Back to Journals » Cancer Management and Research » Volume 10
Liver-enriched activator protein 1 as an isoform of CCAAT/enhancer-binding protein beta suppresses stem cell features of hepatocellular carcinoma
Authors Yang LH, Wang Y , Qiao S, Wang MJ , Chen F, Zi XY , Li JX, Zhang HB, Yu B , Hu YP
Received 18 December 2017
Accepted for publication 9 February 2018
Published 26 April 2018 Volume 2018:10 Pages 873—885
DOI https://doi.org/10.2147/CMAR.S160172
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Lu-Zhe Sun
Li-Hua Yang,1,2,* Ye Wang,1,3,* Shi Qiao,1,* Min-Jun Wang,1 Fei Chen,1 Xiao-Yuan Zi,1 Jian-Xiu Li,1 Hai-Bin Zhang,4 Bing Yu,1,4 Yi-Ping Hu1
1Department of Cell Biology, Center for Stem Cell and Medicine, Second Military Medical University, Shanghai, People’s Republic of China; 2Department of Cell Biology and Neurobiology, Xuzhou Medical University, Xuzhou, People’s Republic of China; 3Department of Urology, Changhai Hospital, Second Military Medical University, Shanghai, People’s Republic of China; 4Department of Hepatic Surgery V, Eastern Hepatobiliary Surgery Hospital, Shanghai, People’s Republic of China
*These authors contributed equally to this work
Purpose: Liver cancer stem cells (CSCs) are known to be associated with the development, survival, proliferation, metastasis, and recurrence of liver tumors. The aim of this study was to investigate the association of liver-enriched activator protein 1 (LAP1) with hepatocellular carcinoma (HCC) and liver CSCs (LCSCs) and explore the impact of LAP1 on LCSCs.
Materials and methods: Differences in LAP1 expression in liver cancer tissues versus matched para-tumoral liver tissues and LCSCs versus non-CSCs were analyzed by Western blotting, real-time polymerase chain reaction, immunohistochemistry, and flow cytometry. The effect of LAP1 on liver cancer cells was evaluated by the expression of CSC markers, oncosphere formation, proliferation, migration, and invasion in vitro. Cell cycle distribution and the number of apoptotic cells were analyzed to assess cell cycle and cell apoptosis. Furthermore, a mouse subcutaneous tumor implant model was established to explore the role of LAP1 in the development of HCC in vivo. Finally, the expression of CSC markers in paraffin-embedded sections was evaluated by immunofluorescence.
Results: LAP1 was weakly expressed in HCC tumors and cell lines and even weaker in LCSCs. LAP1 inhibited the expression of stem cell–associated genes and reduced the abilities of oncosphere formation, proliferation, migration, and invasion in vitro. Cell cycle assay revealed that LAP1 induced G1/G0 arrest. Furthermore, LAP1 decreased subcutaneous tumor-formation ability and the expression of CSC markers and Ki67 in vivo.
Conclusion: LAP1 suppressed the stem cell features of HCC, indicating that it possessed an antitumor effect in liver cancer, both in vitro and in vivo; therefore, LAP1 may prove to be a potential target in liver CSC-targeted therapy.
Keywords: LAP1, liver-enriched activator protein 1, hepatocellular carcinoma, HCC, liver cancer stem cell
Introduction
Hepatocellular carcinoma (HCC) is one of the most common malignancies in the People’s Republic of China. Histologically, HCC is the most common type of primary liver cancer, accounting for 85%–90% of all primary liver cancers.1 The incidence of liver cancer ranks fifth, and mortality from HCC ranks third among all malignancies.1–3 Although continuous progress has been made in the diagnosis and surgical and biological therapies of liver cancer, the overall prognosis of patients with liver cancer remains unsatisfactory due to recurrence, metastasis, chemoresistance, and radiation resistance.4 Therefore, more efforts are required to explore novel and more effective treatments for liver cancer.
