Back to Journals » Cancer Management and Research » Volume 12
LINC00518 Interference Inhibits Non–Small Cell Lung Cancer by Upregulating miR216b-5p Expression
Received 30 June 2020
Accepted for publication 3 September 2020
Published 2 November 2020 Volume 2020:12 Pages 11041—11050
DOI https://doi.org/10.2147/CMAR.S270087
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Sanjeev K. Srivastava
Yuanyuan Ren,1,2,* Huadong Zhu,3,* Song Han4
1Department of Oncology, People’s Hospital of Taizhou, Taizhou, Jiangsu Province 225300, China; 2Department of Oncology, Hospital 5, affiliated with Nantong University, Taizhou 225300, China; 3School of Life Science, Nanchang University, Nanchang, Jiangxi, 330031, China; 4Department of Cardiothoracic Surgery, Affiliated Suzhou Science and Technology Town Hospital of Nanjing Medical University, Suzhou, Jiangsu 215153, China
*These authors contributed equally to this work
Correspondence: Song Han
Department of Cardiothoracic Surgery, Affiliated Suzhou Science and Technology Town Hospital of Nanjing Medical University, Suzhou, Jiangsu 215153, China
Tel +86 512-6958-8712
Email [email protected]
Introduction: Non–small cell lung cancer (NSCLC) accounts for the majority of lung cancer cases, and effective treatment for this disease is still lacking. This study aimed to explore the potential role of LINC00518 and miR216b-5p on cell proliferation and tumor growth in NSCLC.
Methods: The expression of LINC00518, miR216b-5p, MMP7, and MMP9 in NSCLC cell lines was determined by RT-qPCR analysis, which was also used to confirm the transfection effects. After transfection, proliferation, clone-formation ability, migration, and invasion of NSCLC cells were detected by CCK8, clone-formation, wound-healing, and transwell assays, respectively. Western blot analysis was used to detect the expression of MMP7, MMP9, Ki67, and PCNA. A xenograft model was constructed by subcutaneous injection of transfected NSCLC cells into nude mice.
Results: The results indicated that LINC00518 expression was increased and miR216b-5p expression decreased in NSCLC cell lines, and A549 cells were chosen for the next experiments. LINC00518 interference inhibited proliferation, invasion, and migration of A549 cells, together with the progression of NSCLC in vivo. In addition, LINC00518 directly targeted miR216b-5p. Downregulation of miR216b-5p weakened the inhibitory effect of LINC00518 interference on proliferation, invasion, and migration of A549 cells, as well as progression of NSCLC in vivo.
Discussion: In conclusion, LINC00518 interference inhibits NSCLC, which is partially reversed by downregulation of miR216b-5p expression.
Keywords: LINC00518, non–small cell lung cancer, miR216b-5p, proliferation, invasion and migration
Introduction
Lung cancer is the most common respiratory malignant cancer worldwide and the most common primary lung malignant cancer, seriously threatening human life and health.1,2 In addition, the mortality of lung cancer ranks first in both males and females, and non–small cell lung cancer (NSCLC) accounts for about 85% of all lung cancer cases.3,4 Although diagnostic techniques and therapeutic strategies have improved, the 5-year overall survival rate is still <15%, with a high rate of relapse.5 Platinum derivative–based chemotherapy has attained a dead end due to its efficacy.6 Therefore, it is urgent that we find oncogenes related to the development and progression of NSCLC and explore underlying mechanisms.
lncRNAs are a type of long RNA molecule longer than 200 nucleotides.7 lncRNAs have been demonstrated to modulate the cell growth, differentiation, and development of cancers.8,9 LICN00518 is significantlyuoregulated in melanoma tissue, and high LICN00518 levels are an independent risk factor in melanoma patients. LICN00518 promotes the invasion and migration of melanoma cells.10 It has been reported that LINC00518 is upregulated in cervical cancer tissue and that knockdown of LINC00518 significantly inhibits the proliferation, migration, and invasion of cervical cancer cells and induces apoptosis in vitro.11 Wang et al12 found that downregulation of LINC00518 inhibited proliferation, invasion, migration, and epithelial–mesenchymal transition and enhanced apoptosis of breast cancer epithelial cells. In addition, LINC00518 is highly expressed in prostate tumors in vitro and in vivo.13 Therefore, further investigations are worthwhile to explore the role and function of LINC00518 in the development and progression of NSCLC.
