Back to Journals » OncoTargets and Therapy » Volume 11
Lentivirus-mediated shRNA targeting MUTYH inhibits malignant phenotypes of bladder cancer SW780 cells
Authors Gao Q , Liu Y , Xie H, Zhong Y, Liao X, Zhan H, Zhou Q, Ding M, Yang K, Li A, Liu Y, Mei HB, Cai Z
Received 15 May 2018
Accepted for publication 23 July 2018
Published 21 September 2018 Volume 2018:11 Pages 6101—6109
DOI https://doi.org/10.2147/OTT.S174223
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Sanjeev K. Srivastava
Qunjun Gao,1–3 Yuhan Liu,2,3 Haibiao Xie,2,3 Yucheng Zhong,2,3 Xinhui Liao,3 Hengji Zhan,2,3 Qun Zhou,2,3 Mengting Ding,2,3 Kang Yang,2,3 Aolin Li,2,3 Yuchen Liu,2,3 Hongbing Mei,3 Zhiming Cai1–3
1Key Laboratory of Medical Reprogramming Technology, Shenzhen Second People’s Hospital, Guangzhou Medical University, Guangzhou 511436, China; 2Key Laboratory of Medical Reprogramming Technology, Shenzhen Second People’s Hospital, First Affiliated Hospital of Shenzhen University, Shenzhen 518039, China; 3Department of Urology, Shenzhen Second People’s Hospital, First Affiliated Hospital of Shenzhen University, Shenzhen 518039, China
Objectives: MUTYH is a protein-coding gene that takes part in base excision repair. Many previous studies have reported that MUTYH is directly related to hereditary adenomatous polyposis and colorectal cancer and is also associated with other cancers. However, the relationship between MUTYH and bladder cancer (BC) is unknown.
Materials and methods: The expression of MUTYH and clinical characteristics of BC were collected from databases including The Cancer Genome Atlas and Cancer Cell Line Encyclopedia. RNA sequencing and quantitative real-time PCR were used to detect MUTYH expression in SW780 BC cells. The level of MUTYH was stably downregulated by lentivirus-mediated vector in SW780 cells. Cell proliferation was evaluated using Cell Counting Kit-8 assay and 5-ethynyl-20-deoxyuridine assay, migration was detected using scratch assay and Transwell assay, and apoptosis was determined using ELISA.
Results: MUTYH was upregulated in BC tissues and SW780 cells and its expression level was positively associated with the stage and grade of carcinomas. MUTYH was successfully downregulated in SW780 cells by transducing with a lentivirus-mediated shRNA targeting MUTYH. MUTYH knockdown inhibited the proliferation and migration and induced apoptosis in SW780 cells.
Conclusion: Our data suggest that MUTYH is a new participant in bladder urothelial carcinoma. MUTYH may play a role as a biomarker and therapeutic target in BC.
Keywords: MUTYH, bladder cancer, cell proliferation, migration, apoptosis
Introduction
Bladder cancer (BC) is the ninth most common malignant tumor worldwide and has the highest mortality rate among urogenital system tumors.1,2 In recent decades, the morbidity and mortality of BC have attracted worldwide attention.3 Although many BC treatments are available, such as surgery, radiation therapy, chemotherapy, and immunotherapy, the 5-year survival rate of BC is still low due to the high recurrence rate and rapid progress of the disease.4–6 The lack of a profound understanding of BC pathogenesis is one of the most important reasons for its high mortality rate. Therefore, investigation of the molecular mechanisms underlying the pathogenesis of BC is critical.
