LDLR level by 3.35-fold and decreased plasma LDL-C by 35%.
Conclusion: GLP has shown a powerful effect on reducing LDL-C, thus providing a potential therapy for atherosclerotic cardiovascular disease.
Keywords: ionizable lipid nanoparticles, bad cholesterol, PCSK9, small interfering RNA, liver-specific delivery
Graphical Abstract:
Introduction
Hyperlipidemia can induce atherosclerosis, which may increase the incidence of atherosclerotic cardiovascular disease (ASCVD),1 including coronary heart disease, stroke, and myocardial infarction.2 Abnormal rise of total cholesterol, low-density lipoprotein cholesterol (LDL-C), and triglycerides (TGs),3 while reduction of high-density lipoprotein cholesterol,4 are common characteristics of hyperlipidemia.
Currently, ASCVD drugs mainly comprise fibrates that promote fatty acid oxidation, statins, inhibitors for the intestinal absorption of cholesterol, and proprotein convertase subtilisin/kexin type 9 (PCSK9) inhibitors.5 Although statins are the most effective lipid-lowering drugs, many patients do not respond well to statin treatment, and adverse drug reactions limit their use.6 PCSK9 adjusts circulating LDL-C levels by inducing endocytosis of low-density lipoprotein receptors (LDLRs) and lysosomal degradation in hepatic cells. Loss-of-function mutations in human PCSK9 lead to significantly lower LDL-C levels, which do not induce significant side effects for the organism.7–9 Monoclonal antibodies that can inhibit PCSK9 have been developed and tested in clinical trials and have proven effective in reducing LDL-C levels and limiting major cardiovascular events.10 However, the short action and high cost of neutralizing antibodies limit for clinical application.11
Since the discovery of RNA interference (RNAi) in mammalian cells, there has been a considerable deal of interest in using this technology to cure illness.12 RNAi has extensive therapeutic promise since it may reversibly silence genes.13,14 Small interfering RNAs (siRNAs), 21–23-nucleotide-long double-stranded RNAs,15 has been widely studied since siRNAs can suppress target genes expression by selective cleavage of complementary mRNA.16 As a result of their ease of suppression of any gene by its base sequence alone, siRNAs have become widespread tools in biological studies.17
RNAi has emerged as a potential tool for the treatment of hyperlipidemia and ASCVD. For example, the Food and Drug Administration approved a new drug, Inclisiran, an siRNA therapeutic that targets PCSK9 mRNA in the liver, reducing circulating LDL-C levels, which are a primary target in the treatment of hypercholesterolemia.18 However, RNAi treatment has run into several difficulties and has fallen short of expectations in terms of clinical translation. In general, difficulties with in vivo distribution, particularly extrahepatic delivery, impede the translation of siRNA therapies to widespread clinical usage.19 Because of its poor bioavailability after systemic delivery, using (unmodified) siRNA as a therapeutic agent is still challenging. Its unsatisfactory stability and poor pharmacokinetic behavior may result in rapid clearance of siRNA from circulation and off-target effects.20 Moreover, its administration can induce (innate) immune responses.21,22 In a cohort of patients at German lipid clinics, the siRNA drug Inclisiran showed a high interindividual variability in LDL-C reduction, with a significant proportion of the population unable to achieve the expected LDL-C reduction.23 Efficient siRNA delivery systems are necessary to improve the therapeutic effects. The construction of a delivery vector that can safely target siRNA delivery into cells, escape capture by lysosomes, and play a role in RNA cleavage is thus critical if the potential of siRNA therapy is to be realized.
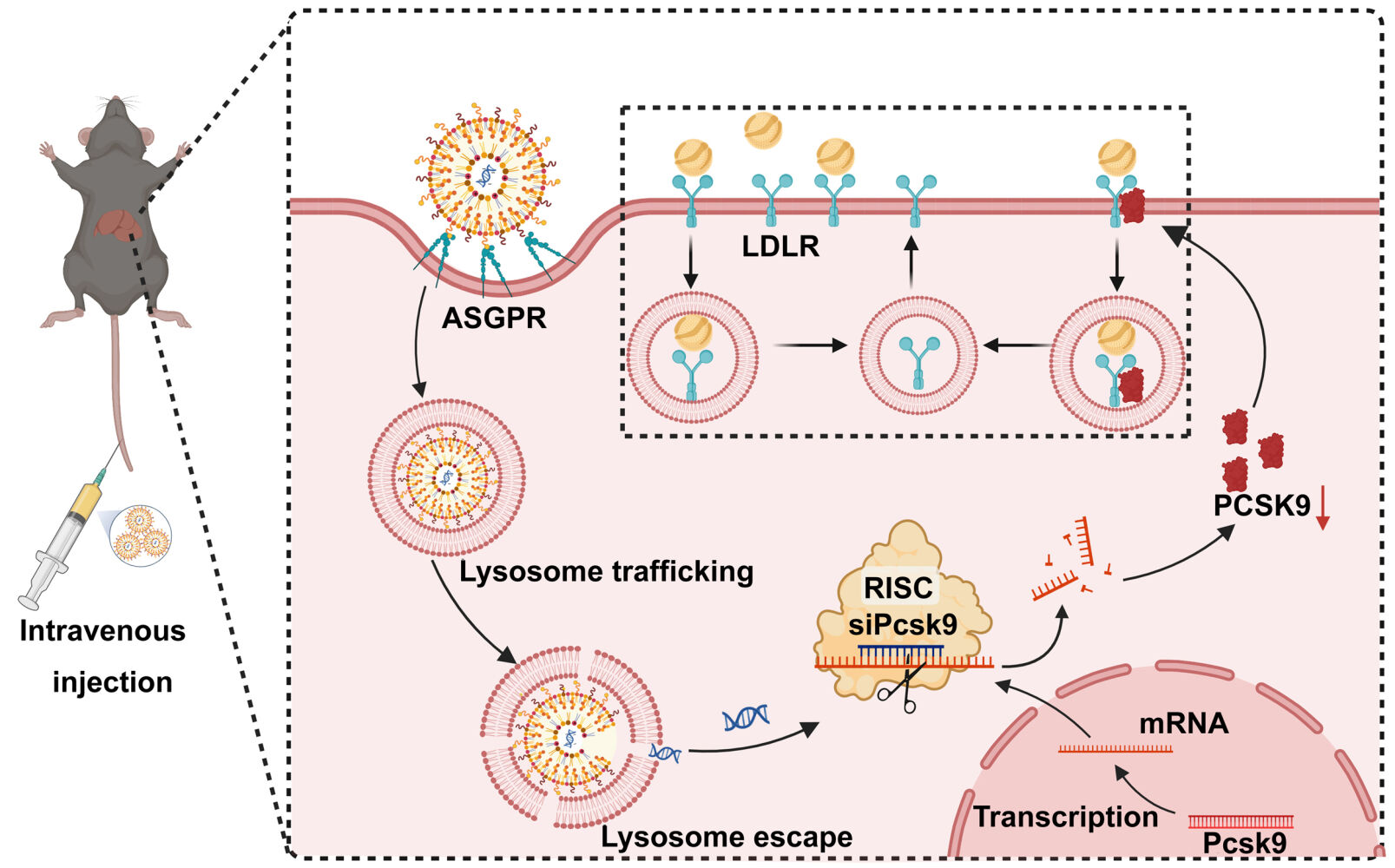
Among all kinds of drug delivery systems, lipid nanoparticles (LNPs) are the most likely approach for intravenous (i.v.) administration of RNAi-based therapeutics for their ionizable amino lipids.24 Ionizable liposomes are designed to respond to pH changes by undergoing charge reversal or conversion. The presence of ionizable lipid components allows for pH-dependent changes in the liposomes’ charge. At physiological pH (pH = 7), the ionizable lipids are typically charged, while in the acidic environment of lysosomes (pH = 5), they undergo protonation, potentially leading to charge reversal.25,26 SM-102 is a type of cationic amino lipid, which is essential in the composition of Moderna COVID-19 vaccine. Also, SM-102 can serve as an available ionizable ingredient for LNPs.27 The components, such as 1.2-distearoyl-sn-glycero-3-phosphocholine (DSPC), cholesterol, and 1.2-dimyristoyl-rac-glycero-3-methoxypolyethylene glycol-2000 (DMG-PEG2000) can also be used as constitutive lipids, and can be modified by different ligands such as antibodies, sugars, and other moieties.28 To prevent the elimination of reticuloendothelial system, polymeric materials, such as DSPE-PEG, can be surface modified to LNPs to escape