Back to Journals » Cancer Management and Research » Volume 13
Knockdown of Long Non-Coding RNA HCP5 Increases Radiosensitivity Through Cellular Senescence by Regulating microRNA-128 in Gliomas
Authors Wang C, Yu G, Xu Y, Liu C, Sun Q, Li W, Sun J, Jiang Y, Ye L
Received 13 January 2021
Accepted for publication 16 April 2021
Published 7 May 2021 Volume 2021:13 Pages 3723—3737
DOI https://doi.org/10.2147/CMAR.S301333
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Chien-Feng Li
Cuihong Wang,1 Guanying Yu,2 Ying Xu,1 Chengfei Liu,1 Qian Sun,1 Wenqing Li,1 Junhua Sun,1 Yuhua Jiang,1 Lan Ye1
1Cancer Center, The Second Hospital of Shandong University, Jinan, Shandong, 250033, People’s Republic of China; 2Department of Gastrointestinal Surgery, Jinan Central Hospital Affiliated to Shandong University, Jinan, Shandong, 250013, People’s Republic of China
Correspondence: Lan Ye
Cancer Center, The Second Hospital of Shandong University, Jinan, Shandong, 250033, People’s Republic of China
Email [email protected]
Introduction: Glioma is the most common malignant brain tumor in adults. Radiation is a key therapy in glioma. However, the radioresistance of glioma was a big challenge. HLA complex P5 (HCP5) has been reported dysregulated in several types of malignant tumor, including glioma. The role of HCP5 in the radiosensitivity of glioma is so far unknown. The present study aimed to investigate the effect of HCP5 on radiosensitivity in gliomas.
Methods: The levels of HCP5 and microRNA (miR)-128 were detected using qRT-PCR. The cell growth curve was used to show the cell proliferation and evaluate the radiosensitivity of glioma cells following exposure to X-ray. Senescence-associated β-galactosidase (SA-β-Gal) staining was used to test the cellular senescence. Luciferase reporter and RNA immunoprecipitation (RIP) assays were performed to determine the correlation between HCP5 and miR-128.
Results: HCP5 level of glioma cells was significantly higher than human astrocytes, whereas miR-128 level was lower in glioma cells. Besides, the HCP5 expression was increased in glioma tissues compared to normal brain tissues (NBTs). Knockdown of HCP5 inhibited cell proliferation and increased radiosensitivity in glioma cells. MiR-128 was predicted to be a target of HCP5. It was demonstrated that HCP5 directly bound to miR-128 and regulated its expression in glioma cells. Furthermore, the effects of HCP5 knockdown on radiosensitivity of glioma cells were attenuated by the inhibitor of miR-128.
Conclusion: These findings suggested that interaction between lncRNA HCP5 and microRNA-128 could regulate the radiosensitivity of glioma cells by intervening in cellular senescence. This might be used as the potential radio-sensitization targets for glioma therapy.
Keywords: glioma, radiosensitivity, HLA complex P5, microRNA-128, cellular senescence
Introduction
Glioma is the most common primary malignant tumor in the brain all over the world.1 The main therapeutic methods for glioma include surgery, radiation, and chemotherapy based on temozolomide. Due to the resistance to apoptosis and radiation, the prognosis of high-grade glioma remains poor.2,3 Recently, novel antiangiogenic therapies such as bevacizumab were used to treat glioma, but it could not benefit overall survival in gliomas and the median survival of glioblastoma (GBM) was only 15 months.4 Nowadays it is still a challenge for researchers to find novel therapeutic strategies for glioma.
Recent studies showed that non-coding RNAs (ncRNAs) are very important for the development of malignant tumors.5 It is known that ncRNAs lack the capacity of protein-coding.6 Basing on their transcription size, ncRNAs are divided into two groups: long non-coding RNAs (lncRNAs) and small ncRNAs. LncRNAs are greater than 200 nucleotides and were reported to be associated with many kinds of malignant tumors.7–9 Recently, the accumulating evidence showed that lncRNA expression was aberrant in gliomas, such as PVT1,10 ZNF281,11 CASC2,12 TUG1,13 PLAC2,14 XIST,15 NEAT116 and so on. This indicates that lncRNA might be a novel therapeutic target for glioma.
LncRNA histocompatibility leukocyte antigen (HLA) complex P5 (HCP5) exists mainly in immune organs, such as spleen, blood, and thymus.17 HCP5 has functional relationships with many other genes within or outside the major histocompatibility complex (MHC) genomic region that is involved with antigen processing and presentation, the interferon regulatory pathway, and epigenomic and ceRNA networks.18 It has been reported that HCP5 was associated with many human malignant tumors, such as cervical cancer,19 prostate cancer,20 bladder cancer,21 thyroid cancer,22,23 breast cancer,24 ovarian cancer25 and hepatocellular carcinoma.26 It is known that HCP5 was involved in a series of oncogenic effects in glioma cells.27 However, the role of HCP5 in the radiosensitivity of glioma remains unclear.
