Back to Journals » Cancer Management and Research » Volume 12
LINC02381 Promoted Cell Viability and Migration via Targeting miR-133b in Cervical Cancer Cells
Authors Chen X, Zhang Z, Ma Y, Su H, Xie P, Ran J
Received 4 November 2019
Accepted for publication 19 April 2020
Published 26 May 2020 Volume 2020:12 Pages 3971—3979
DOI https://doi.org/10.2147/CMAR.S237285
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Sanjeev K. Srivastava
Xiaohua Chen, Zhuxiang Zhang, Yan Ma, Hongxin Su, Peng Xie, Juntao Ran
Department Of Radiation Therapy, First Hospital Of Lanzhou University, Lanzhou City, Gansu Province 730000, People’s Republic of China
Correspondence: Juntao Ran
Department Of Radiation Therapy, First Hospital Of Lanzhou University, No. 1 Donggang West Road, Chengguan District, Lanzhou City, Gansu Province 730000, People’s Republic of China
Tel +86 931-8356435
Email [email protected]
Background: It has been proved that lncRNAs could function as CeRNA for miRNAs in tumor growth and metastasis for cervical cancer. This paper aims to identify the role of LINC02381 in cervical cancer cells.
Materials and Methods: RT-qPCR was utilized to measure the expression levels of LINC02381 in cervical cancer tissues and cells. MTT, colony formation assay, transwell assay, RT-qPCR, and Western blotting were performed to investigate the roles of LINC02381 in cervical cancer cells. RegRNA 2.0 was used to predict the miRNA-binding sites of LINC02381. Luciferase reporter assay and RT-qPCR were employed to confirm the sponging effect between miR-133b and LINC02381.
Results: This study showed that LINC02381 was up-regulated in cervical cancer cells and acted as an oncogene in the development of cervical cancer. LINC02381 promoted cell viability and metastasis via sponging miR-133b. Moreover, miR-133b could target its downstream mediator of RhoA and inhibit its expression.
Conclusion: Overall, our results indicated that LINC02381 functions as an oncogene in cervical cancer and could serve as a novel target for cervical cancer therapies in the future.
Keywords: LINC02381, miR-133b, RhoA, cervical cancer, cell viability
Introduction
Cervical cancer ranges the 3rd most severe female cancer in the United States, and the 2nd most common cause of deaths from cancers in women worldwide, second only to breast cancer.1–3 It has been well established that HPV infections implicated in most cases of cervical squamous cell cancer cases worldwide.4,5 Scientists have made tremendous efforts for the tumorigenesis of cervical cancer, in the fields of long non-coding RNAs (lncRNA) and microRNAs (miRNAs).6–8
It has been proved that lncRNAs could function as CeRNA for miRNAs in the proliferation and migration of tumor cells.9,10 For cervical cancer, many types of ceRNAs have been explored, including RSU1P2,11 PVT1,12 MALAT1,13 and MEG3.14 LINC02381 is an important member of long intergenic non-protein coding RNAs (lincRNAs). As a link to cancer biology, lincRNAs were found to control transcriptional alteration, implying that they are strongly associated with cancer progression. Previous reports demonstrated that lincRNAs were differentially regulated and expressed in the development of tumors, including breast cancer,15 human colorectal cancer,16 and pancreatic cancer.17 There are more than 3000 lincRNAs existing in the human body, however, only less than 1% of them have been characterized.18–20 Although some lincRNAs were revealed for their impacts on certain types of human cancer, the underlying molecular mechanisms are still largely unknown. In this paper, we aimed to investigate the functions of LINC02381 in cervical cancer.
Mature microRNAs (miRNAs, 21–25 nucleotides) are playing important regulatory roles in cell growth, proliferation, differentiation and cell death.21 Previous studies have confirmed that miRNAs are aberrantly expressed in several types of human cancer, and could act as either oncogenes or tumor suppressors.22–24 miR-133b was found to differentially expressed in the tumor suppression in esophageal squamous cell carcinoma,25 colorectal cancer,26 and gastric cancer.27 One study found that miR-133b was the main promoter for cervical cancer development via the activation of ERK and AKT1 pathway.28 Up-regulated miR-133b in cervical tumors might work as an oncogene through negative regulation of tumor suppressive genes.28 These findings indicated that mRNA-133b would be a useful biomarker for cervical cancer diagnosis and prognosis. Besides lncRNAs and mRNAs, researchers have found that overexpression of RhoA promoted the proliferation and migration of cervical cancer cells.29 However, further analysis is needed to clarify the role of miR-133b and RhoA in cervical cancer.