miRNAs are a kind of non-coding RNAs 17–22 nucleotides in length.14 They participate in several biological and pathological processes of cancers, and serve as oncogenes or tumor suppressors.15,16 StarBase predicts that LINC00518 can target miR216b-5p. miR216b-5p expression is significantly reduced in human breast cancer tissue and cell lines, and overexpression of miR216b-5p inhibits the proliferation and progression of breast cancer cells.17 You et al18 demonstrated that overexpression of miR216b-5p inhibited the proliferation of pancreatic cancer cells in vitro, induced cell-cycle arrest and apoptosis, and inhibited tumorigenesis in vivo. To the best of our knowledge, there has been no other study to focus on the role of miR216b-5p in NSCLC. As such, in this study we aimed to study the expression of LINC00518 and miR216b-5p in NSCLC cell lines, with a specific objective of assessing the potential role of LINC00518 and miR216b-5p in cell proliferation and tumor growth in NSCLC.
Methods
Cell Culture
NSCLC cell lines (A549, H23, NCI-H460, and H1975) were brought from American Type Culture Collection (Rockville, MD, USA). PC9 cells were purchased from iCell Bioscience (Shanghai, China). 16HBE cells were purchased from Procell Life Science and Technology(Wuhan, China). All cells were cultured in RPMI 1640 medium (Thermo Fisher Scientific) supplemented with 10% (v:v) FBS) and maintained in a humidified atmosphere with 5% CO2 at 37°C.
Cell Transfection
NSCLC cells were seeded into six-well plates and transfected with si-NC, si-LINC00518-1, si-LINC00518-2, inhibitor NC, miR216b-5p inhibitor 1, and miR216b-5p inhibitor 2 in different combinations. Lipofectamine 2000 reagent (Invitrogen, Shanghai, China) was used in the transfection process. Cells were harvested 48 hours after transfection and transfection effects determined by RT-qPCR analysis.
RT-qPCR Analysis
Total RNA was extracted from NSCLC cells with Trizol reagent (Invitrogen), and genomic DNA was removed from total RNA after treatment with DNase I (Thermo Fisher Scientific). cDNA was attained using a Transcriptor first-strand cDNA–synthesis kit (Roche). Reverse transcription was conducted with a TaqMan micro-RNA reverse-transcription kit (Applied Biosystems). Detection of LINC00518 using SsoAdvanced Universal SYBR green supermix (BioRad) and miR216b-5p with the TaqMan kit was conducted on a CFX96 real-time thermocycler (BioRad). GAPDH and U6 were considered endogenous controls of LINC00518 and miR216b-5p, respectively. Relative expression levels were calculated using the 2–ΔΔCT method.
CCK8 Assays
After transfection for 24 hours, suspensions (103 cells/well) of NSCLC cells were planted into 96-well plates and cultured at 37°C for 24 hours. After the addition of 10 μL of CCK8 (5 mg/mL) in each well, NSCLC cells were incubated for 4 hours. Then, absorbance at 450 nm of each well was determined with a Thermomax microplate reader (Thermo Fisher Scientific).
Colony-Formation Assays
After transfection for 24 hours, NSCLC cells were seeded into a six-well plate. When the colonies were obvious in the visible spectrum, cell colonies were fixed with 4% paraformaldehyde, followed by staining with 0.1% crystal violet. Cell colonies were then photographed.
Wound-Healing Assays
After transfection for 24 hours, NSCLC cells were seeded into a six-well plate to grow into a confluent monolayer. A 200 μL sterile pipette tip was utilized to scratch the confluent monolayer. Then, cells were washed with PBS three times and cultured in serum-free RPMI 1640 for 24 hours. The scratched areas at 0 and 24 hours were photographed by microscopy (Olympus, Tokyo, Japan).
