Back to Journals » OncoTargets and Therapy » Volume 11
LINC00460 modulates KDM2A to promote cell proliferation and migration by targeting miR-342-3p in gastric cancer
Authors Wang F, Liang S, Liu X, Han L, Wang J, Du Q
Received 26 March 2018
Accepted for publication 26 May 2018
Published 2 October 2018 Volume 2018:11 Pages 6383—6394
DOI https://doi.org/10.2147/OTT.S169307
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Samir Farghaly
Fang Wang,1 Shaobo Liang,2 Xiaowei Liu,1 Lei Han,1 Junye Wang,1 Qin Du1
1Department of Oncology, Affiliated Hospital of Jining Medical College, Jining 272029, China; 2Department of Gastroenterology, Central Hospital of Shanxian County, Heze, 274399, China
Background: Increasing evidence has shown that long non-coding RNAs (lncRNAs) play important roles in the occurrence and development of human cancers. LINC00460, a novel tumor-related lncRNA, has been reported to be involved in several types of human malignancies. However, the role of LINC00460 in gastric cancer (GC) is still unclear. The present study aimed at exploring the biological role of LINC00460 in GC and illuminating the potential molecular mechanisms.
Methods: In this study, qRT-PCR, western blotting, MTT assay, and Transwell invasion assay were used to conduct relevant experimental analysis.
Results: Here, we found that LINC00460 was highly expressed in GC tissues and cell lines. Moreover, LINC00460 over-expression was found to promote GC cell proliferation, migration and invasion, whereas LINC00460 down-regulation significantly inhibited these processes. Notably, we confirmed that LINC00460 could up-regulate KDM2A expression by competitively binding to miR-342-3p in GC cells. Furthermore, the suppressive effects of LINC00460 down-regulation on GC cell proliferation, migration and invasion were partially reversed by a miR-342-3p inhibitor.
Conclusion: In summary, our findings provide evidence for LINC00460 as a potential therapeutic target in GC.
Keywords: LINC00460, KDM2A, miR-342-3p, gastric cancer
Introduction
Gastric cancer (GC), one of the most common human malignancies, is a leading cause of cancer-related deaths worldwide, with approximately one million cases diagnosed annually.1–3 Over 700,000 deaths are estimated to occur from GC around the globe every year.4,5 Drug resistance and distant metastasis partially account for the high mobility of GC.6–8 Although great progress has been made in the diagnosis and treatment for GC, its long-term prognosis is still unfavorable. Therefore, development of effective therapeutic strategies is urgently required.
Long non-coding RNAs (lncRNAs) are a group of RNA transcripts longer than 200 nucleotides that do not act as templates for protein synthesis.9–11 Increasing evidence has shown that lncRNAs play vital roles in the occurrence and development of a wide range of human cancers.12–15 Numerous studies have demonstrated that lncRNAs may function as competing endogenous RNAs (ceRNAs) to exert their roles in a variety of human tumors.16–18 Previous studies have demonstrated that LINC00460, a novel cancer-related lncRNA, is deregulated and involved in several types of human malignancies, including nasopharyngeal carcinoma, lung cancer and esophageal squamous cell carcinoma.19–21 However, the role of LINC00460 in GC is still unclear.
This study aimed to explore the biological role of LINC00460 in GC and determine the potential mechanisms. Here, we found that LINC00460 was highly expressed in GC tissues and cell lines and it enhanced GC cell proliferation, migration and invasion. Furthermore, we found that LINC00460 exerted its oncogenic role in GC by sponging miR-342-3p.
Materials and methods
Tissue samples collection
GC tissues and corresponding non-cancerous tissues were obtained from 60 patients who underwent surgical treatment between March 2011 and December 2015 at the Affiliated Hospital of Jining Medical College, Jining, China. Tissue samples were snap frozen in liquid nitrogen immediately after surgical resection and stored at −80°C. All patients enrolled in this study gave written informed consents. This study was approved by the Medical Ethics Committee of the Affiliated Hospital of Jining Medical College.