This paper aims at investigating the functions of LINC02381, miR-133b and RhoA, and their interactions in cell proliferation and migration in cervical cancer cells. We conducted a series of characterization experiments, including qRT-PCR assay, cell proliferation analysis, cell invasion and migration analysis, dual-luciferase reporter assay, RNA pull-down assay, and Western blot to examine their effects on cervical tumor cell growth and migration. Our findings might provide valuable insights into the discovery of novel therapeutic targets of cervical cancer.
Materials and Methods
Tissue Samples
Human cervical tumor tissues were sectioned and obtained from the First Hospital of Lanzhou University. There were thirty groups of cervical tumor tissues and normal tissues, all of which were taken from the patients under surgeries. Cervical cancer patients did not receive any chemotherapy, immunotherapy, or radiotherapy before the surgery. The experiment protocols were approved by the ethics board of the hospital.
qRT-PCR Assay
Total RNAs from the tumor tissues, normal tissues or cell lines were extracted using TRIzol (Invitrogen, USA). cDNA was synthesized using the reverse transcription kit (Takara, Japan). FTC-3000 real-time quantitative thermal cycler (Funglyn Biotech INC, Shanghai, China) was used to measure expression levels of mRNA. GAPDH was used as the endogenous control. The 2−ΔΔCt method was used for gene expression quantification. The primer sequences were as follows: LINC02381, 5ʹ-CTGATGGCCACTCACGCTAT-3ʹ (forward) and 5ʹ- GATCCGGAGGGAGAGCATTC-3ʹ (reverse); GAPDH, 5ʹ-TCCTGTGGCATCCACGAAACT-3ʹ (forward) and 5ʹ-GAAGCATTTGCGGTGGACGAT-3ʹ (reverse).
Cell Culture and Transfection
Ect1/E6E7, HeLa, CaSki, and SIHA were obtained from the Cell Bank of The Chinese Academy of Sciences (Shanghai, China). Cells were cultured in DMEM with 10% fetal bovine serum at 37 °C with 5% CO2. Cells were plated onto six-well dishes at 24 h and then transfected with a concentration of 25 nM control or experimental siRNA for LINC02381 per dish at 80% confluence using the lipofectamine 2000 (Invitrogen Life Technology, Carlsbad, CA, USA) according to the manufacturer’s protocol. The concentration of NC-inhibitor or miR-133b-inhibitor was 50 nM. The LINC02381 plasmids were transfected using the same protocol with 400 ng/well.
Cell Viability Analysis
After transfection with siRNAs for twenty-four hours, cells were seeded in a 96-well plate (5000 cells/well). Cell viability was measured every four hours in 5 mg/mL MTT solution and OD value were read at 570 nm. HeLa or SIHA cells (~300) were planted in a 6-cm petri dish. Colonies were pictured and counted after 14 d. All experiments were performed in 3 groups with 4 duplicates.
Cell Invasion and Migration Analysis
A total of 2×105 cells were put in 8-μm Transwell inserts (BD Biosciences, USA) with a Matrigel-coated membrane. And 20% fetal bovine serum in medium was put to the lower chamber. After 2 days, the residuals on the upper were wiped away. Invasive cells were fixed with methanol and stained with 1% crystal violet. Cells were pictured and counted. A total of 3.5×105 cells were cultured in a 6-well plate. A scratch was made using a 200-μL pipette tip. After washing, cells were pictured on day 0 and day 1. It should be noted that no growth inhibitor was added to the scratch test.
Dual-Luciferase Reporter Assay
LINC02381 and miR-133b, miR-133b mimic, NC mimic, miR-133b-inh, and inh-NC were co-transfected with pmirGlo-NC, pmirGlo-LINC02381-mut or pmirGlo-LINC02381-wt to HeLa and SIHA cells. miR-133b and RhoA, miR-133b mimic, NC mimic, miR-133b-inh and inh-NC were co-transfected with pmirGlo-NC, pmirGlo- RhoA-3ʹUTR-wt or pmiRGlo-RhoA-3ʹUTR-mut to HeLa and SIHA cells. The relative luciferase activities were measured using the dual-luciferase reporter assay kit (Promega, USA).