MUTYH encodes a DNA glycosylase involved in oxidative DNA damage repair. MUTYH is a vital DNA repair enzyme that protects cells from oxidative DNA damage and is critical for a proper cellular response to DNA damage. The enzyme excises adenine bases from the DNA backbone at sites where adenine is inappropriately paired with guanine, cytosine, or 8-oxo-7,8-dihydroguanine, a major oxidatively damaged DNA lesion.7,8 When the MUTYH gene product is damaged by a double allelic germline mutation, the mutation related to cancer, such as APC and/or KRAS gene, is transformed to G through T.9 Previous studies have shown that the MUTYH gene is highly correlated with colorectal cancer and hereditary adenomatous polyposis,7,10 and this gene is also related to lung adenocarcinoma, breast cancer, and gastric cancer.11–13 However, the relationship between the gene and disease of the genitourinary system is still unknown, and the role of MUTYH in the development of BC is completely unclear and needs to be studied. Thus, we hypothesized that MUTYH may play roles in BC.
In this study, we identified MUTYH expression patterns in urinary bladder urothelial carcinoma and investigated the effects of lentivirus-mediated MUTYH knockdown on the growth, invasion, and apoptosis of a urinary bladder urothelial carcinoma cell line (SW780). According to the results, we demonstrated that MUTYH was upregulated in BC tissues and cell line (SW780). Moreover, silencing MUTYH significantly inhibited the proliferation and migration and induced apoptosis of BC cells. Our data show that MUTYH is a powerful tumor marker, and highlight its potential clinical application as a promising prognostic marker and therapeutic target.
Materials and methods
Cell lines and cell culture
The human normal urothelial cell line (SV-40-immortalized human uroepithelial cell line, SV-HUC-1), human embryonic kidney cells (293T), and human bladder transitional cell carcinoma cell line (SW780) were purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA). The SV-HUC-1 cells were cultured in F12K (Thermo Fisher Scientific, Waltham, MA, USA) plus 1% antibiotics (100 U/mL penicillin and 100 μg/mL streptomycin sulfate) and 10% FBS. The SW780 and 293 T cells were grown in DMEM (Thermo Fisher Scientific), supplemented with 10% FBS and 1% antibiotics. All cells were cultured at 37°C in a humidified atmosphere with 5% CO2.
Construction of stable transfected cell lines
A lentiviral vector encoding MUTYH shRNA was designed and synthesized by SyngenTech (Beijing, China). The sequence of the shRNA negative control with no significant homology with the human gene sequence was 5′-TAATTGTCAAATCAGAGTGCTT-3′. The shRNA sequence targeting MUTYH was 5′-GCCAGGAGATTTCAACCAAGC-3′. The fragments of these shRNAs were then cloned into pLV-hU6-hEF1a-EYFP-2A-Puro to construct the lentivirus vectors. Then the lentivirus vector and auxiliary plasmid liposomes (pMD2.G and psPAX.2) were transfected into 293 T cells using Lipofectamine 3000 (Thermo Fisher Scientific) according to the manufacturer’s protocol, to produce lentivirus vectors. The supernatant was collected at 48 and 72 hours after transfection, then centrifuged at 1,500×g to remove cell debris, and finally filtered through a 0.45 μm polyvinylidene difluoride filter. The supernatant was concentrated using a lentivirus concentration solution (1:5; BioGeek) and then incubated overnight at 4°C. The concentrated virus particles were collected and resuspended according to the manufacturer’s protocol. The human bladder transitional cell carcinoma cell line (SW780) was infected by the viral suspension with polybrene (8 μg/mL; HanBio Biotechnology, Pudong, China). At 48 hours after infection, the positive stably transduced cell lines were screened using puromycin (1 μg/mL; Sigma-Aldrich Co., St Louis, MO, USA).