Transwell Assays
After transfection for 24 hours, NSCLC cells were added into the upper chamber coated with Matrigel membrane. In the lower chamber, RPMI 1640 containing 10% FBS was added. After 24 hours, a cotton swab was used to wipe the cells in the upper surface of the membrane, while cells attached to the lower surface were dyed with 0.1% crystal violet for 10 minutes. Finally, the complete filters were washed with PBS once and observed on microscopy (Olympus).
Western Blot Analysis
Total protein was extracted from cultured cells or tumor tissue using an RIPA protein-lysis buffer (Beyotime, Shanghai, China). A BCA protein-assay kit (Thermo Fisher Scientific) was used to measure the concentration of total protein. Then, each lane had 40 μg protein uploaded, which was separated by 10% SDS-PAGE, followed by transfeal to polyvinylidene fluoride (PVDF) membranes. Next, PVDF membranes were blocked with 5% skim milk for 1 hour at room temperature. Then, the primary antibodies — anti-MMP7, anti-MMP9, anti-Ki67, and anti-PCNA — were added to the PVDF membranes for incubation at 4°C overnight. Subsequently, PVDF membranes were washed with TBST solution and added to horseradish peroxidase–conjugated secondary antibodies for 1 hour at room temperature. Finally, protein bands were observed with an enhanced-chemiluminescence system (Bio-Rad Clarity Western ECL).
Dual Luciferase–Reporter Assays
Luciferase assays were performed to confirm the interaction between LINC00518 and miR216b-5p. Wild-type (WT) or mutant LINC00518 were transfected into NSCLC cells with miR216b-5p mimic or negative control (vector) with Lipofectamine 2000 Reagent). Luciferase activity was detected after transfection using dual luciferase–reporter assays (Promega).
Xenograft Model
Male BALB/c nude mice (aged 4–6 weeks, specific pathogen-free) purchased from Shanghai Jiesijie Experimental Animal were randomly divided into four group: si-NC, si-LINC00518, si-LINC00518 + inhibitor NC, and si-LINC00518 + miR216b-5p inhibitor (five mice in each group). Suspensions (100 μL) of transfected NSCLC cells (1.5×106 cells) were inoculated subcutaneously in the right side of the back of nude mice and raised for 21 days. During the experiment, body weight and tumor volume of mice were recorded every 3 days. After 21 days, the mice were killed with anesthesia and tumors removed and photographed. Animal experiments were conducted following the Guide for the Care and Use of Laboratory Animals (NIH publication 86–23, revised 1986) and local regulations. All animal studies were approved by the Animal Ethics Committee of Hospital 5 affiliated with Nantong University.
Statistical Analysis
All experimental data are presented as means ± SD, which were analyzed by one-way ANOVA. Statistical analyses were conducted with SPSS version 18.0, and p<0.05 was considered statistically significant.
Results
Expression Levels of LINC00518 in NSCLC Cell Lines
LINC00518 levels increased in A549, H23, NCI-H460, and H1975 cells and decreased in PC9 cells compared with LINC00518 expression in the 16HBE group. In addition, LINC00518 expression was the highest in A549 cells, so these were selected for further study (Figure 1).
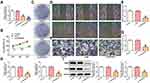
LINC00518 Interference Inhibited Proliferation, Invasion, and Migration of NSCLC Cells
LINC00518 expression decreased in A549 cells transfected with si-LINC00518-1 or si-LINC00518-2 compared with the control group. Furthermore, LINC00518 expression in the si-LINC00518-2 group was lower than in the si-LINC00518-1 group, and thus si-LINC00518-2 was chosen for the next experiments (Figure 2A). Figure 2B indicates that LINC00518 interference inhibited the proliferation of A549 cells at three time points. As shown in Figure 2C, the clone-formation ability of A549 cells declined in the si-LINC00518 group. It was shown that LINC00518 interference suppressed the migration (Figure 2D and E) and invasion (Figure 2F and G) of A549 cells. RT-qPCR analysis indicated that mRNA expression of MMP7 and MMP9 decreased in the si-LINC00518 group (Figure 2H), and protein expression showed a similar change with mRNA expression (Figure 2I).