RNA Pull-Down Assay
HeLa cells were transfected with negative control, miR-133b and miR-133b-Mut for 2 days. These RNAs were labeled with biotin. Cells were incubated with magnetic beads (M-280 streptavidin, Invitrogen, USA) at 4 °C for four hours. Co-precipitated RNA was isolated via lysis buffer with proteinase K (Invitrogen) and 10% SDS. The expression levels of RNAs were detected by qRT-PCR.
Western Blot
Total proteins were extracted at 2 d post-transfection and separated via SDS-PAGE. Antibodies to RhoA (Cat# 2117) and GAPDH (Cat#5174) were bought from Cell Signaling Technology (Leiden, Netherlands). Protein bands were visualized and quantified.
Statistical Analysis
Data are expressed as the mean ± standard deviation (SD) from > 3 independent results. Statistical analysis was performed using Graph Pad Prism 7.0 software. Unpaired Student’s t-test and one-way ANOVA with Tukey’s HSD test was used for comparing differences between two groups or among multiple groups, respectively. *P < 0.05, **P < 0.01 and ***P < 0.001.
Results
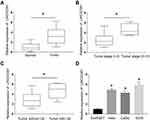
LINC02381 Was Up-Regulated in Cervical Cancer Tissues and Cell Lines
Figure 1A illustrates that the expression of LINC02381 was remarkably up-regulated in cervical cancer tissues (p < 0.05). The patients with higher expression levels of LINC02381 had advanced tumor stage and lymph nodes metastasis (Figure 1B and C). As shown in Figure 1D, LINC02381 was significantly up-regulated in cervical cancer cell lines of Hela, SIHA, and CasKi, compared to the normal Ect/E6E7 cervical cells (p < 0.05). All these results confirmed that LINC02381 was up-regulated in cervical cancer tissues and cell lines.
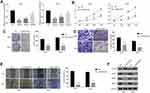
Knockdown of LINC02381 Inhibited Cell Viability, Migration, and Invasion of Cervical Cancer
The transfection of si-LINC02381 remarkably down-regulated the expression of LINC02381 in Hela and SIHA, compared to that in the NC group (p < 0.05) (Figure 2A). SiRNA1 was selected for function analysis due to the best knockdown efficacy. Knockdown of LINC02381 greatly suppressed cell viability (p < 0.05), migration (p < 0.01) and invasion (p < 0.05) in Hela and SIHA than cells transfected with miR-NC (Figure 2B–E). Furthermore, knockdown of LINC02381 down-regulated the expression of epithelial cell marker E-cadherin and up-regulated the expression of mesenchymal cell marker N-cadherin (Figure 2F). MMP2 and MMP9, involved in the degradation of extracellular matrix in tumor cell invasion, were also down-regulated with the knockdown of LINC02381 (Figure 2F). These data demonstrated that LINC02381 played important roles in regulating cervical cancer progression.
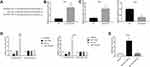
miR-133b Was a Direct Target of LINC02381
Figure 3A reveals the potential binding site between LINC02381 and miR-133b, suggesting miR-133b might be a target of LINC02381. Silencing of LINC02381 elevated the expression levels of miR-133b in HELA (Figure 3B), while overexpression of LINC02381 had an opposite effect (Figure 3C). Figure 3D demonstrates that miR-133b mimic greatly suppressed the luciferase activity of Wt-LINC02381 in Hela and SIHA (p < 0.05). But miR-133b inhibitor increased the activity (p < 0.05). RNA pulldown assay showed that Bio-miR-133b-wt could remarkably elevate the expression levels of LINC02381 but Bio-miR-133b-mut could not (p < 0.05) (Figure 3E). These findings suggested that miR-133b was a direct target of LINC02381 in cervical cancer cells.
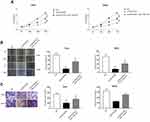
miR-133b Reversed the Suppression Effects of LINC02381 Knockdown on Cervical Cancer Cells
4B and 4C demonstrate the MTT assay (Figure 4A), wound healing assay (Figure 4B) and transwell invasion assay (Figure 4C), respectively. Through these results, we confirmed that knockdown of LINC02381 significantly suppressed cell viability, migration and invasion in Hela cells and SIHA cells. However, miR-133b inhibitor reversed the suppressive effects by knockdown the expression of LINC02381. These data indicated that LINC02381 could regulate cervical cancer cell progression by mediating miR-133b.