RNA extraction and quantitative real-time PCR (qRT-PCR) analysis
Total RNA was extracted from normal SW780 cells, SV-HUC-1 cells, or infected SW780 cells using TRIzol reagent (Thermo Fisher Scientific) according to the manufacturer’s instructions. A Nano Drop spectrometer (Thermo Fisher Scientific) was used to measure the concentration and purity of total RNA according to the 260/280 absorbance ratio. A Revertra Ace qPCR RT Kit (Toyobo, Osaka, Japan) was used to transform RNA to cDNA. The qRT-PCR was performed using SYBR® Premix Ex Taq™ (TaKaRa Bio Inc., Shiga, Japan) according to the manufacturer’s protocols. GAPDH was chosen as the internal control to normalize the data. The primer sequences were as follows: MUTYH primers forward: 5′-ACGACCAAGAGAAACGGGAC-3′, reverse: 5′-GGCCACGAGAATAGTAGCCC-3′. GAPDH primers forward: 5′-AATCCCATCACCATCTTCCAG-3′, reverse: 5′-TTCACACCCATGACGAACAT-3′. The PCR mixtures were prepared according to the manufacturer’s protocols with PCR conditions of 40 cycles of 15 seconds at 95°C, 20 seconds at 55°C, and 30 seconds at 70°C on an ABI PRISM 7300 Fluorescent Quantitative PCR System (Thermo Fisher Scientific). The average value of each triplicate was used to calculate the relative amount of MUTYH using the 2−ΔΔCt method.
Cell proliferation assay
Cell proliferation was examined using Cell Counting Kit-8 assay (CCK-8; TransGen, Beijing, China) and a 5-ethynyl-20-deoxyuridine (EdU) assay kit (Ribobio, Guangzhou, China) according to the manufacturer’s instructions. For the CCK-8 assay, 5×103 stably transfected MUTYH shRNA or shRNA-NC SW780 cells were seeded into a 96-well plate and cultured in a 37°C incubator. At 24, 48, or 72 hours, 10 μL of CCK-8 reagent was added to each well and incubated for 1 hour. An automatic microplate reader (Bio-Rad Laboratories Inc., Hercules, CA, USA) was used to measure the absorbance at 450 nm. For the EdU assay, stably transfected cell lines were seeded into a 24-well flat-bottomed plate and incubated with 300 μL of 50 μM EdU per well for 2 hours at 37°C. Then, the cells were fixed at room temperature for 30 minutes using 150 μL fixative (PBS with 4% paraformaldehyde). Next, 150 μL of 2 mg/mL glycine was added to each well, and the decolorization plate was incubated for 5 minutes followed by washing with 300 μL of PBS. After incubation, 300 μL of 0.5% Triton X-100 was added to each well and incubated for 10 minutes for permeabilization. Then the cells were washed with PBS and reacted with 300 μL of 1× Apollo solution for 30 minutes at room temperature in the dark. Three hundred microliters of osmotic agent (0.5% Triton X-100 in PBS) was added to the cells in each well and the plate was washed and shaken three times; then the plate was washed with 300 μL of methanol twice, followed by one wash in PBS. Subsequently, the cells were incubated with 300 μL of Hoechst 33342 for 30 minutes at room temperature in the dark followed by washing with PBS. The cells were then observed under a fluorescence microscope. All experiments were repeated at least three times.
Cell migration assay
Cell migration ability was measured by scratch assay and Transwell assay. For the scratch assay, cells were seeded into six-well plates and incubated until they reached ~90% confluence. Clear lines in the wells were created using a sterile 200 μL pipette tip. A digital camera system was used to quickly take photographs of each well immediately after scratch creation, and then 24 hours later, the same areas were re-photographed. Migration distance was measured at the time points of 0 and 24 hours using the software program HMIAS-2000. For the Transwell assay, Transwell inserts with 8.0 μm pores (COStor, Kennebunk, ME, USA) were placed into wells of a 24-well plate. Cells were first starved in 200 μL serum-free medium and then 3×104 cells were placed into wells of the uncoated dishes. The bottom chambers were filled with 500 μL medium supplemented with 10% FBS. Cells were cultured at 37°C in a humidified atmosphere with 5% CO2 for 24 hours, after which the cells that had migrated to the bottom surface of the filter membrane were fixed in 4% paraformaldehyde for 20 minutes and stained with 0.1% crystal violet (Sigma-Aldrich Co.) for 25 minutes. The superfluous dye on the surface of the polycarbonate membrane was removed with sterile double-distilled water (ddH2O) and a cotton swab. The invaded cells were observed and photographed using an inverted microscope. Next, the chambers were soaked in 1 mL of 33% glacial acetic acid for 10 minutes to wash out the crystal violet. Then, 100 μL of 33% acetic acid was added to each well of the 96-well plate and the absorbance was measured at a wavelength of 570 nm using a microplate reader (Bio-Rad Laboratories Inc.). All experiments were performed at least three times.