MicroRNAs (miRNAs) are small ncRNAs shorter than 200 bps. MiRNAs inhibit translation or promote degradation of target messenger RNA by binding to their 3′-untranslated region (3′-UTR).28 The recent studies demonstrated that miRNA was involved in the development of various malignant tumors, including gliomas.29 MicroRNA-128 (miR-128) is enriched in the brain, which has a developmental-specific expression pattern, mainly in neurons rather than in atrocities.30 MiR-128 plays an important role in the development of the nervous system and maintains the normal physical functions of the brain.31 Our previous study has reported that miR-128 plays an important role in the proliferation and radiosensitivity of glioblastoma cells by regulating B lymphoma Mo-MLV insertion region 1 homolog (Bmi-1).32–34 By the bioinformatics online tool Starbase,35 it was predicted that miR-128 has putative binding sites with HCP5. A recent study has shown that HCP5-miR128 was important in anaplastic thyroid cancer.23 However, it is unknown that whether HCP5 could regulate miR-128 to affect the radiosensitivity of gliomas.
The major aim of this study was to investigate the expression of HCP5 and miR-128 in glioma tissues and cell lines, and the roles of the interaction between HCP5 and miR-128 in regulating the radiosensitivity of glioma cells.
Materials and Methods
Clinical Specimens
This study was approved by the Ethics Committee of the Second Hospital of Shandong University. Glioma samples and normal brain tissues were obtained from the Second Hospital of Shandong University. Each subject was informed that his/her discard tissue would be used for scientific research and signed consent forms before surgery. All specimens were immediately frozen in liquid nitrogen after surgical resection. According to the WHO classification of tumors in the central nervous system (2007), glioma tissues were divided into three groups: low-grade glioma (LGG, Grade I–II, n = 5), Grade III (n = 7), glioblastoma multiforme (GBM, Grade IV, n=8). Human normal brain tissues (NBTs) were obtained from brain trauma cases as a negative control in the present study (n = 5). The study involving clinical samples was conducted in accordance with the Declaration of Helsinki.
Public Database and Online Tools
The public data of glioma were obtained from the TCGA database, including clinical profiles, survival data and the expression of HCP5 and miR-128. The online tool Gepia236 was used to analyze the expression of HCP5 in gliomas. The Oncolnc database (www.oncolnc.org) was used to analyze the survival data of gliomas correlated with the expression of HCP5 and miR-128. The online bioinformatics tool Starbase (http://starbase.sysu.edu.cn) was used to predict the possible miRNA binding site for LncRNA HCP5.35
Cell Culture
Glioma cell lines (A172, U87, U251, SHG44) were obtained from the National Collection of Authenticated Cell Cultures (Shanghai, China), and T98G cells were obtained from American Type Culture Collection (ATCC; Manassas, VA, USA). A172 and U251 cells were cultured in Dulbecco’S modified eagle medium (DMEM; Gibco, Rockville, MD, USA) with 10% fetal bovine serum (FBS; GE Healthcare Life Sciences, Logan, UT, USA). SHG44 cells were cultured in RPMI-1640 medium (Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 10% FBS. T98G, U87 cells were maintained in Minimum Essential Medium (MEM; GE Healthcare Life Sciences) with 10% FBS. Normal human astrocytes (NHAs) were purchased from Sciencell Research Laboratories (Carlsbad, CA, USA) and cultured according to the instructions provided by the manufacturer. All cells were incubated at 37°C in a humidified atmosphere containing 5% CO2.
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
Total RNA was extracted from tissues or cells using Trizol reagent (Invitrogen, Carlsbad, CA, USA) by following the protocols provided by suppliers. For HCP5, total RNA was used to synthesize cDNA using specific primers and High Capacity cDNA Reverse Transcription kit (Applied Biosystems, Foster City, CA). The PCR amplification was conducted using SYBR Green Taq Mix (Takara) on a Bio-Rad Real-Time PCR System (Bio-Rad Laboratories, Hercules, CA, USA). TaqMan MicroRNA Reverse Transcription kit and TaqMan Universal Master Mix II (Applied Biosystems) were used to detect miR-128 expression. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and U6 were used as endogenous controls for HCP5 and miR-128 expression. For HCP5 PCR cycling conditions were as follows: 95°C for 5 min, followed by 35 cycles of 95°C for 5 s, 60°C for 20 s, 70°C for 10 s. For miR-128, reverse transcription was set as follows: 30 minutes at 16°C, 30 minutes at 42°C, and 5 minutes at 85°C. PCR conditions were set as follows: 2 minutes at 50°C, 10 minutes at 95°C, 40 cycles of 15 seconds at 95°C, and 1 minute at 60°C. Fold change in gene expression was calculated by relative quantification as 2−ΔΔCT. The primers used in the present study were as follows: HCP5, forward 5′-CCG CTG GTC TCT GGA CAC ATA CT-3′, reverse 5′-CTC ACC TGT CGT GGG ATT TTG C-3′; miR-128, forward 5′-GGT CAC AGT GAA CCG GTC-3′, reverse 5′-GTG CAG GGT CCG AGG T-3′; U6, forward 5’- GCT TCG GCA GCA CAT ATA CTA AAA T −3’; reverse 5′-GGA ACG CTT CAC GAA TTT G-3′; GAPDH, forward 5′-CAT TGA CCT CAA CTA CAT GGT T-3′, reverse 5′-CCA TTG ATG ACA AGC TTC CC-3′.