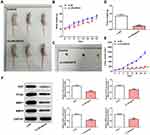
LINC00518 Interference Inhibited the Progression of NSCLC In Vivo
In Figure 3A and B, the weight of mice in the si-LINC00518 and si-NC groups in presented. With increased time, the inhibitory effect of LINC00518 interference on tumor weight was more evident (Figure 3C and D), and the tumors in the si-LINC00518 group were smaller than in the si-NC group (Figure 3E). Ki67, PCNA, MMP7 and MMP9 expression in tumor tissue was suppressed by LINC00518 interference (Figure 3F).
LINC00518 Directly Targeted miR216b-5p
As shown in Figure 4A, miR216b-5p was predicted to combine with LINC00518. miR216b-5p expression in A549, H23, NCI-H460, H1975, and PC9 cells was downregulated compared with 16HBE cells, and miR216b-5p expression in A549 cells showed the lowest levels (Figure 4B). Relative luciferase activity obviously declined when A549 cells had been cotransfected with miR216b-5p mimic and WT-LINC00518, which indicated that LINC00518 directly targeted miR216b-5p (Figure 4C). Also, miR216b-5p expression was significantly upregulated in A549 cells transfected with si-LINC00518 (Figure 4D).
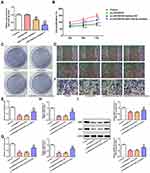
Downregulation of miR216b-5p Weakened the Inhibition Effect of LINC00518 Interference on Proliferation, Invasion, and Migration of NSCLC Cells
Transfection effects were verified by RT-qPCR analysis, which showed that miR216b-5p expression decreased in the miR216b-5p inhibitor 1 group and miR216b-5p inhibitor 2 group. A sthe miR216b-5p inhibitor 2 group showed lower levels of miR216b-5p than the miR216b-5p inhibitor 1 group, miR216b-5p inhibitor 2 was chosen for the next experiment (Figure 5A). As shown in Figure 5B, downregulation of miR216b-5p weakened the inhibitory effect of LINC00518 interference on the proliferation of A549 cells. The clone-formation ability of A549 cells transfected with si-LINC00518 and miR216b-5p inhibitor was improved compared with that of A549 cells transfected with si-LINC00518 or si-LINC00518 and inhibitor NC (Figure 5C). Downregulation of miR216b-5p also enhanced the migration (Figure 5D and E) and invasion (Figure 5F and G) of A549 cells transfected with si-LINC00518. As shown in Figure 5H and I, mRNA and protein expression of MMP7 and MMP9 in the si-LINC00518 + miR216b-5p inhibitor group increased compared to the si-LINC00518 group and si-LINC00518 + inhibitor-NC group.
Downregulation of miR216b-5p Weakened the Inhibition Effect of LINC00518 Interference on the Progression of NSCLC in vivo
As shown in Figure 6A and B, the difference in mousee weights between the si-LINC00518 + miR216b-5p inhibitor group and the si-LINC00518 + inhibitor-NC group was not obvious. Tumor weight increased gradually in the si-LINC00518 + miR216b-5p inhibitor group compared with the si-LINC00518 + inhibitor-NC group (Figure 6C and D), and tumor volume was increased (Figure 6E). Ki67, PCNA, MMP7, and MMP9 expression was enhanced in tumor tissue of mice transfected with si-LINC00518 and miR216b-5p inhibitor compared with those transfected with si-LINC00518 and inhibitor NC (Figure 6F).
Discussion
Most NSCLC patients miss the chance to be cured, because the symptoms of NSCLC are atypical in the early stage.19 Although diagnosis and treatment of NSCLC have improved, NSCLC patients still suffer long-term adverse outcomes.20 Recently, studies have shown that abnormal expression of lncRNAs is related to the progression of cancers, including NSCLC.21 These lncRNAs may serve as diagnostic, therapeutic, and prognostic markers for NSCLC.22 Therefore, it is important to clarify molecular mechanisms of lncRNAs for the prognosis of NSCLC.