Apoptosis assays
Apoptosis was measured by caspase-3 ELISA. The caspase-3 activity of stably transfected cell lines was detected using a human caspase 3 (Casp-3) ELISA Kit (Cusabio Technology LLC., Houston, TX, USA) according to the manufacturer’s protocol. A microplate reader was used to measure the OD values at 450 nm. Experiments were repeated at least three times.
Large database information mining and RNA sequencing
The expression levels and pathological prognosis of MUTYH were validated in The Cancer Genome Atlas (TCGA) dataset.23 Relevant mRNA expression data and clinical data of BC in TCGA cohort (TCGA-BLCA) were downloaded from UCSC Xena (https://xenabrowser.net/heatmap/#, up to March 27, 2018). MUTYH mRNA expression data in different urinary tract cancer cell lines were obtained from the Cancer Cell Line Encyclopedia.14 We also performed RNA sequencing to identify the differences in MUTYH expression in the SVHUC1 and SW780 cell lines. RNA sequencing was carried out by Shanghai Genminix Information Technology Co., Ltd. (Shanghai, China).
Statistical analyses
Each experiment was independently performed in triplicate. Data are presented as mean ± SD. SPSS 8.0 software (SPSS Inc., Chicago, IL, USA) was used to perform the statistical analysis, and GraphPad Prism 6 (GraphPad Software, Inc., La Jolla, CA, USA) was used to deal with all data. Differences between two groups were compared by Student’s t-test, and analysis of variance was used to compare the differences among the means of multiple groups. The nonparametric rank sum test was used to compare data that do not obey normal distribution and homogeneity of variance. P<0.05 was considered to be statistically significant.
Results
MUTYH was upregulated in BC tissues and SW780 cells
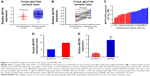
Initially, in TCGA database, data from 426 pairs of specimen tissues, of which 19 were pairs of matched BC tissues (Figure 1A and B) showed that MUTYH expression in tumor tissues was higher than in normal tissues (P<0.01). Data on the MUTYH expression level in various BC cell lines were obtained from the Cancer Cell Line Encyclopedia (Figure 1C). RNA sequencing of cell lines demonstrated that the MUTYH expression level in the BC cell line SW780 was higher than that in the normal urothelial cell line SV-HUC-1 (Figure 1D). (The original results of cell RNA sequencing are presented in the Supplementary material.) The expression levels of MUTYH in SW780 and SV-HUC-1 cells were also detected with qRT-PCR. The results were consistent with the sequencing results, confirming that MUTYH was significantly upregulated in SW780 cells (P<0.01; Figure 1E). Moreover, the MUTYH expression level was significantly correlated with patients’ gender (P<0.001), pathologic stage (P=0.015), and histologic grade (P=0.017). However, the MUTYH expression level was not remarkably correlated with age or lymphatic vessel invasion (Table 1). These data demonstrate that MUTYH is upregulated in BC and may function as an oncogene in BC.