the recognition of macrophages.29 The ligands then conjugate with DSPE-PEG to produce tailored liposomes that can efficiently target drugs to a specific tissue after binding to specific receptors.30 Galactose-PEG-DSPE that can modify liposomes and increase liver-targeting has been previously synthesized.31 Thus, we designed and synthesized LNPs that contain SM-102, DSPC, cholesterol, DMG-PEG2000, and Galactose-PEG-DSPE (as a liver-targeted lipid) for siRNA-targeting PCSK9 knockdown (Scheme 1). The surface of iLNPs are positively charged under acidic pH,32 which aids the galactose (Gal)-modified ionizable lipid Galactose-PEG-DSPE/SM-102/DSPC/DMG-PEG2000/cholesterol nanoparticles (GL) to effectively encapsulate negatively charged siRNA.33 GL were expected to improve the siPcsk9 delivery (GL/siPcsk9, also named as GLP) to the liver and reduce the LDL-C level effectively, and they may be considered as a promising platform for siRNA delivery (Scheme 1).
 |
Scheme 1 Scheme detailing the preparation of siRNA-loaded Gal-iLNPS and its effects in reducing hyperlipidemia. |
Materials and Methods
Materials
1,2-Distearoyl-sn-glycero-3-phosphocholine (DSPC), cholesterol, and 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-poly(ethylene glycol) (DMG-PEG2000) were supplied by Avanti Polar Lipids Inc. (AL, USA). Galactose-PEG-DSPE was obtained from Xi’an Ruixi Biological Technology Co. Ltd. (Xi’an, China). (8-[(2-Hydroxyethyl)[6-oxo-6-(undecyloxy)hexyl]amino]-octanoic acid, 1-octylnonyl ester (SM-102) was provided by MedChemExpress (New Jersey, USA). The siPCSK9, siControl, and Cy5-siPCSK9 were purchased from RiboBio (Guangzhou, China). Sequences of sense and antisense siRNA strands that target PCSK9 and control siRNAs were synthesized, with 5′-UUCCGAAUAAACUCCAGGCdTdT-3′ used as the sense strand and 5′-GCCUGGAGUUUAUUCGGAAdTdT-3′ used as the antisense strand for siPCSK9. The sense and antisense strands 5′-CUUACGCUGAGUACUUCGAdTdT-3′ and 5′-UCGAAGUACUCAGCGUAAGdTdT were used for the siControl. Citrate buffer was purchased from Teknova (Carlsbad, USA). The live-dead cell staining kit, LysoTracker Green, and DAPI were obtained from Invitrogen (Massachusetts, USA). Lipofectamine™ 3000 transfection reagent was purchased from Invitrogen (Massachusetts, USA). WB detection reagents were purchased from Thermo Fisher Scientific (Waltham, USA). SYBR Green qPCR and Evo M-MLV RT kits were purchased from Accurate Biotechnology (Changsha, China). The antibodies targeting PCSK9, LDLR, and tubulin were provided by Sigma-Aldrich (St. Louis, USA). HRP-conjugated antibodies were purchased from Proteintech (Wuhan, China).
Preparation of GLP
GL were prepared for siRNA delivery, and their sizes and zeta potentials were evaluated. SM-102 (an ionizable lipid with a positive charge under acidic conditions) was used as the lipid component for the encapsulation of siRNA into GL, which were composed of SM-102, DSPC, cholesterol, DMG-PEG2000, and Galactose-PEG-DSPE and were prepared according to our previous work.34,35 To endow the GL with an effective siRNA delivery profile and an ideal safety performance, the optimization of molar ratio of components deserves a higher priority. The molar ratio of PEG lipid was altered by varying the amount of cholesterol, which has a relatively small effect on endosomal release, compared to other lipids.36,37 Galactose-PEG lipid was incorporated into iLNPs at 0.5% to 4.5%, producing 1.5%, 3.0%, and 5.0% PEG-lipids (including DMG-PEG2000 and Galactose-PEG-DSPE). SM-102, DSPC, cholesterol, DMG-PEG2000, and Galactose-PEG-DSPE were dissolved in chloroform/methanol (2/1) to produce 10 mM total lipid at a molar ratio as below:
SM-102/DSPC/cholesterol/DMG-PEG2000/Galactose-PEG-DSPE=50/10/(40-X1-X2)/X1/X2, X1 and X2 represented the molar of DMG-PEG2000 and Galactose-PEG-DSPE, respectively. X1 + X2 = 5, X2 = 0, 0.5, 1, 1.5, 2, 2.5, 3, 3.5, 4, and 4.5.
The solvents in the lipid mixtures were removed by a RV 10 rotary evaporation (IKA, Germany) until a liposome film was formed at the bottom of the bottle and evacuated in vacuum desiccators for 2 h to remove the organic solvent. The liposome film was hydrated by adding 1 mL of 10 mM pH 3.0 citrate buffer for 30 min to keep the final concentration 1 mg/mL. Finally, the lipid mixture was extruded through a LiposoFast manual extruder (Avestin Inc., Canada) to form the cationic liposomes (GL). siPcsk9 was also dissolved in 1mL RNase-free citrate buffer (10 mM, pH 3.0) to obtain a final concentration of 1 mg/mL. The siPcsk9 was complexed to the GL at a weight ratio of 1:30 and incubated at room temperature (RT) for 15 min to form GLP. GLP was dialyzed overnight against a PBS buffer using membranes with a molecular weight cut-off (MWCO) of 3.5 kDa and the pH restored to neutral. GLC or LP used as control was prepared as mentioned above. Unmodified iLNPs without Galactose-PEG-DSPE (LP) were produced with SM-102, DSPC, cholesterol, and DMG-PEG2000 at a ratio of 50:10:37:3. Lipo3000/siPcsk9 was prepared by gently mixing Lipo3000 and siPcsk9 in a weight ratio of 2:1.