Cancer stem cells (CSCs) play important roles in the development, metastasis, drug resistance, and many other biological processes of tumors.5–7 The concept of CSCs was first proposed by Mackillop et al,8 who proposed that there may be a small number of cells in cancer that had similar functions as stem cells. In 1997, Bonnet and Dick9 identified CD34+/CD38− in leukemia as CSCs. Subsequent research further identified CSCs in various solid tumors, including cancers of the breast, lung, prostate, brain, and pancreas.10–13 Meanwhile, many studies confirmed that CSCs also exist in HCC and could be enriched by several defined surface markers such as CD133, CD13, CD44, OV6, epithelial cell adhesion molecule (EpCAM), CD24, and CD90.6,14,15 According to the CSC theory, CSCs contribute to tumorigenesis, metastasis, recurrence, and chemoresistance through cell symmetry and asymmetric division;16 therefore, CSCs could be used as the target in tumor therapy.17,18
CCAAT/enhancer-binding protein beta (CEBPβ) – a liver-enriched transcription factor – contains three isoforms, including two liver-enriched activating protein isoforms, LAP1 and LAP2, and a liver-enriched inhibitory protein isoform called LIP.19 CEBPβ not only regulates the differentiation, proliferation, and survival of hepatocytes but also plays a critical role in inflammatory response and tumorigenesis.20–22 Abnormal expression of CEBPβ is closely associated with the malignancy of a variety of tumors such as glioma, Wilm’s tumor, renal cell tumor, and ovarian cancer.23–25 LAP1 – the longest isoform – is the most highly expressed isoform of CEBPβ. LAP1 is implicated in cancer progression. LAP1 was reported to repress migration and invasion of HCC cells in vitro by binding to the ORM2 promoter and activating its expression.26 However, little is known about the function of LAP1 in liver CSCs (LCSCs).
In the present study, we explored the association of LAP1 with HCC and LCSCs and the effect of LAP1 on LCSCs. CEBPβ and LAP1 were found to be weakly expressed in HCC and even weaker in LCSCs. LAP1 suppressed the expression of marker genes of LCSCs and reduced capacities of oncosphere formation, proliferation, wound healing, migration, and invasion in vitro. More importantly, LAP1 decreased subcutaneous tumorigenicity in vivo. These data suggest that LAP1 could be used as an indicator of the diagnosis and prognosis of HCC and may be developed as a target for liver cancer therapy in the future.
Materials and methods
Cell lines and tumor specimens
Human liver cancer cell lines Huh7, Hep3B, and HepG2 were purchased from the American Type Culture Collection and stored in our laboratory. LM3 was purchased from the Cell Bank of the Type Culture Collection of the Chinese Academy of Sciences. All liver cancer cell lines were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% fetal bovine serum (FBS; HyClone), 100 mg/mL penicillin, and 100 U/mL streptomycin. The primarily cultured human liver cancer cell line CLC13 from liver cancer samples was kindly provided by Dr Lijian Hui from the Chinese Academy of Sciences (Shanghai, People’s Republic of China).27 CLC13 cells were cultured in RPMI1640 (HyClone), supplemented with 10% FBS (Gibco), 110 µg/mL sodium pyruvate, 10 µg/mL insulin, 5.5 µg/mL transferrin, 40 ng/mL epidermal growth factor (EGF), and 6.7 ng/mL sodium selenite (Gibco). The use of CLC13 was approved by the ethical committees of Shanghai Xuhui District Central Hospital and Shanghai East China Hospital. Human liver cancer specimens were obtained from the Eastern Hepatobiliary Surgery Hospital/Institute (Shanghai, People’s Republic of China), with written informed consent from the patients and approval from the Institutional Review Board of Shanghai Eastern Hepatobiliary Surgery Hospital.
Western blot
Tumor cells or tumor tissues were lysed in radio immunoprecipitation assay buffer for Western blotting. The proteins were separated on 10% SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and electrophoretically transferred onto polyvinylidene difluoride membranes (Millipore), which were then blocked in tris-buffered saline and Tween 20 (TBST) containing 5% defatted milk for 1 h at room temperature and incubated with primary antibodies overnight at 4°C with mouse anti-tubulin (1:10,000; Proteintech Group, Chicago, IL, USA) and rabbit anti-CEBPβ (1:1,000; Santa Cruz Biotechnology, Santa Cruz, CA, USA). After being washed with TBST and incubated with either horseradish peroxidase (HRP)-conjugated goat anti-rabbit (1:5,000; Proteintech Group) or HRP-conjugated goat anti-mouse (1:5,000; Proteintech Group) secondary antibody, immune complexes were visualized by using Pierce™ enhanced chemiluminescent (ECL) Western Blotting Substrate (Thermo Fisher Scientific).