  | Table 1 Correlation between MUTYH expression and clinicopathologic characteristics of bladder cancer patients based on TCGA-BLCA cohorta |
MUTYH was suppressed by lentivirus-mediated shRNA infection in SW780 cells
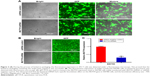
The SW780 cell line was derived from a grade I transitional cell carcinoma patient (www.atcc.org.), and in our previous work, we successfully constructed a stably transfected cell line using a lentivirus-mediated vector in SW780 cells.15 Therefore, to further explore the biological role of MUTYH in BC cells, we employed the SW780 cell line as a model to construct the stably transfected cell line. Figure 2A shows the fluorescence expression in 293 T cells during lentivirus packaging. SW780 cells were infected with virus solution packaged by 293 T cells. The virus consistently expressed MUTYH shRNA or shRNA negative control. The successful infection of SW780 cells was determined using fluorescence microscopy to observe GFP expression (Figure 2B). The results showed that the lentiviral shRNA had a specific knockdown effect on the MUTYH gene in SW780 cells, and the knockdown efficiency was ~70% (P<0.01; Figure 2C).
MUTYH knockdown inhibited the proliferation of SW780 cells
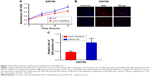
In order to investigate the possible effect of MUTYH on the growth of BC cells, the proliferative activity of SW780 cells stably transfected with MUTYH shRNA or shRNA negative control was determined by CCK-8 assay and Edu assay. First, the CCK-8 assay results showed that the proliferative ability of the MUTYH shRNA group cells was significantly inhibited compared with the negative control cells over a 72-hour period (Figure 3A). The EdU assay also demonstrated that the number of EdU-positive SW780 cells was significantly decreased in the MUTYH shRNA group compared with the shRNA negative control group (Figure 3B). Moreover, the quantitative results indicated that the number of EdU-positive cells in the MUTYH shRNA group was significantly reduced in SW780 cells (P<0.05; Figure 3C). These data demonstrated that MUTYH knockdown significantly inhibited the proliferation of SW780 cells.
MUTYH knockdown inhibited the migration of SW780 cells
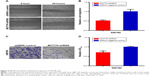
Next we investigated whether MUTYH suppressed cell migration in BC cells. The scratch assay and Transwell assay were carried out to measure the migration of SW780 cells stably transfected with MUTYH shRNA or shRNA negative control. Compared with the negative control group, cell migration ability was obviously inhibited in the MUTYH shRNA group (P<0.01; Figure 4A and B). Likewise, the Transwell assay was used to further verify our experimental results. These data demonstrated that the migration distances in the negative control group were distinctly greater than in the MUTYH shRNA group (P<0.01; Figure 4C and D). These results suggest that MUTYH knockdown inhibits the migration of SW780 cells.
MUTYH knockdown induced apoptosis of SW780 cells
To investigate whether MUTYH induces apoptosis of BC cells, a caspase-3 ELISA was performed to measure the relative caspase-3 activity of SW780 cells stably transfected with MUTYH shRNA or shRNA negative control. From the results shown in Figure 5, we found that the relative activity of caspase-3 was significantly increased in the MUTYH shRNA group compared with the negative control group (P<0.01). These results indicated that MUTYH knockdown induces apoptosis of SW780 cells.
  | Figure 5 Apoptosis was induced in SW780 cells stably transfected with MUTYH shRNA using caspase-3 ELISA. |
Discussion
Protein-coding genes are genes that encode proteins with specific functions. Differences in coding gene expression result in different functional proteins, which participate in a variety of biological actions, including tumor development. Previous studies have shown that protein-coding genes have a significant relationship with the development and prognosis of different tumors.16–19 MUTYH, one of the protein-coding genes, encodes the DNA glycosylated enzyme involved in basal resection and repair.11 MUTYH is a post-replicative DNA glycosylase that is highly conserved throughout the evolution process and is involved in the correction of mismatching caused by the misreplication of 8-hydroxyguanine.8 Several MUTYH mutations have been found to be responsible for mutyh-related polyposis, an autosomal recessive genetic disease characterized by the susceptibility of adenomatous polyps in the colon and rectum.20 It is known that mutation of mismatched repair genes will increase the susceptibility to cancer in organs other than the colon, and previous studies have found that MUTYH is highly related to colorectal cancer, breast cancer, lung adenocarcinoma, and gastric cancer, but the relationship between MUTYH and BC has not been revealed.7,11–13,21 In addition, by sorting out TCGA database, sequencing, and qPCR data, we found that MUTYH expression was relatively high in BC tissues compared with adjacent noncancerous tissues. MUTYH expression was also relatively high in SW780 cells compared with SV-HUC-1 cells. The MUTYH expression level was positively correlated with patients’ gender, pathologic stage, and histologic grade. Consequently, it was very meaningful and necessary to explore the relationship between MUTYH and BC.