Characterization of GL, LP, and GLP
Morphology, Particle Size, and Zeta Potential
The morphologies of the GL, LP, and GLP were captured under transmission electron microscopy (JEM-2100, JEOL, Japan). Dispersed LNPs were dropped onto a carbon-coated grid and dried overnight at 25 °C before observation under an electron microscope. The particle size and zeta potential of the GL, LP, and GLP were measured in distilled water using DLS.
Stability Analysis
To evaluate the stability profile, the GL, LP, and GLP were dissolved in an incubation buffer for 1, 2, 3, 4, 5, 6, and 7 d before measuring variations in the nanoparticle size using DLS.
Agarose Gel Electrophoresis and Stability Analysis
The GLP was prepared as described previously. Weight ratios of 2.5:1, 5:1, 10:1, 20:1, 30:1, and 40:1 (GL to siRNA) were used. Agarose gel electrophoresis was performed to observe the siRNA encapsulation ability of GLP by measuring the unloaded siRNA in the supernatant. Unloaded siRNA was collected by centrifugation at 16000 rpm for 30 min at 4 °C. GLP (10 µL) at various weight ratios and naked siRNAs were loaded onto 1% agarose gels containing 2 μL of 6× loading buffer and run with 1× Tris-acetate-EDTA running buffer at 100 V for 30 min. The results were recorded using a gel documentation imaging system (GE Healthcare Life Sciences, USA).
To evaluate the protection of the LNPs against siRNA degradation by serum nucleases, GLP was incubated with 0.1 mg mL−1 RNase A at 37 °C for 0, 0.25, 0.5, 0.75, 1, and 6 h. Free siRNA exposed to RNase A was set as control. Triton X-100 (0.1%) was used to dissociate the combined siRNA. Subsequently, a total of 0.2 μg of siRNA of each group was loaded to the agarose gel, respectively. The samples were assessed using a gel document imaging system, and siRNA degradation was detected.
In vitro siRNA Release
Detectable GLs were fabricated using Cy5-siRNA as payload. The manner in which Cy5-siRNA was released from Cy5-GLP was examined by fluorescence analysis. PBS (10 mM) at pH 7.4 or pH 5 was adopted as release medium. An aliquot comprising 1 mL of Cy5-GLP that was containing a total of 20 μg of siRNA was added into the dialysis membrane (molecular weight cut off, 3500) and immersed in 20 mL of PBS at pH 7.4 or 5 and incubated for different time points (0, 2, 4, 6, 8, 10, 12, 24, 48, 60, and 72 h) at 25 °C. One milliliter of dialysate was then collected and replaced with fresh PBS of the same volume. The fluorescence intensity of the collected dialysate was measured at Ex = 633 nm and Em = 670 nm using a multimode plate reader (EnSpire, Perkin Elmer, USA).
Cell Culture
Hepa 1–6 cells, 3T3 cells, and H9C2 cells (ATCC, Manassas, USA) were nourished by complete DMEM. Mle-12 cells were cultured in a complete F12 medium. These cells were placed in a humidified 5% CO2 atmosphere with a constant temperature of 37°C for proliferation.
In vitro Evaluation of Galactose-Dependent Uptake
Hepa 1–6 cells were cultured to an 80% confluency before transfection. Different proportions of galactose (0.5%, 1%, 1.5%, 2%, 2.5%, and 3%) were formulated into Cy5-GLP. Cy5-GLP with a Cy5-siRNA concentration of 2 μg mL−1 were produced and incubated for 9 h. The cells were stained with DAPI and images obtained by CLSM (Zeiss 880, Germany). For further quantitative analysis, cellular uptake was performed by FACS (Amnis Corporation, Seattle, USA).
Cellular Uptake Study
Cellular uptake of Cy5-GLP was evaluated over time by CLSM or FACS. Nanoparticles with a Cy5-siRNA concentration of 2 μg mL−1 was incubated for 1, 3, 6, 9, and 12 h. Cells were then stained with DAPI and the images were obtained with CLSM. Besides, FACS was also utilized to quantitatively analyze the cellular uptake.
Hepa 1–6 cells were seeded into the confocal culture dishes at 1×105 cells/well. After the incubation of 24 h, the cells were washed and replaced with a fresh medium containing Cy5-GLP with Cy5-siRNA concentrations of 0.5, 1, 1.5, 2, and 2.5 μg mL−1. After approximately 9 h, the supernatant was discarded, and the cells were washed and stained with DAPI. Images were obtained using a CLSM with a 63 × objective. For FACS analysis, cells were digested with trypsin, washed three times with PBS, suspended in 100 μL of PBS, and analyzed using FACS. The cellular uptake of a series of formulations, including siRNA, LP, and GLP with siRNA equivalent to 2 μg mL−1 was also evaluated. The preceding steps are performed as described above.
Cell Line Selective Uptake Study
Cy5-GLP at a Cy5-siRNA concentration of 2 μg mL−1 were incubated with different cell lines (NIH 3T3, MLE-12, and H9C2) for 9 h at 37 °C. CLSM and FACS were also used for visual observation and quantitatively analysis, respectively.
Lysosomal Escape
Cy5-siRNA was used to assess lysosomal escape. Hepa 1–6 cells were incubated with GLP (Cy5-siRNA equivalent to 2 μg mL−1) for 3 and 9 h. Cells were then washed three times, stained with LysoTracker Green for 30 min, washed again with PBS, and imaged using CLSM. Excitation wavelengths of 488 and 633 nm were utilized for LysoTracker Green and Cy5-siRNA, respectively.
Total RNA Isolation, cDNA Synthesis, and Quantitative PCR (qPCR)
Hepa 1–6 cells were transfected with Cy5-GLP at Cy5-siRNA concentrations of 0.25, 0.5, 1, 2, 3, and 4 μg mL−1. To compare the transfection effects of different groups, Hepa 1–6 cells were also transfected with GLP at an siRNA dose of 2 μg mL−1. Naked siRNA, GL, GLC, LP, Lipo3000/siPcsk9 (siRNA equivalent to 2 μg mL−1) and PBS were set as control. Total RNA was extracted using TRIzol reagent after 48 h of incubation. cDNA synthesis was performed using 2 μg total RNA and SYBR Green. Real-time PCR quantification was performed with PCSK9 using SYBR Green qPCR. A 2−ΔΔCt value was determined to estimate PCSK9 mRNA expression (forward primers 5’-GAGACCCAGAGGCTACAGATT-3’ and reverse primers 5’-AATGTACTCCACATGGGGCAA-3’), and GAPDH expression (forward primers 5’-TGCACCACCAACTGCTTAGC-3’ and reverse primers: 5’-GGCATGGACTGTGGTCATGAG-3’) was used for reference.