LINC02381 Modulated the Expression of RhoA by Regulating miR-133b in Cervical Cancer Cell Lines
In order to determine whether miR-133b exerts its function through regulating the expression of downstream protein, we searched its potential target protein on targetscan. Figure 5A reveals the shared binding cites between RhoA and miR-133b. miR-133b mimic remarkably suppressed the luciferase activity of Wt-RhoA in Hela and SIHA (Figure 5B). However, miR-133b inhibitor increased this activity. RhoA GTPase has been reported to function as a key regulator of cell polarity and tight junction formation and stability in cells. Knockdown of LINC02381 resulted in a significant decrease of the expression levels of RhoA (p < 0.05) (Figure 5C), but miR-133b inhibitor reversed the effects. In addition, the expression of LINC02381 was negatively correlated with the expression of miR-133b, but positively correlated with the expression of RhoA in cervical cancer tissues (Figure 5D). These results indicated that LINC02381 modulated RhoA via regulating miRNA-133b in cervical cancer.
Discussion
Recent studies have shown that lincRNAs are improperly regulated and expressed in human cancers.20 Many lincRNAs were found differentially expressed between primary breast carcinomas.30 It has been found that some lincRNAs can control the transcriptional alteration, suggesting that the abnormal lincRNA profiling between normal cells and cancer cells is strongly related to cancer progression. To the best of our knowledge, no lincRNAs have been profiled to aid the cervical cancer diagnosis, prognosis, or the selection of potential therapeutic methods. Although lincRNAs have impacts on many other human diseases, the basis of their molecular mechanisms in cervical cancer are largely unknown.
The effects and expressions of lincRNAs were only discovered in several types of cancer such as lincRNA-RoR in breast cancer,30 and lincRNA-p21 in colorectal cancer.16 Previous studies have revealed that its development was largely related to the up-regulation of related long non-coding RNAs like GAS5,31 PVT1,12 and MEG314 in cervical cancer. All the data from these literature found that the levels of lncRNAs were elevated during the development of cervical tumors, as well as cervical cancer cell lines.32,33 Our qRT-PCR analysis investigated the LINC02381 expression profiles in 30 cervical cancer tissues and 30 adjacent normal tissues. The expression of LINC02381 was significantly upregulated in cervical cancer tissues, as well as the cervical cancer cell lines of Hela, SIHA, and CaSki. These results are consistent with the findings from other studies.
In 2016, HJ. Kim examined the role of lncRNA HOXA11 in regulating cervical cancer, in the aspect of tumor progression and stemness maintenance.34 They found that knockdown of HOXA11 decreased cell proliferation in cervical cancer cells.34 In our research, we also noticed that the knockdown of LINC02381 significantly inhibited cell proliferation, migration and invasion cervical cancer cell lines. Our results demonstrated that LINC02381 plays crucial roles in the regulation of cervical cancer progression.
LncRNA could serve as competing endogenous RNAs and exert its regulatory functions in different human cancers via sponging of miRNAs.35,36 One study found that lincRNA-RoR targeted and sponged miR-145, which regulated the cell invasion in triple-negative breast cancers.30 For cervical cancer, we successfully predicted miR-133b was a potential target of LINC02381, and knockdown of LINC02381 decreased the expression of miR-133b, while overexpression of LINC02381 played opposite effects. Our wound healing and transwell invasion assays revealed that miR-133b inhibitor could reverse the effect of knockdown of LINC02381, which significantly inhibited cell proliferation migration and invasion. Our results showed that LINC02381 regulated cervical cancer cell progression through mediating miR-133b, which were in agreement with previous studies.
miR-133b was reported to be an important promoter for cervical cancer cell proliferation.26 The elevation of miR-133b could indicate the progression of activated cells to develop into cervical cancer. They also found that miR-133b had an oncogenic effect to promote tumorigenesis and metastasis in cervical cancer cells.26 RhoA was widely known as an oncogene that promotes cancer progression in various types of cancer.37,38 It was identified as a downstream effector of miR-133b in cervical cancer.28 We predicted the common binding sites between RhoA and miR-133b. The dual-luciferase reporter assays revealed that transfection with miR-133b mimics significantly inhibited the luciferase activity of Wt-RhoA in both Hela and SIHA cells, whereas miR-133b inhibitor increased the activity. In cervical cancer tissues, the expression of LINC02381 was negatively correlated with the expression of miR-133b, but positively correlated with RhoA. Limited researches have been conducted on the effects of lincRNA, miR-133b and RhoA on cervical cancer development. Our findings could serve as a bridge between the gaps.