This is the first study to investigate the relationship between MUTYH and BC. In the study of gene function, it is important to knock down the target gene without affecting other genes, and RNAi mediated by shRNA is a special gene silencing technology.22 Thus, to further study the role of MUTYH in BC, we used lentiviral vectors to construct stably transfected cell lines of SW780 cells that stably expressed MUTYH shRNA or shRNA negative control. Then, we examined the proliferation, migration, and apoptosis of these two cell lines in order to further explore the biological functions of MUTYH. The results showed that MUTYH knockdown, in the MUTYH shRNA group, significantly inhibited cell proliferation and migration and induced apoptosis of BC SW780 cells compared with the shRNA negative control group, suggesting that inhibition of MUTYH expression could suppress the development and progression of BC.
Limitations
In this study, although we report our preliminary findings, there are still some limitations. First, we only verified the biological function of MUTYH in the BC cell line SW780; verification of this relationship between the gene and tumor development and progression would increase the impact of our findings. Second, tumor growth and metastasis are complex multilevel multifactorial processes that require more detailed studies to confirm our projections. In our future work, we will focus on mouse tumor formation experiments to verify whether the findings are consistent at the whole body level, and whether the tumor growth can be inhibited after knocking down MUTYH. We will also further investigate the mechanism of the relationship between MUTYH and BC. More information should be obtained by comparing RNA sequencing and gene expression in SW780 cells before and after MUTYH knockdown.
Conclusion
This study demonstrated that MUTYH is upregulated in BC, and that high MUTYH expression is associated with advanced stage and high-grade carcinomas. In addition, we first revealed the important role of MUTYH in regulating the proliferation, apoptosis, and cell migration of the BC SW780 cells. The therapeutic targeting of MUTYH may be a promising approach against BC development and metastasis.
Acknowledgments
We are indebted to the contributors whose names are not included in the author list, but who participated in this program. This work was supported by the National Key Basic Research Program of China (973 Program) (2014CB745201), the Chinese High-Tech (863 Program) (2014AA020607), the National Natural Science Foundation of China (81772737), the Shenzhen Municipal Government of China (JCYJ20170413161749433, JSGG20160301161836370), the Sanming Project of Shenzhen Health and Family Planning Commission (SZSM201412018 and SZSM201512037), and the high-level university’s medical discipline construction (2016031638). In addition, we thank International Science Editing (http://www.internationalscienceediting.com) for editing this manuscript.
Disclosure
The authors report no conflicts of interest in this work.