Cell culture
One normal human gastric epithelial cell line GSE1 and three GC cell lines (MGC803, BGC823 and SGC7901) were purchased from the Chinese Academy of Sciences Cell Bank (Shanghai, China). All cells were cultured in RPMI-1640 (Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS) (Thermo Fisher Scientific) and grown in humidified 5% CO2 at 37°C. MiR-5095 mimics, inhibitor and relative controls were obtained from Genepharma (Shanghai, China).
Cell transfection
The transfection was conducted by using Lipofectamine 2000 (Thermo Fisher Scientific) as described previously. LINC00460 mimics and si-LINC00460 were obtained from Genepharma (Shanghai, China).
Quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was extracted from tissues and cells using the Trizol reagent (Invitrogen) according to the manufacturer’s instructions. For microRNA analysis, qRT-PCR was performed using the TaqMan MicroRNA Reverse Transcription Kit, TaqMan Universal PCR Master Mix (Thermo Fisher Scientific), and the corresponding primers. For mRNA analysis, qRT-PCR was performed using the TaqMan High-Capacity cDNA Reverse Transcription Kit, TaqMan Fast PCR Master Mix (Thermo Fisher Scientific) and the corresponding primers. β-actin was used as an internal control to normalize KDM2A expression. qRT-PCR was performed in triplicate on a RealPlex4 real-time PCR detection system from Eppendorf Co. Ltd (Hamburg, Germany).
Cell proliferation
Cells were seeded at 5,000 cells/well in 96-well plates at 24 hours after transfection. Cell proliferation was measured using an MTT Cell Proliferation and Cytotoxicity Assay Kit (Sigma-Aldrich Co., St Louis, MO, USA). Following incubation at 37°C for different periods of time (0, 24, 48 and 72 hours), the culture medium was removed and MTT (20 μL; 5 mg/mL) was added to each well. After incubation at 37°C for another 4 hours, MTT solution was removed and replaced with dimethyl sulfoxide (DMSO; 150 μL, 4%; Sigma-Aldrich). Absorbance was measured at 560 nm after using a microplate spectrophotometer (Thermo Fisher Scientific, Vantaa, Finland).
Wound healing assays
Cell migration was evaluated using a wound healing assay. In brief, transfected cells were cultured in six-well plates (5×104 cells per well). At 90%–95% confluence, the monolayer of cells was scratched by a sterile plastic micropipette tip, and then cells were cultured under standard conditions for 24 hours. Following several washes, wound recovery of the wound was observed and photographed using an X71 inverted microscope (Olympus, Tokyo, Japan).
Transwell invasion assays
The Transwell invasion assay was performed to determine cell invasion potential. Transfected cells (1×105) were seeded into the upper chamber of Matrigel-coated inserts with free-serum medium. Medium with 10% FBS was added to the lower chamber as chemoattractant. The cells were allowed to invade for 48 hours at 37°C with 5% CO2. Cells invaded to the lower surface of the filter were fixed in 70% ethanol for 30 minutes and stained with 0.1% crystal violet for 10 minutes. The number of cells that invaded to the lower side was counted in five randomly selected fields under an X71 inverted microscope (Olympus Corporation, Tokyo, Japan).
Cell cycle analysis
Cells were harvested at 48 hours after transfection. The cells were washed using PBS and fixed in ethanol at −20°C. The cells were then washed with PBS, rehydrated and resuspended in propidium iodide (PI)-RNase A solution (Sigma, Finland) at 37°C for 30 minutes. The stained cells (1×105) were then analyzed for DNA content using a flow cytometer (BD Biosciences, San Jose, CA, USA).
RNA-binding protein immunoprecipitation (RIP)
RIP assays were performed using an EZ-Magna RIP™ RNA-Binding Protein Immunoprecipitation Kit (EMD Millipore, Billerica, MA, USA) according to the manufacturer’s protocol. Briefly, RIP buffer containing magnetic beads conjugated with human anti-Ago2 antibody (EMD Millipore, Billerica, MA, USA) or negative control IgG (EMD Millipore) was added to the cell lysate and incubated overnight at 4°C. Proteinase K was used to digest the protein and the co-precipitated RNAs were then isolated. The purified RNAs were subject to RT-PCR analysis.