Recent studies have shown that LINC00518 expression is increased in multiple cancers.23 Yang et al24 studied a coexpression network, finding that LINC00518 was obviously upregulated in triple-negative breast cancer. LINC00518 serves as a sensitive and effective target and is also increased in cutaneous melanoma.25 In the present study, LINC00518 expression increased in NSCLC cell lines. Also, LINC00518 interference suppress the proliferation, clone-formation ability, migration, and invasion of A549 cells and suppressed tumor growth in vivo. miRcode (http://www.mircode.org) predicted that miR216b-5p could not be the only target of LINC00518. We also searched the literature and found that some miRNAs have been explored in lung cancer. There have been several studies concentrating on the role of miR216b-5p in multiple cancers, while the role of miR216b-5p in lung cancer has not been explored. Therefore, we chose miR216b-5p for our study. Also, the underlying correlation between LINC00518 and miR216b-5p was demonstrated by luciferase-reporter assays, which indicated that LINC00518 directly targeted miR216b-5p.
Studies have shown the regulatory effects of lncRNAs on the function of miR216b-5p. Dai et al26 showed that TUG1 expression was increased and miR216b-5p expression decreased in hepatocellular carcinoma tissue and cell lines, while TUG1 knockdown repressed the growth of hepatocellular carcinoma cells and promoted apoptosis by upregulating miR216b-5p. Here, we found that miR216b-5p expression notably declined in NSCLC cell lines. Moreover, LINC00518 negatively regulated miR216b-5p expression in A549 cells. Zheng et al27 demonstrated that miR216b-5p was downregulated in cervical cancer tissue and cell lines, and a negative relationship between miR216b-5p with LINC00152 expression in cervical cancer tissue was presented. miR216b-5p was sponged by LINC00152 to regulate the proliferation of cervical cancer cells. In this experiment, miR216b-5p expression declined in NSCLC cell lines. LINC00518 interference suppressed the proliferation, clone-formation ability, migration, and invasion of A549 cells, as well as tumor growth in vivo, by upregulating miR216b-5p. However, this effect was partially reversed by knockdown of miR216b-5p.
In conclusion, our research shows that LINC00518 plays an essential role in NSCLC progression by regulating miR216b-5p expression. Importantly, LINC00518 was found to regulate cell proliferation, clone-formation ability, invasion, and migration via regulating miR216b-5p. Therefore, targeting the LINC00518–miR216b-5p axis might be a new direction for the treatment of NSCLC. Of course, there are also some limitations in our study. In future, we will analyze the expression pattern of LINC00518 and miR216-5p in lung cancer using bioinformatics and detect the expression of LINC00518 and miR216b-5p in NSCLC patients. At least two cell lines will be used to verify the research findings from functional experiments.
Funding
There is no funding to report.
Disclosure
The authors declare that they have no competing interests.
References
1. Chen W, Zheng R, Baade P, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. doi:10.3322/caac.21338
2. Torre LA, Freddie B, Siegel RL, Jacques F, Joannie LT, Ahmedin J. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi:10.3322/caac.21262
3. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7–34.
4. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424.
5. Didkowska J, Wojciechowska U, Mańczuk M, Łobaszewski J. Lung cancer epidemiology: contemporary and future challenges worldwide. Ann Transl Med. 2016;4:150. doi:10.21037/atm.2016.03.11
6. Schiller JH, Harrington D, Belani CP, et al. Comparison of four chemotherapy regimens for advanced non-small-cell lung cancer. N Engl J Med. 2002;346(2):92–98. doi:10.1056/NEJMoa011954
7. Jarroux J, Morillon A, Pinskaya M. History, discovery, and classification of lncRNAs. 1008. Adv Exp Med Biol. 2017;1–46.
8. Peng W-X, Koirala P, Mo Y-Y. LncRNA-mediated regulation of cell signaling in cancer. Oncogene. 2017;36.
9. Anna SC, Yumi K, Yusuke Y, Fumitaka T, Takahiro O. Emerging roles of long non-coding RNA in cancer. Cancer Sci. 2018;109:2093–2100. doi:10.1111/cas.13642
10. Luan W, Ding Y, Ma S, et al. Long noncoding RNA LINC00518 acts as a competing endogenous RNA to promote the metastasis of malignant melanoma via miR-204-5p/AP1S2 axis. Cell Death Dis. 2019;10(11):855. doi:10.1038/s41419-019-2090-3
11. Wang DW, You D, Dong J, Liu TF. Knockdown of long non-coding RNA LINC00518 inhibits cervical cancer proliferation and metastasis by modulating JAK/STAT3 signaling. Eur Rev Med Pharmacol Sci. 2019;23(2):496–506.