WB Assay
WB was conducted to evaluate the translation efficiency of PCSK9 mRNA. Hepa 1–6 cells were transfected with GLP (siRNA equivalent to 0.25, 0.5, 1, 2, 3, and 4 μg mL−1, respectively), as described above. Naked siRNA, GL, GLC, LP, Lipo3000/siPcsk9 (siRNA equivalent to 2 μg mL−1) and PBS was also performed in this study for comparison. After incubation for approximately 48 h, the total protein in the cells was extracted by 100 μL lysis buffer in each well. The six-well plates were shaken in an ice bath for 30 min. Cell lysates were collected and centrifuged at 13000 rpm at 4 °C for 20 min. The supernatant was harvested and quantified by BCA assay. SDS-PAGE gel was used to separate the proteins. The protein bands were then transferred onto PVDF membranes and blocked with 5% nonfat milk at room temperature for approximately 1 h. The membrane was incubated at 4 °C overnight with antibodies against PCSK9 and tubulin and then incubated with relevant secondary antibody for 1.5 h. An Amersham Imager 600 system (GE, USA) was used to scan and photograph the membranes.
Hemolysis Experiment
The erythrocyte hemolysis assay was performed as described in our previous work.31 A red blood cell (RBC) standard dispersion was obtained via C57-mouse whole-blood centrifugation at 2000 rpm for 5 min and washed several times with saline. Dispersed RBCs (20 µL) were then added to 1 mL of saline containing GLP with siRNA concentrations of GLP 1, 2, 3, 4, and 5 μg mL−1, respectively. 20 μL RBC dispersion was also added to 1 mL of saline containing GL, naked siRNA, GLC, LP, GLP, and Lipo3000/siPcsk9 (siRNA equivalent to 2 μg mL−1). After 1 h, the samples were treated by centrifugation at 1×104 rpm for 10 min and 100 µL of the supernatant of each group was taken out for absorbance measurement at 540 nm. The hemolysis rate was calculated through the (Equation 1):
where At, A0, and Ac represents the absorbance of the sample, blank control, and positive control, respectively.
Cytotoxicity Assay
The in vitro cytotoxicity of the GLP was evaluated using a live/dead cell staining kit. Hepa 1–6 cells were treated with DMEM containing GLP with a series of concentrations of siRNA ranging from 1 to 5 μg mL−1. In addition, the cytotoxicity of GL, naked siRNA, GLC, LP, GLP, and Lipo3000/siPcsk9 with siRNA equivalent to 2 μg mL−1 was also evaluated. After 48 h incubation, cells were stained with a live/dead cell staining kit and imaged with CLSM.
In vivo Biodistribution
C57BL/6 6-8-week-old male mice were obtained from the Vital River Laboratory Animal Center (Beijing, China). All animal studies were conducted in accordance with the guidelines in the Guide for the Care and Use of Laboratory Animals published by the US National Institutes of Health. The experiments were performed under the supervision of Experimental Animal Ethics Committee of Guangzhou Medical University. The C57BL/6 mice were fed in a pathogen- free barrier facility with a regulated environment, a 12-h light dark cycle, a comfortable temperature, and unlimited access to food and water. C57BL/6 mice were randomly divided into four groups and treated with PBS, Cy5-siRNA, Cy5-LP, or Cy5-GLP (Cy5-siRNA equivalent to 20 μg) via tail vein injection. Cy5 fluorescence signals were tracked from the whole body of mice by an imaging system (CRi Maestro, USA) at 1, 6, 12, 24, and 48 h. After in vivo tracking, the main organs were isolated and imaged.
The Regulation of PCSK9 in vivo
C57BL/6 mice were randomly divided into seven groups with five animals in each group for saline, GL, siRNA, GLC, LP, GLP, and Lipo3000/siPcsk9 administration. The mice in the saline group were injected with 100 μL saline as a control. For the experimental groups, 100 μL of GL, siRNA, GLC, LP, GLP, or Lipo3000/siPcsk9 (siRNA equivalent to 20 μg) was injected into the tail vein every 5 d. The mice were weighed on an electronic scale every 5 d. Blood samples were collected for biochemical analysis after final administration. Serum was isolated for a blood lipid test to evaluate the PCSK9 protein, LDL-C, high-density lipoprotein cholesterol, TG, total cholesterol, and hepatic functions (AST and ALT). PCSK9 levels were measured using an ELISA assay kit from Bio-Techne China Co. Ltd. (Catalog number, MPC900). Biochemical indexes including LDL-C, CHO, HDL, TG, ALT, and AST were detected with an Automatic biochemical analyzer LB-11ABA (Labotronics Ltd., UK). The mice were then euthanized to obtain the main organs for histological analysis, and partial liver samples were isolated and analyzed by WB.
Statistical Analysis
All assays were independently repeated at least thrice, and data were presented as mean ± SEM. GraphPad Prism (version 8, San Diego, CA, USA) were used to analyze statistically significant differences between two groups with the Student’s two-tailed t-test, or multiple groups by one-way analysis of variance. p ≤ 0.05 were regarded as a significant difference.
Results and Discussion
Preparation and Characterization of GLP
The obtained GL were comprised of an ionizable amino lipid (SM-102), DSPC, cholesterol, PEG-DMG, and Galactose-PEG-DSPE. GLP were prepared via film dispersion method by mixing and extruding through polycarbonate membranes according to our previous work.30 The particle size and zeta potential of the GLP were determined by dynamic light scattering (DLS, Malvern, UK). The zeta potential of GLP with a weight ratio of 30:1 was nearly neutral at pH 7.4 (Table S1). No significant difference was observed in the size of LNPs formed using different proportions of lipids. siRNA was found to be encapsulated into GL with an efficiency of 93.5% at a Galactose-PEG-DSPE proportion of 2.5%. Thus, an SM-102/DSPC/DMG-PEG2000/Galactose-PEG-DSPE/cholesterol ratio of 50/10/0.5/2.5/37 was selected for subsequent experimental studies. Similarly, SM-102, DSPC, DMG-PEG2000, and cholesterol were utilized at a ratio of 50/10/3/37 to carry siPcsk9 (LP, without Galactose-PEG-DSPE) as a control. The morphologies of GL, LP, and GLP were characterized by transmission electron microscopy. All nanoparticles were observed to have a near-spherical shape (Figure 1A). DLS analysis confirmed that the sizes of the GL, LP, and GLP were less than 100 nm (Figure 1B), and GLP were slightly larger than both LP and GL. Interestingly, the three different liposomes remained stable at 4 °C for at least 1 week with no significant changes (Figure 1C).
The decomplexation of siRNA from GL is of vital importance for enhancing gene-silencing efficiency. The cumulative analysis indicated that GLP showed a release rate of approximately 80% after 48 h at pH 5.0 (Figure 1D). In contrast, the cumulative release of GLP was only approximately 50% after 48 h at pH 7.4, indicating that GL possess pH-sensitive property, suggesting that siRNA can be efficiently decomposed from GLP within the cellular environment. The pH-responsive property of the liposomes implies that they will facilitate the release of siRNA into Hepa 1–6 cells if GLP is captured by endosomes or lysosomes.38,39
Agarose gel electrophoresis was performed to test the encapsulation and protection by the liposomes. The siRNA in the supernatant was 6.1% with the weight ratios of GL to siRNA >30:1, indicating the effective encapsulation of GL to siRNA was over 93%. Subsequent studies were thus performed at a weight ratio of 30/1 (Figure 1E). We also evaluated the protection from RNase degradation by GL. Compared with naked siRNA, siRNA entrapped by GL was protected from degradation at all time points, while naked siRNA began to degrade 15 min after treatment with RNase A and was completely degraded after 30 min (Figure 1F). The galactose-modified lipid structure enabled to encapsulate siRNA efficiently and protect the siRNA from degradation by RNase.