Quantitative real time-PCR (qRT-PCR)
Total RNA was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA, USA), and reverse transcription was undertaken using M-MLV Reverse Transcriptase (Promega, M530A) according to the manufacturer’s instructions. Quantitative PCR was conducted using LightCycler 480 (Roche) with SYBR Premix Ex Taq reagent (TaKaRa). All samples were examined in triplicate. Data were normalized to control samples. The primers are shown in Table S1.
Hematoxylin and eosin (H&E), immunohistochemistry, and immunofluorescence staining
Paraffin-embedded HCC sections from 14 patients were deparaffinized with xylene and rehydrated. For H&E staining, the rehydrated sections were stained with H&E (Sigma-Aldrich). For immunohistochemical (IHC) staining, the rehydrated sections were immersed in citrate buffer (pH 6.0) for antigen retrieval at 121°C for 2 min and then cooled down to room temperature slowly. The endogenous peroxidase activity was blocked with 3% hydrogen peroxide (H2O2) for 10 min. Nonspecific binding was blocked with 1% bovine serum albumin for 20 min at room temperature. The sections were incubated with rabbit anti-CEBPβ antibodies (1:200; Santa Cruz Biotechnology, Santa Cruz, CA, USA) at 4°C overnight in a humidified chamber, then incubated with HRP-conjugated goat anti-rabbit antibodies (1:500; Proteintech Group) at 37°C for 30 min, and then detected with 3,3′-diaminobenzidine (DAB) (Dako, Glostrup, Denmark) and counterstained with Mayer’s hematoxylin solution (Sigma-Aldrich). Seeing that there existed differences from IHC staining, rabbit anti-Ki67 (1:200; Invitrogen, Carlsbad, CA, USA) and mouse anti-CD133 (Proteintech Group, Chicago) and mouse anti-CD13 (Proteintech Group) antibodies were used for immunofluorescence (IF) staining. Nuclei were labeled with the blue fluorescent Hoechst 33342 dye.
Colony-formation assays
For colony-formation assays, 1,000 Huh7 cells or 2,000 CLC13 cells per well were plated on six-well plates and cultured for 7 days. Then, cells were fixed with 4% formaldehyde for 10 min and stained with crystal violet solution. The number of clones (>50 cells) was counted microscopically.
CCK8 assays
For CCK8 assays, 1,200 Huh7 cells or 3,000 CLC13 cells per well were plated on 96-well plates and incubated for 24 h. Then, 10 μL CCK8 reagent was added to each well and incubated for 2 h at 37°C. The optical density value was measured at a wavelength of 450 nm by using a microplate reader every day for 7 days.
Flow cytometry and cell sorting
Cells were dissociated into single cells by trypsinization and labeled with BV421-CD13 human antibody (301716, Biolegend, San Diego, CA, USA) at 4°C for 15 min. The concentration of the antibody used was according to the manufacturer’s recommendation. The stained cells were analyzed or collected with the FACS Calibur machine (BD Biosciences).
Cell cycle and cell apoptosis
Cell cycle and cell apoptosis were assessed by the Cell Cycle Assay Kit (CCS012, MULTI Sciences) and the Annexin V-APC/7-AAD Apoptosis Detection Kit (AP105, MULTI Sciences), respectively, following the manufacturer’s instructions.
Sphere-formation assay
Hepatoma cells were plated on six-well ultra-low attachment culture plates (Corning) in DMEM/F12 supplemented with B27, N2, 20 ng/mL EGF and 20 ng/mL basic fibroblast growth factor (bFGF), and cultured in a CO2 incubator for 7 days. The number of spheroids was counted under a microscope.
For separation of oncospheres and nonsphere cells, we collected sphere-formation medium containing both nonsphere and sphere cells in a 15-mL centrifuge tube. Pellets formed after 5-min tranquilization were recognized as spheres. The supernatant was then moved into a new centrifuge tube carefully and centrifuged at 1,500g for 5 min to obtain nonspheric pellets.