Luciferase reporter assays
Wild-type of LINC00460 or KDM2A 3′UTR was amplified from a human cDNA library. Mutations of miR-342-3p binding site were introduced by site-directed mutagenesis using a fast mutation kit (NEB, Beverly, MA, USA). The PCR fragment was cloned into a psiCHECK-2 vector downstream of the firefly luciferase coding region within the sites for XhoI and NotI (Takara, Tokyo, Japan). psiCHECK-2-control was used as an internal control.
Western blotting analysis
Total protein lysates were resolved by 10% SDS-PAGE and transferred to polyvinyl difluoride membranes (EMD Millipore). After blocking in Tris buffered saline containing 0.1% Tween-20 (TBS-T) with 5% nonfat dry milk for 30 minutes, membranes were washed four times in TBS-T and incubated overnight at 4°C with primary antibodies. Primary antibodies were all obtained from Abcam (Cambridge, MA, USA) and used at the following dilutions: anti-KDM2A (ab174693, 1/300), anti-CCND1 (ab40754, 1/300), anti-CDK4 (ab199728, 1/300), anti-MMP2 (ab92536, 1/500), anti-MMP9 (ab76003, 1/500), anti-vimentin (ab45939, 1/500), and anti-β-actin (ab5694, 1/1,000). After extensive washing, membranes were incubated with horseradish peroxidase-linked goat polyclonal anti rabbit IgG secondary antibodies at a dilution of 1:2,000 for 1 hour at room temperature. Immunoreactivity was detected by enhanced chemiluminescence (ECL Kit, Pierce Biotechnology, Waltham, MA, USA) and exposure to radiography film. β-actin served as the loading control.
Statistical analysis
Data were expressed as the mean ± SD from at least three independent experiments. Two-tailed Student’s t-test was applied to compare the differences between two groups and one-way analysis of variance (ANOVA) followed by Dunnett’s multiple comparison was employed to compare the differences among three independent groups. The correlation between LINC00460 expression and miR-342-3p or KDM2A expression in GC tissues was determined using Pearson’s correlation analysis. P<0.05 was considered statistically significant.
Results
LINC00460 is highly expressed in GC tissues and cell lines
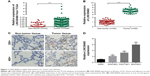
Although LINC00460 has been shown to be involved in several types of human cancers, its biological role in GC is still obscure. Initially, we downloaded LINC00460 expression data from the TCGA database and found that LINC00460 was highly expressed in GC tissues compared with normal tissues (Figure 1A). Subsequently, we determined the expression of LINC00461 in 60 pairs of GC tissues and corresponding non-cancerous tissues and discovered that LINC00460 was highly expressed in GC tissues (Figure 1B and C). Additionally, we found that LINC00460 was significantly upregulated in GC cell lines (Figure 1D). MGC803 (the lowest endogenous LINC00460 expression) and MBGC823 (highest endogenous LINC00460 expression) cells were chosen for subsequent studies. Taken together, these results suggest that LINC00460 is highly expressed in GC tissues and cell lines.
LINC00460 promotes GC cell proliferation and cell cycle progression
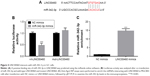
To investigate the role of LINC00460 in GC, we conducted over-expression studies in MGC803 cells and knockdown studies in BGC823 cells. Transfection efficacy was analyzed by qRT-PCR (Figure 2A). As presented in Figure 2B and C, LINC00460 mimics significantly promoted GC cell proliferation and cell cycle progression, whereas LINC00461 knockdown notably repressed GC cell proliferation and induced cell cycle arrest. These results indicate that LINC00460 promotes GC cell proliferation and cell cycle progression.