We prepared a type of siRNA loaded liposomes with galactose-modified lipid components, which showed pH responsiveness, effective loading efficiency, and protection from RNase degradation.
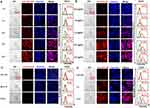
Hemolysis and Cytotoxicity
Biocompatibility is critical for the application of therapeutic systems.40 We investigated the hemolysis rate and cytotoxicity induced by GLP. GLP did not cause any noteworthy hemolytic toxicity when its actual siRNA concentrations were no more than 5 μg mL−1 (Figure 2A and B), in accordance with the results of live/dead assay under confocal laser scanning microscope (CLSM), which was evidenced by the ignorable dead cells (red fluorescence) induced by GLP (Figure 2C). In comparison, the positive control Triton X-100 resulted in nearly all cells emitting red fluorescence, implying these cells were dead (Figure 2C). Different formulations were also analyzed by hemolysis and live/dead assay The formulations, such as GL, siPCSK9, GLC, LP, and GLP, induced no significant hemolysis (Figure 2D and E). Notably, the commercial reagent Lipofectamine 3000 (Lipo3000) encapsulated siRNA resulted in significant hemolysis (Figure 2D and E). The live/dead assay confirmed the results as described above (Figure 2F). The results confirmed that GLP causes low hemolysis and cytotoxicity, providing the possibility for their further use.
Cellular Uptake in vitro
The modification with galactose to nanoparticles is reported to improve the cellular uptake of liver cells by binding to the asialoglycoprotein receptor.35 We thus formulate iLNPs with different galactose-PEG lipid contents to test whether the introduction of different lipids can alter the cellular uptake of iLNPs. Galactose-PEG lipid was incorporated into iLNPs at 0.5–3%. CLSM and flow cytometry (FACS) analysis indicated that the increase of the galactose-PEG lipid contents led to an increase in the cellular uptake, until the molar ratio of the galactose-PEG lipid reached 2.5% (Figure S1). This confirms the formulation chosen for GLP in this study.
Efficient cellular uptake is a prerequisite for effective gene silence. In this study, transfection conditions were optimized by adjusting the concentration and transfection time of the GLP. Cyanine5 (Cy5) labeled siRNA targeting to Pcsk9 (Cy5-siPcsk9) was used to assess the transfection efficiency. CLSM and FACS analysis showed that a 9-h transfection duration met a cellular uptake of up to 96.7%, whereas a longer transfection time did not surpass this peak (Figure 3A). Furthermore, the cellular uptake was increased with increasing Cy5-GLP concentration, and reached its maximum value at a Cy5-GLP concentration of 2 μg mL−1 (Figure 3B). Further Cy5-GLP increases did not induce additional transfection efficiency. Thus, a 2-μg Cy5-GLP concentration and a 9-h transfection time were selected for subsequent experiments.
The cellular uptake of each formulation by Hepa 1–6 cells was then evaluated. The uptake of GLP was evaluated using FACS and compared with those of naked siRNA and LP. The strong fluorescence observed in Hepa 1–6 cells by CLSM analysis indicates that GL facilitate siRNA uptake (Figure 3C). The Cy5 fluorescence in the GLP group was stronger than that observed in the LP group. However, only ignorable red fluorescence was observed after incubating Hepa 1–6 cells with naked siRNA. Thus, the encapsulation of siRNA by GL can be considered to increase the transduction of siRNA. We find that the cellular uptake of GLP is cell-line dependent. CLSM and FACS analysis indicated that the cellular uptake by NIH 3T3, MLE-12, and H9C2 cells reached only 3.2%, 2.4%, and 2.6%, respectively (Figure 3D). The FACS analysis is consistent with the confocal results. The galactose-modified iLNPs improved the cellular uptake in liver cells. The cellular uptake was time- and dosage-dependent, and the optimized transfection condition was 9 h incubation with the GLP at a siPcsk9 at 2 μg/mL.
Lysosome Escape and the Biological Effects Induced by GLP
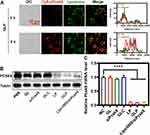
The rapid and efficient escape of siRNA from endosomes and lysosomes indicates successful formation of RNA-induced silencing complex in the cytosol, which is critical for gene silencing.41 CLSM analysis demonstrated that the fluorescence signals representing lysosomes (green fluorescence) overlap with siRNAs (red fluorescence) within 3 h (Figure 4A), implying that the GLP was uptaken by the cells and located in the lysosomes. A substantial amount of red fluorescence was found outside the lysosomes after 9 h, suggesting that the siRNA had escaped from the lysosomes. The effective lysosomal escape of ionizable amino lipids, SM-102, is realized mainly through endo/lysosomal instability.42 The amino group is protonated in the weak acidic environment (eg, pH 5.0) since this pH is lower than the pKa of SM-102. This characteristic can be utilized for the encapsulation of siRNA since the positive group in the nanoparticles can bind to the negatively charged nucleic acids via electrostatic interaction. For physiological conditions (eg, pH 7.4) where the pH is higher than the pKa of SM-102, the surface of iLNP exhibits neutral charge, which is conducive to the extension of blood circulation and the reduction of systemic toxicity. When the nanoparticles are uptaken into endo/lysosomes, the amino group of SM-102 becomes positively charged in the acidic environment.43 The interaction between cationic nanoparticles and anionic endosomal lipids leads to the deconstruction of endosomal membranes, which indirectly contributes to the release of siRNA into the cytosol.44 Based on cell specificity and lysosomal escape analysis, efficient cellular uptake into Hepa1-6 cells was observed for GLP, and the use of GLP protected the siRNA from degradation by enzymes, facilitating the release of siRNA from the lysosomes. These results demonstrate that GLP can be considered as an effective siRNA delivery system.
The effect of GLP on PCSK9-silencing in Hepa 1–6 cells was evaluated in vitro. Western blot (WB) assay was conducted to semi-quantify the protein level of PCSK9 in the Hepa 1–6 cells. As the siRNA concentration increased, the level of PCSK9 protein gradually decreased and reached a low level by GLP with the siRNA concentration equivalent to 2 μg mL−1, and the increase of siRNA concentration did not show further decrease of PCSK9 (Figure S2A), which was also confirmed by real-time PCR analysis (Figure S2B). Compared with the control group, GLP induced a significant reduction of PCSK9 protein levels following transfection for 48 h (Figure 4B and C), which was similar to the commercial reagent, Lipofectamine 3000. In contrast, there were no significant changes in the control, siRNA, scramble control (GL/siControl, GLC), and phosphate-buffered saline (PBS) groups.