In vitro migration and invasion assays
Cell migration was assessed by wound-healing and Transwell assays. For the wound-healing assay, a scratch was made in the center of the culture dish using a sterile 20-μL pipette tip before cells were fully confluent. The wound was observed using a bright-field microscope at 24- and 48-h after scratching. The migration rate = (scratch widths before migration – scratch widths after migration)/scratch widths before migration ×100%. Transwell migration assay was conducted using 24-well Transwell Chambers without Matrigel (BD Biosciences) according to the manufacturer’s instructions. Briefly, 1×105 Huh7 cells or 1×105 CLC13 cells in medium with 10% FBS were added to the top chamber, and the medium containing 20% FBS was added to the bottom chamber as an attractant. After 48-h incubation, the migrated cells were fixed and stained with crystal violet. The number of migrated cells was counted in 10 fields under a 10× objective lens.
Transwell invasion assay was conducted using 24-well BioCoat Matrigel Invasion Chambers (BD Biosciences) according to the manufacturer’s instructions. Briefly, 4×105 Huh7 cells in medium with 10% FBS were added to the top chamber, and the medium containing 20% FBS was added to the bottom chamber as an attractant. The remaining steps are the same as those described for the Transwell migration assay.
In vivo xenograft experiments
For tumor growth assays, 5×105 Huh7 cells or 1×106 CLC13 cells were subcutaneously injected into 6-week-old male BALB/c nude mice. Every 3 days, tumor volume was calculated by the formula V=0.5W2L (V, volume; L, length; and W, width). BALB/c nude mice were obtained from the Animal Centre of the Chinese Academy of Medical Sciences, they were fed in standard pathogen-free conditions, and all animal experiments were in accordance with the Guide for the Care and Use of Laboratory Animals and were approved by the Institutional Animal Care and Use Committee of the Second Military Medical University.
Statistical analysis
All data are presented as the mean±SD. Statistical methods were indicated in figure legends, and statistical computations were conducted using GraphPad Prism version 6.0. P<0.05 was considered statistically significant; *, **, and *** indicate P<0.05, P<0.01, and P<0.001, respectively.
Results
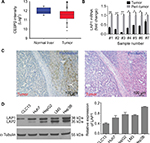
LAP1 is weakly expressed in HCC
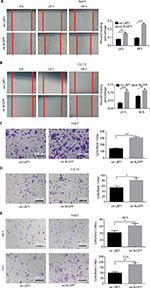
The result of an online-available HCC transcriptome dataset (GSE62232) analysis showed that CEBPβ was weakly expressed in HCC tumors (Figure 1A, Student’s t-test, *P<0.05). In addition, qRT-PCR showed that the expression level of LAP1 mRNA in HCC tissues was significantly lower than that in seven pairs of matched para-tumoral liver tissues (Figure 1B, Student’s t-test, *P<0.05). These findings were further validated by IHC staining. At the same time, H&E staining of the same tissue samples was done to confirm the pathological type of HCC (Figure 1C). The result showed that LAP1 was significantly downregulated in HCC tumor tissues as compared with para-tumoral tissues. Then, we measured the expression level of CEBPβ in five human hepatoma cell lines (Huh7, Hep3B, HepG2, LM3, and CLC13) and found that Huh7 and CLC13 cell lines contained the lowest amounts of LAP1 among these cell lines (Figure 1D) and, therefore, these two cell lines were used in our further experiments.
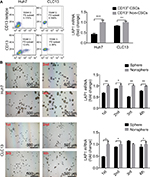
LAP1 is weakly expressed in LCSCs
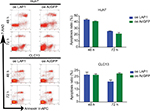
Knowing that CSCs play complex roles in the development and progression of cancer, we further investigated the expression of LAP1 in LCSCs. CD13 is known to be a target for identifying CSCs in a proportion of HCC; therefore, we sorted CD13+ cells from Huh7 and CLC13 HCC cell lines as LCSCs. Then, we tested the LAP1 expression in CD13+ CSCs and CD13− non-CSCs of the two cell lines and found that the LAP1 expression in CD13+ CSCs was significantly lower than that in CD13− non-CSCs (Figure 2A). Furthermore, we conducted an oncosphere experiment, and the results showed that LAP1 expression in CSC-enriched spheres was significantly lower than that in nonspheres. Importantly, through serial passage of Huh7 and CLC13 sphere cells, similar observations were obtained in the following three generations of oncosphere assay (Figure 2B). Altogether, LAP1 was weakly expressed in HCC tumor tissues and LCSCs.