LINC00460 promotes GC cell migration and invasion
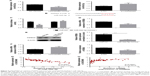
To investigate whether LINC00460 influences the motility of GC cells, we performed wound healing assays and Transwell invasion assays. As shown in Figure 3A and B, LINC00460 mimics significantly promoted GC cell migration and invasion, whereas LINC00460 knockdown notably inhibited GC cell migration and invasion. In addition, we analyzed the expression of cell cycle- and cell motility-associated proteins and discovered that LINC00460 mimics notably increased the protein expression of CCND1, CDK4, vimentin, MMP2 and MMP9, while the expression of these proteins was significantly reduced by LINC00460 knockdown (Figure 3C). These results suggest that LINC00460 promotes GC cell migration and invasion.
LINC00460 interacts with miR-342-3p in GC cells
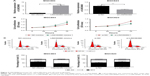
Mounting evidence has revealed that lncRNAs may function as competing endogenous RNAs (ceRNAs) to exert their roles in a variety of human tumors. To explore the potential mechanisms by which LINC00460 promotes GC cell proliferation, migration and invasion, we applied miRanda online software to predict the potential targets of LINC00460. MiR-342-3p drew our attention due to its importance in several types of human cancers and was selected as a candidate target of LINC00460 (Figure 4A). As obvious from luciferase reporter assays, co-transfection of wild-type LINC00460 and miR-342-3p significantly repressed the luciferase activity, while co-transfection of mutant LINC00460 and miR-342-3p did not induce significant alterations in the luciferase activity (Figure 4B). To verify the interaction between LINC00460 and miR-342-3p, we performed Anti-Ago2 RIP assays and found that miR-342-3p was significantly enriched by LINC00460 mimics (Figure 4C). These results indicate that LINC00460 interacts with miR-342-3p in GC cells.
LINC00460 up-regulates KDM2A expression by competitively binding to miR-342-3p
To further explore the potential mechanisms involved, we analyzed the expression of miR-342-3p and discovered that LINC00460 mimics significantly inhibited miR-342-3p expression, while miR-342-3p expression was notably enhanced by LINC00460 knockdown (Figure 5A). Additionally, we performed bio-informatics analysis via miRanda online software to predict the possible targets of miR-342-3p. KDM2A drew our attention for its significance in the oncogenesis and tumor progression of human tumors and was selected as a candidate target of miR-342-3p (Figure 5B). As presented in Figure 5C, co-transfection of miR-342-3p and wild-type KDM2A 3′UTR significantly decreased the luciferase activity, while co-transfection of miR-342-3p and mutant KDM2A 3′UTR did not trigger significant changes in the luciferase activity. Moreover, miR-342-3p mimics were found to notably inhibit KDM2A mRNA and protein expression (Figure 5D and E). Furthermore, LINC00460 mimics were found to significantly increase the mRNA expression of KDM2A, while LINC00460 knockdown decreased KDM2A expression (Figure 5F). As shown in Figure 5G and H, LINC00460 expression was negatively correlated with miR-342-3p expression in GC tissues, but was positively correlated with KDM2A mRNA expression in GC tissues. These results indicate that LINC00460 up-regulates KDM2A through sponging miR-342-3p.
MiR-342-3p inhibitor alleviates the inhibitory effects of LINC00460 down-regulation on GC cell proliferation, migration and invasion
To investigate whether the effects of LINC00460 on GC cell proliferation, migration and invasion are mediated by miR-342-3p, we down-regulated the expression of miR-343-3p in the BGC823 cells treated with si-LINC00460. As shown in Figure 6A–C, the miR-342-3p inhibitor alleviated the inhibitory effects of LINC00460 knockdown on GC cell proliferation, migration and invasion. Moreover, the miR-342-3p inhibitor was noticed to elevate the protein expression levels of KDM2A in BGC823 cells treated with si-LINC00460 (Figure 6D). Additionally, miR-342-3p was discovered to alleviate the inhibitory effects of LINC00460 knockdown on the protein expression of CCND1, CDK4, MMP2, MMP9 and vimentin (Figure 6E). These results suggest that the effects of LINC00460 on GC cell proliferation, cell migration and invasion are mediated by miR-342-3p.