Biodistribution and Pharmacokinetic Analysis in vivo
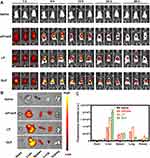
To evaluate the biodistribution of the GLP, Cy5-siRNA was encapsulated in GL and administered to mice by i.v. tail injection. We found that the Cy5-GLP were systemically distributed in mice 1 h after injection (Figure 5A). After 6 h, the Cy5-GLP were found to accumulate predominantly in the liver. After 48 h, the red fluorescence representing Cy5-GLP was still significantly stronger than that representing the Cy5-LP or siPcsk9, indicating that GL promoted the accumulation and retention of siRNA in the liver (Figure 5A). Ex vivo imaging of the major organs suggested that the GLP was mostly absorbed by the liver (Figure 5B). The quantitative analysis of isolated organs indicated that the Cy5-GLP-treated mice showed approximately 30% higher liver accumulation than the Cy5-LP-treated mice (Figure 5C). In addition, the fluorescence signal was significantly reduced 48 h after the injection of bare siPcsk9, indicating rapid metabolization under normal circulation (Figure 5C). The modification of the iLNPs with galactose may contribute to the improved accumulation in the liver by binding to the asialoglycoprotein receptor in the liver cells. In vivo tracking indicated that GLP improved siRNA delivery to the liver.
The Silence of Pcsk9 in vivo
Here, we follow the treatment regimen outlined in Figure 6A to test whether enhancing LDLR expression by reducing PCSK9 expression could reduce LDL-C levels. During the administration period, the body weight of each group continued to rise steadily, indicating favorable systematic safety (Figure 6B). Changes in the PCSK9 levels of the GLP and control groups were confirmed by enzyme-linked immunosorbent assay and WB. The results showed serum PCSK9 levels of 47.07 ± 7.45 ng mL−1 in the GLP-treated group, which was substantially lower than that in the saline group (133.93 ± 11.22 ng mL−1) (Figure 6C). However, the positive control group Lipo3000/siPcsk9-treated mice (101.35 ± 16.52 ng mL−1) and LP-treated mice (89.37 ± 6.43 ng mL−1) showed approximately 20% lower PCSK9 levels than the saline-, GL-, and siRNA-treated mice, and no difference was observed in the GLC group comparing with the saline group (Figure 6C). Meanwhile, the serum LDL-C levels of the GLP-treated mice were reduced by approximately 35% compared to the saline group. However, Lipo3000/siPcsk9 and LP also led to non-significant reductions in the LDL-C levels. The PCSK9 and LDL-C levels induced by LP and Lipo3000/siPcsk9 were not statistically significant. It is well known that PCSK9 inhibits LDL absorption in hepatocytes by promoting LDLR degradation,45 which affects serum LDL-C levels.46 However, the serum biochemical parameters cholesterol (CHO), high-density liptein cholesterol (HDL-C), and triglyceride (TG) in the GLP did not change significantly compared to other groups. Safety in vivo was a concern when drug formulations were administered. We found that the serum biochemical parameters, alanine aminotransferase (ALT) and aspartate aminotransferase (AST), in the GLP remained within the normal range and showed no significant difference in comparison to saline, whereas the opposite was observed in the Lipo3000/siPcsk9 group (Figure 6C). In the GLP group, PCSK9 protein was downregulated in the GLP group, while the LDLR levels got a significant promotion compared to those in the control group (Figure 6D and E). The LDLR expression was 3.35-fold (p<0.05) higher than that in the saline group following injection with GLP (Figure 6D and E). The results demonstrated that GLP was safe in vivo and induced efficient PCSK9 inhibition to lower LDL-C. We further evaluated the safety of GLP for the in vivo administration. Major organs, including the heart, liver, spleen, lungs, and kidneys, were collected from animals treated with saline, GL, siRNA, LP, or GLP, and histopathological sections were prepared. Hematoxylin and eosin staining of the extracted tissues indicated no significant toxicity as a result of GLP treatment in mice (Figure S3). These data indicate that GLP have no immunostimulatory effect and can be used for the safe and effective delivery of nucleic acids in vivo.
Discussion
The findings described in this study strongly correlate with the results obtained from the experiments conducted. The use of GLP, a novel nanoparticle-based delivery system, in combination with siPCSK9, has shown promising results in the treatment of hyperlipidemia. The study demonstrated that the GLP formulation, consisting of five lipid components including SM-102, DSPC, cholesterol, DMG-PEG2000, and galactose-DSPE-PEG2000 effectively delivered siPCSK9 to target cells. One of the key advancements and innovations of GLP lies in its unique composition. Among the lipid components used, SM-102 is an ionizable lipid that plays a crucial role in facilitating the efficient encapsulation and delivery of siPCSK9.47 The inclusion of other lipid components, such as DSPC, cholesterol, DMG-PEG2000, and galactose-DSPE-PEG2000, endows GLP with enhanced stability, targeting ability, and cellular uptake.48,49 The relevance of these findings is significant in the context of hyperlipidemia and ASCVD treatment. Current therapeutic approaches, such as statins and other lipid-lowering medications, have limitations in terms of efficacy and tolerability.50 GLP offers a promising strategy to address these limitations by specifically silencing PCSK9, which plays a critical role in LDL cholesterol metabolism. Compared to conventional lipid-based delivery systems, GLP demonstrates several advantages.51 The presence of SM-102 as an ionizable lipid enables the efficient encapsulation and release of siPCSK9 within the target cells.52 This ionizable property allows for enhanced endosomal escape and cytoplasmic delivery, increasing the efficacy of gene silencing. Additionally, the use of a galactose-DSPE-PEG2000 lipid component facilitates active targeting of hepatocytes, a primary cell line expressing PCSK9, further enhancing the specificity and efficacy of GLP.49 By directly targeting PCSK9 gene expression, GLP offers a more precise and targeted approach compared to conventional medications that may have broader effects on lipid metabolism. This targeted gene silencing can lead to a substantial reduction in LDL cholesterol levels, making it a potentially valuable therapeutic strategy for patients with hyperlipidemia and ASCVD.
However, it is important to acknowledge the limitations of this study. The findings are based on preclinical experiments conducted in animal models, and further research is needed to validate the safety, efficacy, and long-term effects of GLP in humans. Additionally, the study primarily focused on the reduction of LDL cholesterol levels, and it would be valuable to investigate the effects of GLP on other lipid parameters and clinical outcomes. In terms of clinical implications, our findings suggest that GLP for PCSK9 silencing holds promise as a potential therapeutic strategy for hyperlipidemia and ASCVD. The ability to target and specifically silence the PCSK9 gene using GLP nanoparticles offers a novel approach for reducing LDL cholesterol levels and cardiovascular risk. If successfully translated to human studies, GLP could potentially provide an alternative or adjunctive treatment for patients who are intolerant to statins or have inadequate responses to traditional therapies. Future research directions may include optimizing the composition and formulation of GLP to further enhance their stability, specificity, and efficiency. Additionally, studies investigating the long-term safety, potential off-target effects, and durability of GLP-mediated gene silencing are necessary. Furthermore, to explore the combination of GLP with other lipid-lowering agents or novel therapies could provide synergistic effects and further improve treatment outcomes for hyperlipidemia and ASCVD.
The findings presented in this study highlight the advanced and innovative nature of the GLP technology in the context of hyperlipidemia and ASCVD treatment. The use of a nanoparticle-based delivery system, incorporating SM-102 and other lipid components, demonstrates promising results in the targeted delivery of siPCSK9 and subsequent gene silencing. These findings have significant implications for the development of novel therapeutic strategies for hyperlipidemia and ASCVD, offering a more precise and targeted approach compared to current treatments.