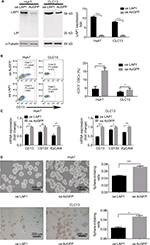
LAP1 inhibits the stemness of LCSCs
To explore the function of LAP1 in HCC, we constructed Huh7 and CLC13 cells stably expressing LAP1 by using the lentivirus infection system, and an pLVX-AcGFP1-N1 (AcGFP)-overexpressing empty vector was used as the control (Figure 3A). Notably, flow cytometric analysis revealed that LAP1 overexpression decreased the number of CD13+ liver CSCs markedly (Figure 3B). Compared with the control, the mRNA level of CSC markers CD133, CD13, and EpCAM was significant downregulated in both Huh7 and CLC13 cells infected with LAP1 (Figure 3C). Furthermore, sphere-formation experiments showed that LAP1 overexpression dramatically decreased oncosphere formation as compared with the AcGFP-overexpressing group (Figure 3D). These data indicate that LAP1 overexpression suppressed the stemness features of LCSCs.
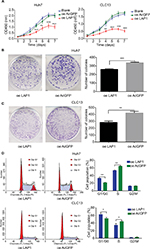
LAP1 suppresses the proliferation of HCC cell lines in vitro
Seeing that LAP1 inhibited the stemness features of LCSCs, we explored whether LAP1 played a critical role in the expansion of HCC cell lines. We conducted CCK8 cell-proliferation and colony-formation assays to explore the effect of LAP1 expression on the proliferation of HCC cells. Compared with the control, LAP1 overexpression suppressed the proliferation of Huh7 and CLC13 cells markedly (Figure 4A). The colony-formation assay showed that LAP1 overexpression could also markedly reduce the frequency of colony formation in Huh7 and CLC13 cells (Figure 4B and C).
To distinguish between the possibilities that LAP1 overexpression inhibited growth or caused cell death in hepatoma cells, we undertook flow cytometry to quantitate the population of apoptotic cells and cell cycle distribution. Compared with AcGFP overexpression, LAP1 overexpression promoted G1/G0 phase arrest in Huh7 and CLC13 cells (Figure 4D), whereas no cell apoptosis was observed in these two cell lines (Figure S1).
LAP1 inhibits the migration and invasion of HCC
To explore the function of LAP1 on the migration and invasion of HCC cells, wound-healing and Transwell migration assays were conducted. It was found that LAP1 suppressed the migratory ability of Huh7 and CLC13 cells efficiently (Figure 5A–D). The result of a Matrigel invasion assay showed that LAP1 suppressed the invasive ability of Huh7 cells (Figure 5E). These findings demonstrate that LAP1 possesses antitumor activity in HCC cells.
LAP1 suppressed the tumor proliferation and stemness of HCC in vivo
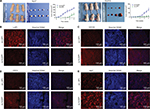
For the in vivo tumor-formation assay, tumor formation was observed in nude mice injected with LAP1-transfected and AcGFP empty vector-transfected Huh7 and CLC13 cells. In addition, the tumor size and tumor volume in the LAP1 overexpression group were significantly smaller than those in the AcGFP empty vector-transfected cell group. The tumor size was measured every 3 days and, after 1 month, the mice were sacrificed, and the tumors were removed and weighed (Figure 6A). Then, the tumors were paraffin embedded and IF stained. Using a rabbit CEBPβ antibody, the overexpression of LAP1 was confirmed (Figure 6B). Furthermore, the result showed that the intensity of CD13, CD133, and Ki67 was obviously lower in LAP1-overexpressing CLC13 cells than in the control, indicating that LAP1 overexpression was negatively correlated with the LCSC markers CD13 and CD133 as well as Ki67 (Figure 6C–E). Taken together, LAP1 overexpression inhibited xenograft tumor growth and stemness markedly, suggesting that LAP1 plays a tumor suppressor role in HCC.