Discussion
GC, one of the most common gastrointestinal neoplasms worldwide, has imposed enormous pressures on human health and life.22 Although great progress has been made in the diagnosis and treatment for GC, its long-term prognosis is still poor. There is enough evidence showing that lncRNAs play important roles in the tumorigenesis and development of a wide range of human cancers. Previous studies have shown that LINC00460 is involved in several types of human tumors. Kong et al found that LINC00460 was upregulated in nasopharyngeal carcinoma and acted as an oncogene through sponging miR-149-5p.19 Li et al reported that LINC00460 promoted epithelial-mesenchymal transition and lung cancer cell migration.20 Liang et al reported that LINC00460 promoted tumorigenesis and tumor progression in esophageal squamous cell carcinoma.21 However, the role of LINC00460 in GC is still unknown. Therefore, it may be useful for developing effective therapeutic strategies to gain a better understanding of the role of LINC00460.
In the present study, we initially downloaded LINC00460 expression data from the TCGA database and found that LINC00460 was highly expressed in GC tissues. Consistently, LINC00460 was found to be highly expressed in GC tissues obtained from patients and in GC cell lines. To explore the biological role of LINC00460 in GC, we carried out functional studies. LINC00460 over-expression was noticed to promote GC cell proliferation, cell cycle progression, cell migration and invasion. Moreover, LINC00460 mimics significantly elevated the protein expression levels of CDK4, CCND1, MMP2, MMP9 and vimentin. In addition, LINC00460 knockdown was observed to inhibit cell proliferation, cell cycle progression, cell migration, cell invasion and the expression of the above-mentioned proteins. CDK4 and CCND1, two important cell cycle-associated proteins, play crucial roles in the regulation of cell cycle progression.23,24 It is well known that high levels of MMP2, MMP9 and vimentin contribute to tumor cell motility.25,26 Our findings thus suggest that LINC00460 may act as an oncogene.
Numerous studies have demonstrated that lncRNAs may function as competing endogenous RNAs to exert their roles in various types of human cancers. To explore the potential molecular mechanism by which LINC0460 promotes GC cell proliferation, migration and invasion, we conducted mechanistic studies. LINC00460 was found to upregulate KDM2A expression to promote GC cell proliferation, migration and invasion through sponging miR-342-3p. MiR-342-3p has been reported to be down-regulated and involved in the occurrence and development of cervical cancer, non-small-cell lung cancer, hepatocellular carcinoma and osteosarcoma.27–30 Histone lysine methylation, an important epigenetic regulatory process, plays vital roles in controlling gene expression and maintaining chromosome structure. KDM2A has been confirmed to exert an oncogenic role in a wide range of human tumors, including breast cancer, gastric cancer and lung cancer.31–33 It is documented that KDM2A could enhance cancer stemness to promote tumor progression.34,35
It is documented that LINC00460 exerts its oncogenic roles in different human malignancies via distinct molecular mechanisms, including nasopharyngeal carcinoma, lung cancer, and esophageal squamous cell carcinoma.19–21 To the best of our knowledge, this is the first study to elucidate the molecular mechanisms by which LINC00460 functions as an oncogene in GC. In the present study we investigated the effects of LINC00460 on GC cell proliferation, migration and invasion in vitro. Nonetheless, some limitations exist in this work. In the future studies, we will carry out further research to analyze the effect of LINC00460 on GC tumor growth in vivo and validate whether LINC00460 exerts an inhibitory effect on tumor growth in vivo.
In summary, LINC00460 is highly expressed in GC tissues and cell lines and exerts an oncogenic role through sponging miR-342-3p in GC. This study provides new insights into the potential molecular mechanisms underlying GC tumorigenesis and progression. Thus, our data indicate that LINC00460 may be used as a novel therapeutic target for GC.
Disclosure
The authors report no conflicts of interest in this work.