Conclusions
The findings in this study proved that LINC02381 promoted cell viability and migration via the sponging of miR-133b in cervical cancer cells. LINC02381 may be used as a novel prognostic tool in cancer management.
Ethical Statement
The research has been carried out in accordance with the World Medical Association Declaration of Helsinki. All patients and healthy volunteers provided written informed consent prior to their inclusion within the study.
Author Contributions
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Waggoner SE. Cervical cancer. lancet. 2003;361(9376):2217–2225. doi:10.1016/S0140-6736(03)13778-6
2. Clifford G, Smith J, Plummer M, Munoz N, Franceschi S. Human papillomavirus types in invasive cervical cancer worldwide: a meta-analysis. Br J Cancer. 2003;88(1):63. doi:10.1038/sj.bjc.6600688
3. Peng L, Yuan X, Jiang B, Tang Z, Li G-C. LncRNAs: key players and novel insights into cervical cancer. Tumor Biol. 2016;37(3):2779–2788. doi:10.1007/s13277-015-4663-9
4. Munoz N, Bosch FX, de Sanjosé S, et al. Epidemiologic classification of human papillomavirus types associated with cervical cancer. New Eng j Med. 2003;348(6):518–527. doi:10.1056/NEJMoa021641
5. Castellsagué X. Natural history and epidemiology of HPV infection and cervical cancer. Gynecol Oncol. 2008;110(3):S4–S7. doi:10.1016/j.ygyno.2008.07.045
6. Wang Y-D, Cai N, Wu X, Cao H, Xie L, Zheng P. OCT4 promotes tumorigenesis and inhibits apoptosis of cervical cancer cells by miR-125b/BAK1 pathway. Cell Death Dis. 2013;4(8):e760. doi:10.1038/cddis.2013.272
7. Stanley MA, Browne HM, Appleby M, Minson AC. Properties of a non-tumorigenic human cervical keratinocyte cell line. Int j Cancer. 1989;43(4):672–676. doi:10.1002/ijc.2910430422
8. Ma -Y-Y, Wei S-J, Lin Y-C, et al. PIK3CA as an oncogene in cervical cancer. Oncogene. 2000;19(23):2739. doi:10.1038/sj.onc.1203597
9. Li J-H, Liu S, Zhou H, Qu L-H, Yang J-H. StarBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein–RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42(D1):D92–D97. doi:10.1093/nar/gkt1248
10. Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146(3):353–358. doi:10.1016/j.cell.2011.07.014
11. Liu Q, Guo X, Que S, et al. LncRNA RSU1P2 contributes to tumorigenesis by acting as a ceRNA against let-7a in cervical cancer cells. Oncotarget. 2017;8(27):43768. doi:10.18632/oncotarget.10844
12. Iden M, Fye S, Li K, Chowdhury T, Ramchandran R, Rader JS. The lncRNA PVT1 contributes to the cervical cancer phenotype and associates with poor patient prognosis. PLoS One. 2016;11(5):e0156274. doi:10.1371/journal.pone.0156274
13. Yang L, Bai H, Deng Y, Fan L. High MALAT1 expression predicts a poor prognosis of cervical cancer and promotes cancer cell growth and invasion. Eur Rev Med Pharmacol Sci. 2015;19(17):3187–3193.