Discussion
Several studies have highlighted the crucial role of CEBPβ in suppressing tumor features. For example, forced expression of CEBPβ in HepG2 hepatocarcinoma cells induced arrest of cells at or near the G1–S boundary.28 CEBPβ knockout mice developed a lymphoproliferative disorder, suggesting that CEBPβ inhibited the expansion of the lymphoid cell compartment.29 CEBPβ was found to be expressed at a low level in hematological malignancies in chronic myelogenous leukemia. CEBPβ played a role in the BCR/ABL of bone marrow progenitor cells by activating the transcription and expression of the downstream target gene GADD45A, thus promoting cell differentiation and inhibiting cell proliferation.30
However, growth arrest induced by CEBPβ is highly context specific because CEBPβ was found to display growth-promoting activity in several cases. CEBPβ can function as a pro-oncogenic transcription factor that promotes proliferation and/or survival of some tumor cells. For example, deletion of the CEBPβ gene rendered mice totally resistant to carcinogen-induced skin tumor development.31 Shimizu et al32 reported that CEBPβ inhibited apoptosis and improved the survival rate of pancreatic cancer cells. Gardiner et al33 found that CEBPβ-1 overexpression increased transformation and upregulated the expression of the CSC marker ALDH1A1, leading to chemoresistance in Ewing sarcoma. Therefore, CEBPβ can function as a pro-oncogenic transcription factor that promotes proliferation and survival of some tumor cells. How it stimulates mitotic growth and why it elicits a completely opposite effect on proliferation in different cellular contexts are intriguing questions for future investigation.
The incidence and mortality of HCC are at the forefront of all malignancies.1,3 Although great progress has been made in the diagnosis and treatment of liver cancer, tumor recurrence and metastasis in patients with liver cancer remain the main reasons for poor prognosis.4 CSCs have some characteristics including self-renewal, differentiation, and the ability to form a new tumor.34 Studies have confirmed that CSCs may be responsible for cancer development, relapse, and metastasis. Human HCC is driven and maintained by LCSCs. Thus, targeting CSCs may become a promising therapeutic strategy to cure deadly malignancies.
Furthermore, CEBPβ expression has been linked to a variety of rodent and human cancers.20 CEBPβ appears to play a critical role in the development of both the mammary gland and cancers through its involvement in the development, differentiation, and proliferation of mammary epithelial cells.35–37
LAP1 is the most highly expressed isoform of CEBPβ, which is a liver-enriched transcription factor involved in cellular metabolism, development, proliferation, and differentiation. In this study, our observations were confirmed by Park’s cohort dataset GSE62232. We found that LAP1 was weakly expressed in HCC tumors and LCSCs, indicating that LAP1 may play a negative regulatory role in CSCs. However, no study has explored the function of LAP1 in LCSCs and other kinds of CSCs.
The self-renewal capacity of CSCs is known to be one of the important features for the development of cancer. To explore the function of LAP1 on the self-renewal capacity of LCSCs, we first measured the effect of LAP1 on the proliferative capacity of HCC cell lines. The results showed that LAP1 overexpression was able to inhibit the proliferative capacity of HCC cells, and that LAP1 could attenuate the stemness of HCC. Moreover, stem cell–associated membrane markers can be used to measure the stemness of HCC cells. Knowing that CD133, CD13, and EpCAM are the most useful stemness biomarkers, we conducted qRT-PCR and IF to measure their expressions, and found that LAP1 could decrease the expression of CD133, CD13, and EpCAM, thus suggesting that LAP1 could attenuate the stemness of HCC and inhibit the self-renewal ability of LCSCs. Furthermore, the sphere-formation experiment was conducted, knowing that it is an important experiment to measure the self-renewal capacity of HCC. We found that LAP1 overexpression could inhibit the sphere formation of HCC in terms of the size and number of the spheres. According to the theory of CSCs, the subcutaneous tumor-formation ability is an important method to measure the self-renewal capacity of CSCs in mice. To explore the influence of LAP1 on the stemness of LCSCs, we transplanted HCC to the mice and built a subcutaneous tumor mice model. After approximately 1 month, differences between the LAP1 overexpression and control groups were observed. It was found that LAP1 could significantly inhibit the formation and growth of subcutaneous tumors; furthermore, when the tumors were paraffin embedded and IF stained, we found the intensities of CD13, CD133, and Ki67 were obviously lower in LAP1 overexpressing CLC13 cells than in the control, which strongly supports the conclusion that LAP1 can attenuate the stemness of LCSCs and HCCs. At the same time, we measured other tumor biological functions of LAP1 by the Transwell assay and scratch test, and found that LAP1 overexpression could inhibit the migration and invasion of HCC in vitro. Furthermore, LAP1 overexpression could cause cell cycle arrest in the G1/G0 phase, which also indicates the tumor suppressor gene function of this LAP1.