Conclusions
In this study, a type of galactose-modified ionizable lipid-based siRNA delivery system was developed, in which siRNA was efficiently encapsulated by GL to form liver-specific nanoparticles. GLP were shown to be biocompatible and non-toxic, with excellent cellular uptake and gene-silencing efficiency in a range of in vitro and in vivo evaluations. GLP can effectively reduce LDL-C, with an indelible effect on the treatment of hyperlipidemia and prevention of ASCVD. In conclusion, this study implies that GLP may be an effective product for the reduction of LDL-C.
Acknowledgments
This work was financially supported by the National Natural Science Foundation of China (82072047, 81700382), the Research Foundation of Education Bureau of Guangdong Province (2021ZDZX2004), the Guangzhou Science and Technology Project (202201011593), Plan on enhancing scientific research in GMU (02-410-2302068XM), and the open research funds from the Sixth Affiliated Hospital of Guangzhou Medical University, Qingyuan People’s Hospital (202201-303). The authors appreciated the help from Schematic illustrations were supported by BioRender.com.
Disclosure
The authors declare that they have no competing interests in this work.
References
1. Berberich AJ, Hegele RA. The complex molecular genetics of familial hypercholesterolaemia. Nat Rev Cardiol. 2019;16(1):9–20. doi:10.1038/s41569-018-0052-6
2. Bauer RC, Tohyama J, Cui J, et al. Knockout of Adamts7, a novel coronary artery disease locus in humans, reduces atherosclerosis in mice. Circulation. 2015;131(13):1202–1213. doi:10.1161/CIRCULATIONAHA.114.012669
3. Liu S, Long Y, Yu S, et al. Borneol in cardio-cerebrovascular diseases: pharmacological actions, mechanisms, and therapeutics. Pharmacol Res. 2021;169:105627. doi:10.1016/j.phrs.2021.105627
4. Bergeron N, Phan BA, Ding Y, Fong A, Krauss RM. Proprotein convertase subtilisin/kexin type 9 inhibition: a new therapeutic mechanism for reducing cardiovascular disease risk. Circulation. 2015;132(17):1648–1666. doi:10.1161/CIRCULATIONAHA.115.016080
5. Hafiane A, Daskalopoulou SS. Targeting the residual cardiovascular risk by specific anti-inflammatory interventions as a therapeutic strategy in atherosclerosis. Pharmacol Res. 2022;178:106157. doi:10.1016/j.phrs.2022.106157
6. Hadjiphilippou S, Ray KK. Cholesterol-Lowering Agents. Cir Res. 2019;124(3):354–363. doi:10.1161/CIRCRESAHA.118.313245
7. Dong Y, Love KT, Dorkin JR, et al. Lipopeptide nanoparticles for potent and selective siRNA delivery in rodents and nonhuman primates. Proc Natl Acad Sci U S A. 2014;111(11):3955–3960. doi:10.1073/pnas.1322937111
8. Goldstein JL, Brown MS. A century of cholesterol and coronaries: from plaques to genes to statins. Cell. 2015;161(1):161–172. doi:10.1016/j.cell.2015.01.036
9. Wu C, Xi C, Tong J, et al. Design, synthesis, and biological evaluation of novel tetrahydroprotoberberine derivatives (THPBs) as proprotein convertase subtilisin/kexin type 9 (PCSK9) modulators for the treatment of hyperlipidemia. Acta Pharm Sin B. 2019;9(6):1216–1230. doi:10.1016/j.apsb.2019.06.006
10. Cordero A, Santos-Gallego CG, Fácila L, et al. Estimation of the major cardiovascular events prevention with Inclisiran. Atherosclerosis. 2020;313:76–80. doi:10.1016/j.atherosclerosis.2020.09.021
11. Sabatine MS, Giugliano RP, Wiviott SD, et al. Efficacy and safety of evolocumab in reducing lipids and cardiovascular events. New Eng J Med. 2015;372(16):1500–1509. doi:10.1056/NEJMoa1500858
12. Kim B, Park JH, Sailor MJ. Rekindling RNAi therapy: materials design requirements for in vivo siRNA delivery. Adv Mater. 2019;31(49):e1903637. doi:10.1002/adma.201903637
13. Kim HJ, Kim A, Miyata K, Kataoka K. Recent progress in development of siRNA delivery vehicles for cancer therapy. Adv Drug Deliv Rev. 2016;104:61–77. doi:10.1016/j.addr.2016.06.011
14. Kanasty R, Dorkin JR, Vegas A, Anderson D. Delivery materials for siRNA therapeutics. Nat Mater. 2013;12(11):967–977. doi:10.1038/nmat3765
15. Setten RL, Rossi JJ, Han SP. The current state and future directions of RNAi-based therapeutics. Nat Rev Drug Discov. 2019;18(6):421–446. doi:10.1038/s41573-019-0017-4
16. Gomes-da-Silva LC, Fonseca NA, Moura V, Pedroso de lima MC, Simões S, Moreira JN. Lipid-based nanoparticles for siRNA delivery in cancer therapy: paradigms and challenges. Acc Chem Res. 2012;45(7):1163–1171. doi:10.1021/ar300048p
17. Weng Y, Xiao H, Zhang J, Liang XJ, Huang Y. RNAi therapeutic and its innovative biotechnological evolution. Biotechnol Adv. 2019;37(5):801–825. doi:10.1016/j.biotechadv.2019.04.012
18. Ray KK, Kallend D, Leiter LA, et al. Effect of inclisiran on lipids in primary prevention: the ORION-11 trial. Eur Heart J. 2022;43(48):5047–5057. doi:10.1093/eurheartj/ehac615
19. Fu Z, Zhang X, Zhou X, et al. In vivo self-assembled small RNAs as a new generation of RNAi therapeutics. Cell Res. 2021;31(6):631–648. doi:10.1038/s41422-021-00491-z
20. Zhang L, Deng S, Zhang Y, et al. Homotypic targeting delivery of siRNA with artificial cancer cells. Adv Healthc Mater. 2020;9(9):e1900772. doi:10.1002/adhm.201900772
21. Hu B, Zhong L, Weng Y, et al. Therapeutic siRNA: state of the art. Signal Transduct Target Ther. 2020;5(1):101. doi:10.1038/s41392-020-0207-x
22. Kulkarni JA, Witzigmann D, Chen S, Cullis PR, van der Meel R. Lipid nanoparticle technology for clinical translation of siRNA therapeutics. Acc Chem Res. 2019;52(9):2435–2444. doi:10.1021/acs.accounts.9b00368
23. Makhmudova U, Schatz U, Perakakis N, et al. High interindividual variability in LDL-cholesterol reductions after inclisiran administration in a real-world multicenter setting in Germany. Clin Res Cardiol. 2023;112(11):1639–1649. doi:10.1007/s00392-023-02247-8