References
Antoni S, Ferlay J, Soerjomataram I, Znaor A, Jemal A, Bray F. Bladder cancer incidence and mortality: a global overview and recent trends. Eur Urol. 2017;71(1):96–108. | ||
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7–30. | ||
Kamat AM, Hahn NM, Efstathiou JA, et al. Bladder cancer. Lancet. 2016;388(10061):2796–2810. | ||
Abufaraj M, Dalbagni G, Daneshmand S, et al. The role of surgery in metastatic bladder cancer: a systematic review. Eur Urol. 2018;73(4):543–557. | ||
Rose TL, Milowsky MI. Improving systemic chemotherapy for bladder cancer. Curr Oncol Rep. 2016;18(5):27. | ||
Sofra M, Fei PC, Fabrizi L, et al. Immunomodulatory effects of total intravenous and balanced inhalation anesthesia in patients with bladder cancer undergoing elective radical cystectomy: preliminary results. J Exp Clin Cancer Res. 2013;32:6. | ||
de Oliveira AH, da Silva AE, de Oliveira IM, Henriques JA, Agnez-Lima LF. MutY-glycosylase: an overview on mutagenesis and activities beyond the GO system. Mutat Res. 2014;769:119–131. | ||
Mazzei F, Viel A, Bignami M. Role of MUTYH in human cancer. Mutat Res. 2013;743–744:33–43. | ||
Cheadle JP, Sampson JR. MUTYH-associated polyposis – from defect in base excision repair to clinical genetic testing. DNA Repair (Amst). 2007;6(3):274–279. | ||
Nascimento EFR, Ribeiro ML, Magro DO, et al. Tissue expression of the genes mutyh and ogg1 in patients with sporadic colorectal cancer. Arq Bras Cir Dig. 2017;30(2):98–102. | ||
Out AA, Wasielewski M, Huijts PE, et al. MUTYH gene variants and breast cancer in a Dutch case–control study. Breast Cancer Res Treat. 2012;134(1):219–227. | ||
Shinmura K, Goto M, Suzuki M, et al. Reduced expression of MUTYH with suppressive activity against mutations caused by 8-hydroxyguanine is a novel predictor of a poor prognosis in human gastric cancer. J Pathol. 2011;225(3):414–423. | ||
Singh A, Singh N, Behera D, Sharma S. Genetic investigation of polymorphic OGG1 and MUTYH genes towards increased susceptibility in lung adenocarcinoma and its impact on overall survival of lung cancer patients treated with platinum based chemotherapy. Pathol Oncol Res. 2017 Dec 5 [Epub ahead of print]. | ||
Barretina J, Caponigro G, Stransky N, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483(7391):603–607. | ||
Xie H, Zhan H, Gao Q, et al. Synthetic artificial “long non-coding RNAs” targeting oncogenic microRNAs and transcriptional factors inhibit malignant phenotypes of bladder cancer cells. Cancer Lett. 2018;422:94–106. | ||
Li S, Sun X, Miao S, Liu J, Jiao WX. Differential protein-coding gene and long noncoding RNA expression in smoking-related lung squamous cell carcinoma. Thorac Cancer. 2017;8(6):672–681. | ||
Li Z, Yao Q, Zhao S, Wang Z, Li Y. Protein coding gene CRNKL1 as a potential prognostic biomarker in esophageal adenocarcinoma. Artif Intell Med. 2017;76:1–6. | ||
Zhou L-N, Li S-C, Li X-Y, Ge H, Li H-M. Identification of differential protein-coding gene expressions in early phase lung adenocarcinoma. Thorac Cancer. 2018;9(2):234–240. | ||
He X, Hou J, Ping J, Wen D, He J. Opa interacting protein 5 acts as an oncogene in bladder cancer. J Cancer Res Clin Oncol. 2017;143(11):2221–2233. | ||
Al-Tassan N, Chmiel NH, Maynard J, et al. Inherited variants of MYH associated with somatic G:C–>T:A mutations in colorectal tumors. Nat Genet. 2002;30(2):227–232. | ||
Win AK, Dowty JG, Cleary SP, et al. Risk of colorectal cancer for carriers of mutations in MUTYH, with and without a family history of cancer. Gastroenterology. 2014;146(5):e1–e5:1208–1211. | ||
Lambeth LS, Smith CA. Short hairpin RNA-mediated gene silencing. Methods Mol Biol. 2013;942:205–232. | ||
University of California, Santa Cruz. Xena Functional Genomics Explorer. Available from: https://xenabrowser.net/heatmap/#. Accessed September 10, 2018. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.