The above results demonstrated that LAP1 could suppress the self-renewal capability of LCSCs and may prove to be a target for eradicating LCSCs and a novel promising therapeutic approach for inhibiting liver cancer.
Conclusion
LAP1 was weakly expressed in HCC and LCSCs. LAP1 could suppress the self-renewal capability of LCSCs and possessed a tumor-suppressive effect on HCC cells, suggesting that LAP1 may serve as an optimal target in liver CSC-targeted therapy.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (grant numbers 31771511 and 31401166), Shanghai Committee of Science and Technology of China (grant number 15JC1403900), and China Postdoctoral Science Foundation (grant number 2013M542434).
Author contributions
All authors contributed to the conception and design of the study, acquisition of data, or analysis and interpretation of the data, and to the critical revision of the manuscript for important intellectual content, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. | ||
Wang R, Sun Q, Wang P, et al. Notch and Wnt/β-catenin signaling pathway play important roles in activating liver cancer stem cells. Oncotarget. 2016;7(5):5754–5768. | ||
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66(1):7–30. | ||
Maluccio M, Covey A. Recent progress in understanding, diagnosing, and treating hepatocellular carcinoma. CA Cancer J Clin. 2012;62(6):394–399. | ||
Hao J, Zhang Y, Deng M, et al. MicroRNA control of epithelial-mesenchymal transition in cancer stem cells. Int J Cancer. 2014;135(5):1019–1027. | ||
Chiba T, Iwama A, Yokosuka O. Cancer stem cells in hepatocellular carcinoma: therapeutic implications based on stem cell biology. Hepatol Res. 2016;46(1):50–57. | ||
Charafe-Jauffret E, Ginestier C, Iovino F, et al. Breast cancer cell lines contain functional cancer stem cells with metastatic capacity and a distinct molecular signature. Cancer Res. 2009;69(4):1302–1313. | ||
Mackillop WJ, Ciampi A, Till JE, Buick RN. A stem cell model of human tumor growth: implications for tumor cell clonogenic assays. J Natl Cancer Inst. 1983;70(1):9–16. | ||
Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997;3:730–737. | ||
MacDonagh L, Gray SG, Breen E, et al. Lung cancer stem cells: the root of resistance. Cancer Lett. 2016;372(2):147–156. | ||
Tang DG, Patra Wala L, Calhoun T, et al. Prostate cancer stem/progenitor cells: identification, characterization, and implications. Moll Carcinogen. 2007;46(1):1–14. | ||
Lathia JD, Mack SC, Mulkearns-Hubert EE, Valentim CL, Rich JN. Cancer stem cells in glioblastoma. Genes Dev. 2015;29(12):1203–1217. | ||
Lee CJ, Dosch J, Simeone DM. Pancreatic cancer stem cells. J Clin Oncol. 2008;26(17):2806–2812. | ||
Tong CM, Ma S, Guan XY. Biology of hepatic cancer stem cells. J Gastroenterol Hepatol. 2011;26(8):1229–1237. | ||
Ji J, Wang XW. Clinical implications of cancer stem cell biology in hepatocellular carcinoma. Semin Oncol. 2012;39(4):461–472. | ||
Yamashita T, Wang XW. Cancer stem cells in the development of liver cancer. J Clin Invest. 2013;123(5):1911–1918. | ||
Yang Q, Wang Y, Yang Q, et al. Cuprous oxide nanoparticles trigger ER stress-induced apoptosis by regulating copper trafficking and overcoming resistance to sunitinib therapy in renal cancer. Biomaterials. 2017;146:72–85. | ||
Yu B, Wang Y, Yu X, et al. Cuprous oxide nanoparticle-inhibited melanoma progress by targeting melanoma stem cells. Int J Nanomedicine. 2017;12:2553–2567. | ||