12. Wang H-B, Wei H, Wang J-S, et al. Down-regulated expression of LINC00518 prevents epithelial cell growth and metastasis in breast cancer through the inhibition of CDX2 methylation and the Wnt signaling pathway. Biochimica Et Biophysica Acta Mol Basis Dis. 2019;1865(3):708–723. doi:10.1016/j.bbadis.2019.01.003
13. Jin H, Sun M, Geng H, et al. Long non-coding RNA Linc00518 promotes paclitaxel resistance of the human prostate cancer by sequestering miR-216b-5p. Biol Cell. 2019;111(2):39–50. doi:10.1111/boc.201800054
14. Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi:10.1016/j.cell.2009.01.002
15. Gamazon ER, Trendowski MR, Wen Y, et al. Gene and MicroRNA perturbations of cellular response to pemetrexed implicate biological networks and enable imputation of response in lung adenocarcinoma. Sci Rep. 2018;8(1):733. doi:10.1038/s41598-017-19004-3
16. Wienholds E, Plasterk RH. MicroRNA function in animal development. FEBS Lett. 2005;579(26):5911–5922. doi:10.1016/j.febslet.2005.07.070
17. Menbari M, Rahimi K, Ahmadi A, et al. MiR-216b-5p inhibits cell proliferation in human breast cancer by down-regulating HDAC8 expression. Life Sci. 2019;237:116945. doi:10.1016/j.lfs.2019.116945
18. You Y, Tan J, Gong Y, et al. MicroRNA-216b-5p functions as a tumor-suppressive RNA by targeting TPT1 in pancreatic cancer cells. J Cancer. 2017;8(14):2854–2865. doi:10.7150/jca.18931
19. Wang J, Li H. CircRNA circ_0067934 silencing inhibits the proliferation, migration and invasion of NSCLC cells and correlates with unfavorable prognosis in NSCLC. Eur Rev Med Pharmacol Sci. 2018;22:3053–3060.
20. Lu T, Wang Y, Di Chen JL, Jiao W. Potential clinical application of lncRNAs in non-small cell lung cancer. OncoTargets Ther. 2018;11:8045–8052.
21. Wei -M-M, Zhou G-B. Long non-coding RNAs and their roles in non-small-cell lung cancer. Genomics Proteomics Bioinformatics. 2016;14(5):280–288. doi:10.1016/j.gpb.2016.03.007
22. Y J, Lin J, Liu T, et al. Analysis of lncRNA expression profiles in non-small cell lung cancers (NSCLC) and their clinical subtypes. Lung Cancer (Amsterdam, Netherlands). 2014;85(2):110–115. doi:10.1016/j.lungcan.2014.05.011
23. Sun S, Li X, Ma H, et al. Long noncoding RNA LINC00265 promotes glycolysis and lactate production of colorectal cancer through regulating of miR-216b-5p/TRIM44 axis. Digestion. 2019;1–10.
24. Yang F, Liu Y-H, Dong S-Y, et al. Co-expression networks revealed potential core lncRNAs in the triple-negative breast cancer. Gene. 2016;591(2):471–477. doi:10.1016/j.gene.2016.07.002
25. Sobarun P, Hoang VL, Yamada M, Lambie D, Soyer HP, Prow TW. Microbiopsy biomarker profiling in a superficial melanoma resembling a pigmented basal cell carcinoma. JAMA Dermatol. 2017;153(4):334–336. doi:10.1001/jamadermatol.2016.5537
26. Dai Q, Deng J, Zhou J, et al. Long non-coding RNA TUG1 promotes cell progression in hepatocellular carcinoma via regulating miR-216b-5p/DLX2 axis. Cancer Cell Int. 2020;20:8. doi:10.1186/s12935-019-1093-6
27. Zhang JJ, Du XJ, Wang HP, et al. Long non-coding RNA 00152 promotes cell proliferation in cervical cancer via regulating miR-216b-5p/HOXA1 axis. Eur Rev Med Pharmacol Sci. 2019;23(9):3654–3663.
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.