24. Tam YY, Chen S, Cullis PR. Advances in lipid nanoparticles for siRNA delivery. Pharmaceutics. 2013;5(3):498–507. doi:10.3390/pharmaceutics5030498
25. Sun D, Lu Z-R. Structure and function of cationic and ionizable lipids for nucleic acid delivery. Pharm Res. 2023;40(1):27–46. doi:10.1007/s11095-022-03460-2
26. Huo H, Cheng X, Xu J, Lin J, Chen N, Lu X. A fluorinated ionizable lipid improves the mRNA delivery efficiency of lipid nanoparticles. J Mater Chem B. 2023;11(19):4171–4180. doi:10.1039/D3TB00516J
27. Cho HY, Chuang TH, Wu SN. Effective perturbations on the amplitude and hysteresis of Erg-mediated potassium current caused by 1-Octylnonyl 8-[(2-hydroxyethyl)[6-oxo-6(undecyloxy)hexyl]amino]-octanoate (SM-102), a cationic lipid. Biomedicines. 2021;9(10):1367. doi:10.3390/biomedicines9101367
28. Wang F, Li Y, Jiang H, et al. Dual-ligand-modified liposomes co-loaded with anti-angiogenic and chemotherapeutic drugs for inhibiting tumor angiogenesis and metastasis. Int J Nanomed. 2021;16:4001–4016. doi:10.2147/IJN.S309804
29. Tang Y, Wang X, Li J, et al. Overcoming the reticuloendothelial system barrier to drug delivery with a “don’t-eat-us” strategy. ACS Nano. 2019;13(11):13015–13026. doi:10.1021/acsnano.9b05679
30. Li X, Diao W, Xue H, et al. Improved efficacy of doxorubicin delivery by a novel dual-ligand-modified liposome in hepatocellular carcinoma. Cancer Lett. 2020;489:163–173. doi:10.1016/j.canlet.2020.06.017
31. Milan A, Mioc A, Prodea A, et al. The optimized delivery of triterpenes by liposomal nanoformulations: overcoming the challenges. Int J Mol Sci. 2022;23(3):1140. doi:10.3390/ijms23031140
32. Terada T, Kulkarni JA, Huynh A, et al. Characterization of lipid nanoparticles containing ionizable cationic lipids using design-of-experiments approach. Langmuir. 2021;37(3):1120–1128. doi:10.1021/acs.langmuir.0c03039
33. Yang T, Li C, Wang X, et al. Efficient hepatic delivery and protein expression enabled by optimized mRNA and ionizable lipid nanoparticle. Bioact Mater. 2020;5(4):1053–1061. doi:10.1016/j.bioactmat.2020.07.003
34. Zhang L, Wang P, Feng Q, et al. Lipid nanoparticle-mediated efficient delivery of CRISPR/Cas9 for tumor therapy. NPG Asia Mater. 2017;9:e441. doi:10.1038/am.2017.185
35. Zhang L, Wang L, Xie Y, et al. Triple-targeting delivery of CRISPR/Cas9 to reduce the risk of cardiovascular diseases. Angew Chem Int Ed. 2019;58(36):12404–12408. doi:10.1002/anie.201903618
36. Kim M, Jeong M, Hur S, et al. Engineered ionizable lipid nanoparticles for targeted delivery of RNA therapeutics into different types of cells in the liver. Sci Adv. 2021;7(9):eabf4398. doi:10.1126/sciadv.abf4398
37. Kauffman KJ, Dorkin JR, Yang JH, et al. Optimization of lipid nanoparticle formulations for mRNA delivery in vivo with fractional factorial and definitive screening designs. Nano Lett. 2015;15(11):7300–7306. doi:10.1021/acs.nanolett.5b02497
38. Zhang Y, Yang L, Wang H, et al. Bioinspired metal-organic frameworks mediated efficient delivery of siRNA for cancer therapy. Chem Engin J. 2021;426:131926. doi:10.1016/j.cej.2021.131926
39. Zhang Y, Qin Y, Li H, et al. Artificial platelets for efficient siRNA delivery to clear ”bad cholesterol”. ACS Appl Mater Interfaces. 2020;12(25):28034–28046. doi:10.1021/acsami.0c07559
40. Zhang HT, Peng R, Chen S, et al. Versatile nano-PROTAC-induced epigenetic reader degradation for efficient lung cancer therapy. Adv Sci. 2022;9:29.
41. Peng Q, Li H, Deng Q, et al. Hybrid artificial cell-mediated epigenetic inhibition in metastatic lung cancer. J Colloid Interface Sci. 2021;603:319–332. doi:10.1016/j.jcis.2021.06.066
42. Hu B, Li B, Li K, et al. Thermostable ionizable lipid-like nanoparticle (iLAND) for RNAi treatment of hyperlipidemia. Sci Adv. 2022;8(7):eabm1418. doi:10.1126/sciadv.abm1418
43. Yanez Arteta M, Kjellman T, Bartesaghi S, et al. Successful reprogramming of cellular protein production through mRNA delivered by functionalized lipid nanoparticles. Proc Natl Acad Sci U S A. 2018;115(15):e3351–e3360. doi:10.1073/pnas.1720542115
44. Weng Y, Li C, Yang T, et al. The challenge and prospect of mRNA therapeutics landscape. Biotechnol Adv. 2020;40:107534. doi:10.1016/j.biotechadv.2020.107534
45. Akinc A, Querbes W, De S, et al. Targeted delivery of RNAi therapeutics with endogenous and exogenous ligand-based mechanisms. Mol Ther. 2010;18(7):1357–1364. doi:10.1038/mt.2010.85
46. Xu X, Dong Y, Ma N, et al. MiR-337-3p lowers serum LDL-C level through targeting PCSK9 in hyperlipidemic mice. Metabolism. 2021;119:154768. doi:10.1016/j.metabol.2021.154768
47. Sebastiani F, Arteta MY, Lerche M, et al. Apolipoprotein E binding drives structural and compositional rearrangement of mRNA-containing lipid nanoparticles. ACS Nano. 2021;15(4):6709–6722. doi:10.1021/acsnano.0c10064
48. Leung AWY, Chen KTJ, Ryan GM, et al. DMPC/Chol liposomal copper CX5461 is therapeutically superior to a DSPC/Chol formulation. J Control Release. 2022;345:75–90. doi:10.1016/j.jconrel.2022.03.004
49. Wang CY, Chen Z, Tang XF, et al. Influences of galactose ligand on the uptake of TADF liposomes by HepG2 cells. Photodiagnosis Photodyn Ther. 2020;32:102014. doi:10.1016/j.pdpdt.2020.102014
50. Eygeris Y, Gupta M, Kim J, Sahay G. Chemistry of lipid nanoparticles for RNA delivery. Acc Chem Res. 2022;55(1):2–12. doi:10.1021/acs.accounts.1c00544
51. Haddadzadegan S, Dorkoosh F, Bernkop-Schnürch A. Oral delivery of therapeutic peptides and proteins: technology landscape of lipid-based nanocarriers. Adv Drug Deliv Rev. 2022;182:114097. doi:10.1016/j.addr.2021.114097
52. Lee SH, Kim N, Kim M, et al. Single-cell transcriptomics reveal cellular diversity of aortic valve and the immunomodulation by PPARγ during hyperlipidemia. Nat Commun. 2022;13(1):5461. doi:10.1038/s41467-022-33202-2
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.