Descombes P, Schibler U. A liver-enriched transcriptional activator protein, LAP, and a transcriptional inhibitory protein, LIP, are translated from the same mRNA. Cell. 1991;67(3):569–579. | ||
Sebastian T, Johnson PF. Stop and go: anti-proliferative and mitogenic functions of the transcription factor C/EBPbeta. Cell Cycle. 2006;5(9):953–957. | ||
Ramji DP, Foka P. CCAAT/enhancer-binding proteins: structure, function and regulation. Biochem J. 2002;365(Pt 3):561–575. | ||
Saint-Auret G, Danan JL, Hiron M, et al. Characterization of the transcriptional signature of C/EBPbeta isoforms (LAP/LIP) in Hep3B cells: implication of LIP in pro-survival functions. J Hepatol. 2011;54(6):1185–1194. | ||
Li W, Kessler P, Yeger H, et al. A gene expression signature for relapse of primary wilms tumors. Cancer Res. 2005;65(7):2592–2601. | ||
Oya M, Horiguchi A, Mizuno R, Marumo K, Murai M. Increased activation of CCAAT/enhancer binding protein-beta correlates with the invasiveness of renal cell carcinoma. Clin Cancer Res. 2003;9(3):1021–1027. | ||
Homma J, Yamanaka R, Yajima N, et al. Increased expression of CCAAT/enhancer binding protein beta correlates with prognosis in glioma patients. Oncol Rep. 2006;15(3):595–601. | ||
Fang T, Cui M, Sun J, et al. Orosomucoid 2 inhibits tumor metastasis and is upregulated by CCAAT/enhancer binding protein β in hepatocellular carcinomas. Oncotarget. 2015;6(18):16106–16119. | ||
Qiu Z, Zou K, Zhuang L, et al. Hepatocellular carcinoma cell lines retain the genomic and transcriptomic landscapes of primary human cancers. Sci Rep. 2016;6:27411. | ||
Buck M, Turler H, Chojkier M. LAP (NF-IL-6), a tissue-specific transcriptional activator, is an inhibitor of hepatoma cell proliferation. EMBO J. 1994;13(4):851–860. | ||
Screpanti I, Romani L, Musiani P, et al. Lymphoproliferative disorder and imbalanced T-helper response in C/EBP beta-deficient mice. EMBO J. 1995;14(9):1932–1941. | ||
Guerzoni C, Bardini M, Mariani SA, et al. Inducible activation of CEBPB, a gene negatively regulated by BCR/ABL, inhibits proliferation and promotes differentiation of BCR/ABL-expressing cells. Blood. 2006;107(10):4080–4089. | ||
Zhu S, Yoon K, Sterneck E, Johnson PF, Smart RC. CCAAT/enhancer binding protein-beta is a mediator of keratinocyte survival and skin tumorigenesis involving oncogenic Ras signaling. Proc Natl Acad Sci U S A. 2002;99(1):207–212. | ||
Shimizu Y, Kishimoto T, Ohtsuka M, et al. CCAAT/enhancer binding protein-beta promotes the survival of intravascular rat pancreatic tumor cells via antiapoptotic effects. Cancer Sci. 2007;98(11):1706–1713. | ||
Gardiner JD, Abegglen LM, Huang X, et al. C/EBPβ-1 promotes transformation and chemoresistance in Ewing sarcoma cells. Oncotarget. 2017;8(16):26013–26026. | ||
Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414(6859):105–111. | ||
Zahnow CA. CCAAT/enhancer binding proteins in normal mammary development and breast cancer. Breast Cancer Res. 2002;4(3):113–121. | ||
Grimm SL, Rosen JM. The role of C/EBPbeta in mammary gland development and breast cancer. J Mammary Gland Biol Neoplasia. 2003;8(2):191–204. | ||
Zahnow CA. CCAAT/enhancer-binding protein beta: its role in breast cancer and associations with receptor tyrosine kinases. Expert Rev Mol Med. 2009;11:e12. |
Supplementary materials
  | Table S1 Real-time PCR primers used in this study Abbreviations: EpCAM, epithelial cell adhesion molecule; PCR, polymerase chain reaction. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.