References
Wang J, Zhang J, Zhou C, Chen L, Yu Q. An insertion/deletion polymorphism within the proximal promoter of EGLN2 is associated with susceptibility for gastric cancer in the Chinese population. Genet Test Mol Biomarkers. 2014;18(4):269–273. | ||
Guo J, Yan Y, Yan Y, et al. Tumor-associated macrophages induce the expression of FOXQ1 to promote epithelial-mesenchymal transition and metastasis in gastric cancer cells. Oncol Rep. 2017;38(4):2003–2010. | ||
Wang MY, Li QX, He J, et al. Genetic variations in the mTOR gene contribute toward gastric adenocarcinoma susceptibility in an Eastern Chinese population. Pharmacogenet Genomics. 2015;25(11):1–530. | ||
Wang HL, Zhou PY, Liu P, Zhang Y. ALDH2 and ADH1 genetic polymorphisms may contribute to the risk of gastric cancer: a meta-analysis. PLoS One. 2014;9(3):e88779. | ||
Chandanos E, Lagergren J. Oestrogen and the enigmatic male predominance of gastric cancer. Eur J Cancer. 2008;44(16):2397–2403. | ||
Zhao X, Yang L, Hu J. Down-regulation of miR-27a might inhibit proliferation and drug resistance of gastric cancer cells. J Exp Clin Cancer Res. 2011;30(1):55. | ||
Liang J, Ge F, Guo C, et al. Inhibition of PI3K/Akt partially leads to the inhibition of PrP(C)-induced drug resistance in gastric cancer cells. Febs J. 2009;276(3):685–694. | ||
Huang Y, Yang M, Yang H, Zeng Z. Upregulation of the GRIM-19 gene suppresses invasion and metastasis of human gastric cancer SGC-7901 cell line. Exp Cell Res. 2010;316(13):2061–2070. | ||
Jin G, Sun J, Isaacs SD, et al. Human polymorphisms at long non-coding RNAs (lncRNAs) and association with prostate cancer risk. Carcinogenesis. 2011;32(11):1655–1659. | ||
Cimadamore A, Gasparrini S, Mazzucchelli R, et al. Long non-coding RNAs in prostate cancer with emphasis on second chromosome locus associated with prostate-1 expression. Front Oncol. 2017;7:305. | ||
Xu MD, Qi P, Weng WW, et al. Long non-coding RNA LSINCT5 predicts negative prognosis and exhibits oncogenic activity in gastric cancer. Medicine. 2014;93(28):e303. | ||
Zhu M, Chen Q, Liu X, et al. lncRNA H19/miR-675 axis represses prostate cancer metastasis by targeting TGFBI. Febs J. 2014;281(16):3766–3775. | ||
Zheng HT, Shi DB, Wang YW, et al. High expression of lncRNA MALAT1 suggests a biomarker of poor prognosis in colorectal cancer. Int J Clin Exp Pathol. 2014;7(6):3174–3181. | ||
Liu JH, Chen G, Dang YW, Li CJ, Luo DZ. Expression and prognostic significance of lncRNA MALAT1 in pancreatic cancer tissues. Asian Pac J Cancer Prev. 2014;15(7):2971–2977. | ||
Wu C, Luo J, Jing L. Long non-coding RNA (lncRNA) urothelial carcinoma-associated 1 (UCA1) enhances tamoxifen resistance in breast cancer cells via inhibiting mTOR signaling pathway. Med Sci Monit. 2016;22:3860–3867. | ||
Liu D, Yu X, Wang S, et al. The gain and loss of long noncoding RNA associated-competing endogenous RNAs in prostate cancer. Oncotarget. 2016;7(35):57228–57238. | ||
Li J, Zhang Z, Xiong L, et al. SNHG1 lncRNA negatively regulates miR-199a-3p to enhance CDK7 expression and promote cell proliferation in prostate cancer. Biochem Biophys Res Commun. 2017;487(1):146–152. | ||
Wang WT, Sun YM, Huang W, He B, Zhao YN, Chen YQ. Genome-wide long non-coding RNA analysis identified circulating LncRNAs as novel non-invasive diagnostic biomarkers for gynecological disease. Sci Rep. 2016;6:23343. | ||