14. Zhang J, Yao T, Wang Y, Yu J, Liu Y, Lin Z. Long noncoding RNA MEG3 is downregulated in cervical cancer and affects cell proliferation and apoptosis by regulating miR-21. Cancer Biol Ther. 2016;17(1):104–113. doi:10.1080/15384047.2015.1108496
15. Hou P, Zhao Y, Li Z, et al. LincRNA-ROR induces epithelial-to-mesenchymal transition and contributes to breast cancer tumorigenesis and metastasis. Cell Death Dis. 2014;5(6):e1287. doi:10.1038/cddis.2014.249
16. Wang G, LI Z, ZHAO QI, et al. LincRNA-p21 enhances the sensitivity of radiotherapy for human colorectal cancer by targeting the Wnt/β-catenin signaling pathway. Oncol Rep. 2014;31(4):1839–1845. doi:10.3892/or.2014.3047
17. Zhan H-X, Wang Y, Li C, et al. LincRNA-ROR promotes invasion, metastasis and tumor growth in pancreatic cancer through activating ZEB1 pathway. Cancer Lett. 2016;374(2):261–271. doi:10.1016/j.canlet.2016.02.018
18. Pan Y, Li C, Chen J, et al. The emerging roles of long noncoding RNA ROR (lincRNA-ROR) and its possible mechanisms in human cancers. Cell Physiol Biochem. 2016;40(1–2):219–229. doi:10.1159/000452539
19. Chen S, Liang H, Yang H, et al. LincRNa-p21: function and mechanism in cancer. Med Oncol. 2017;34(5):98. doi:10.1007/s12032-017-0959-5
20. Tsai M-C, Spitale RC, Chang HY. Long intergenic noncoding RNAs: new links in cancer progression. Cancer Res. 2011;71(1):3–7. doi:10.1158/0008-5472.CAN-10-2483
21. Hwang H, Mendell J. MicroRNAs in cell proliferation, cell death, and tumorigenesis. Br J Cancer. 2006;94(6):776. doi:10.1038/sj.bjc.6603023
22. Zimmerman AL, Wu S. MicroRNAs, cancer and cancer stem cells. Cancer Lett. 2011;300(1):10–19. doi:10.1016/j.canlet.2010.09.019
23. Esquela-Kerscher A, Slack FJ. Oncomirs—microRNAs with a role in cancer. Nat Rev Cancer. 2006;6(4):259.
24. Hayes J, Peruzzi PP, Lawler S. MicroRNAs in cancer: biomarkers, functions and therapy. Trends Mol Med. 2014;20(8):460–469.
25. Kano M, et al. miR‐145, miR‐133a and miR‐133b: tumor‐suppressive miRNAs target FSCN1 in esophageal squamous cell carcinoma. Int j Cancer. 2010;127(12):2804–2814.
26. Akcakaya P, et al. miR-185 and miR-133b deregulation is associated with overall survival and metastasis in colorectal cancer. Int J Oncol. 2011;39(2):311–318.
27. Qiu T, et al. miR‐145, miR‐133a and miR‐133b inhibit proliferation, migration, invasion and cell cycle progression via targeting transcription factor Sp1 in gastric cancer. FEBS Lett. 2014;588(7):1168–1177.
28. Qin W, et al. MicroRNA-133b is a key promoter of cervical carcinoma development through the activation of the ERK and AKT1 pathways,”. Oncogene. 2012;31(36):4067.
29. Liu X, Chen D, Liu G. Overexpression of RhoA promotes the proliferation and migration of cervical cancer cells. Biosci Biotechnol Biochem. 2014;78(11):1895–1901.
30. Eades G, Wolfson B, Zhang Y, Li Q, Yao Y, Zhou Q. LincRNA-RoR and miR-145 regulate invasion in triple-negative breast cancer via targeting ARF6. Mol Cancer Res. 2015;13(2):330–338.
31. Cao S, Liu W, Li F, Zhao W, Qin C. Decreased expression of lncRNA GAS5 predicts a poor prognosis in cervical cancer. Int J Clin Exp Pathol. 2014;7(10):6776.
32. Sun N-X, et al. Long noncoding RNA-EBIC promotes tumor cell invasion by binding to EZH2 and repressing E-cadherin in cervical cancer. PLoS One. 2014;9(7):e100340.
33. Yang M, Zhai X, Xia B, Wang Y, Lou G. Long noncoding RNA CCHE1 promotes cervical cancer cell proliferation via upregulating PCNA. Tumor Biol. 2015;36(10):7615–7622.
34. Kim HJ, et al. The long noncoding RNA HOXA11 antisense induces tumor progression and stemness maintenance in cervical cancer. Oncotarget. 2016;7(50):83001.
35. Liu K, Yan Z, Li Y, Sun Z. Linc2GO: a human LincRNA function annotation resource based on ceRNA hypothesis. Bioinformatics. 2013;29(17):2221–2222.
36. Wang Y, et al. Endogenous miRNA sponge lincRNA-RoR regulates Oct4, Nanog, and Sox2 in human embryonic stem cell self-renewal. Dev Cell. 2013;25(1):69–80.
37. Gulhati P, et al. mTORC1 and mTORC2 regulate EMT, motility, and metastasis of colorectal cancer via RhoA and Rac1 signaling pathways. Cancer Res. 2011;71(9):3246–3256.
38. Kamai T, et al. Overexpression of RhoA, Rac1, and Cdc42 GTPases is associated with progression in testicular cancer. Clin Cancer Res. 2004;10(14):4799–4805.
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.