Kong YG, Cui M, Chen SM, Xu Y, Xu Y, Tao ZZ. LncRNA-LINC00460 facilitates nasopharyngeal carcinoma tumorigenesis through sponging miR-149-5p to up-regulate IL6. Gene. 2018;639:77–84. | ||
Li K, Sun D, Gou Q, et al. Long non-coding RNA linc00460 promotes epithelial-mesenchymal transition and cell migration in lung cancer cells. Cancer Lett. 2018;420:80–90. | ||
Liang Y, Wu Y, Chen X, et al. A novel long noncoding RNA linc00460 up-regulated by CBP/P300 promotes carcinogenesis in esophageal squamous cell carcinoma. Biosci Rep. 2017;37(5):BSR20171019. | ||
Sakakibara T, Hibi K, Koike M, et al. Plasminogen activator inhibitor-1 as a potential marker for the malignancy of gastric cancer. Cancer Sci. 2006;97(5):395–399. | ||
Aguirre D, Boya P, Bellet D, et al. Bcl-2 and CCND1/CDK4 expression levels predict the cellular effects of mTOR inhibitors in human ovarian carcinoma. Apoptosis. 2004;9(6):797–805. | ||
Nadal A, Jares P, Pinyol M, et al. Association of CDK4 and CCND1 mRNA overexpression in laryngeal squamous cell carcinomas occurs without CDK4 amplification. Virchows Arch. 2007;450(2):161–167. | ||
Liabakk NB, Talbot I, Smith RA, Wilkinson K, Balkwill F. Matrix metalloprotease 2 (MMP-2) and matrix metalloprotease 9 (MMP-9) type IV collagenases in colorectal cancer. Cancer Res. 1996;56(1):190–196. | ||
Satelli A, Li S. Vimentin in cancer and its potential as a molecular target for cancer therapy. Cell Mol Life Sci. 2011;68(18):3033–3046. | ||
Li XR, Chu HJ, Lv T, Wang L, Kong SF, Dai SZ. miR-342-3p suppresses proliferation, migration and invasion by targeting FOXM1 in human cervical cancer. FEBS Lett. 2014;588(17):3298–3307. | ||
Xie X, Liu H, Wang M, et al. miR-342-3p targets RAP2B to suppress proliferation and invasion of non-small cell lung cancer cells. Tumour Biol. 2015;36(7):5031–5038. | ||
Gao Y, Zhang SG, Wang ZH, Liao JC. Down-regulation of miR-342-3p in hepatocellular carcinoma tissues and its prognostic significance. Eur Rev Med Pharmacol Sci. 2017;21(9):2098–2102. | ||
Zhang S, Liu L, Lv Z, Li Q, Gong W, Wu H. MicroRNA-342-3p inhibits the proliferation, migration, and invasion of osteosarcoma cells by targeting astrocyte-elevated gene-1 (AEG-1). Oncol Res. 2017;25(9):1505–1515. | ||
Rizwani W, Schaal C, Kunigal S, Coppola D, Chellappan S. Mammalian lysine histone demethylase KDM2A regulates E2F1-mediated gene transcription in breast cancer cells. PLoS One. 2014;9(7):e100888. | ||
Dhar SS, Alam H, Li N, et al. Transcriptional repression of histone deacetylase 3 by the histone demethylase KDM2A is coupled to tumorigenicity of lung cancer cells. J Biol Chem. 2014;289(11):7483–7496. | ||
Kong Y, Zou S, Yang F, et al. RUNX3-mediated up-regulation of miR-29b suppresses the proliferation and migration of gastric cancer cells by targeting KDM2A. Cancer Lett. 2016;381(1):138–148. | ||
Chen JY, Li CF, Chu PY, et al. Lysine demethylase 2A promotes stemness and angiogenesis of breast cancer by upregulating Jagged1. Oncotarget. 2016;7(19):27689–27710. | ||
Chen JY, Luo CW, Lai YS, Wu CC, Hung WC. Lysine demethylase KDM2A inhibits TET2 to promote DNA methylation and silencing of tumor suppressor genes in breast cancer. Oncogenesis. 2017;6(8):e369. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.