Cell Transfection
The small interfering RNA against HCP5 (si-HCP5) and negative control (con-HCP5), miR-128, miR-128 inhibitor (anti-miR-128), and control inhibitors (con-anti-miRNA) were purchased from GenePharma (Shanghai, China). Lipofectamine 2000 (Invitrogen) was used to perform transfections according to the manufacturer’s instructions. Glioma cells were plated in 24-well plates and cultured 24 hours before transfection. Cells were harvested or used to perform subsequent experiments 48 hours following transfection.
Cell Radiation
Glioma cells were continuously incubated following radiation until the experiments were finished. X-ray radiation was performed by a linear accelerator source (Elekta, Stockholm, Sweden) at a dose rate of 400cGy/min. Before radiation, a radiation plan was designed, and the accuracy of the X-ray radiation doses was verified by a radiation therapy physicist using I’mRT MatriXX 2D-ion chamber array (IBA Dosimetry, Schwarzenbruck, Germany). According to our previous studies,32–34 the dosage of 8 Gy was selected to be used in the present study.
Cell Proliferation Assay
Following 48 hours for transfection, glioma cells were harvested and re-seeded in 24-well plates at a density of 5×104 cells per well and immediately exposed to X-ray radiation if needed according to the experiment design. Every 24 h, the number of cells in three wells was quantified using a cell counter (Inno-AllianceBiotech, Wilmington, DE, USA) and the mean was calculated. The results are presented as the mean ±standard error of three independent experiments.
Cell Senescence Assay
It is reported that senescent cells express a beta-G-galactosidase activity, which is histochemically detectable at pH 6.0.37 This activity is termed the senescence-associated β-galactosidase (SA-β-Gal). SA-β-Gal can be detected by histochemical staining of cells using the artificial substrate X-gal. SA-β-Gal staining was performed to detect the senescence ratio using the SA-β-Gal Kit (Beyotime Institute of Biotechnology) following the manufacturer’s instructions. The cells were considered to be positive when the cytoplasm was blue color stained with SA-β-Gal.
Luciferase Reporter Assay
Fragment of the 3′-untranslated region (UTR) of HCP5 containing the binding sites of miR-128 was amplified by PCR, The mutant type fragment sequences were also prepared. Then the wild type and mutant type fragment of HCP5 3’-UTR were separately synthesized and cloned into the pmirGLO dual-luciferase vector (Promega, Madison, WI), named HCP5-WT and HCP5-MUT, respectively. Vectors were transfected into U87 and U251 cells seeded in 24-well plates when the confluence reached 60–70%, using Lipofectamine 2000 (Invitrogen). Relative luciferase activities were measured by the dual-luciferase reporter assay system (Promega). Firefly luciferase activity was normalized to renilla luciferase activity for each analysis.
RNA Immunoprecipitation (RIP) Assay
Imprint RNA immunoprecipitation kit (Sigma-Aldrich) was used to perform RIP assay according to the manufacturer’s recommended protocol. Glioma cells were collected and lysed in RIP lysis buffer (Solarbio, Beijing, China). Subsequently, the cell lysate was incubated with anti-Argonaute2 (anti-Ago2) or anti-IgG control overnight at 4°C. Then the samples were incubated with proteinase K and immunoprecipitated RNA was isolated. Purified RNA was detected by qRT-PCR.
Statistical Analysis
GraphPad Prism v8.3.0 (GraphPad Software, La Jolla, CA) software was used for statistical analysis. All data were presented as the mean ± SD from at least three independent replicates. Differences between two different groups were analyzed using the Student’s t-test. Multiple groups were analyzed using one-way ANOVA with Dunnett's post-hoc test. Kaplan-Meier curve was used to perform survival analysis. Multiply variance analysis was performed to investigate the correlation between HCP5 or miR-128 expression and clinical indicators. Differences were considered to be statistically significant when p < 0.05.
Results
Expression of LncRNA HCP5 and MiR-128 in Glioma Cell Lines and Tissues
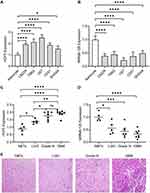
The qRT-PCR was performed to investigate the expression profile of HCP5 and miR-128 in glioma cell lines (LN229, T98G, U87, U251, SHG44), and normal human astrocytes (NHAs) was as normal control. The level of HCP5 was significantly higher in all glioma cell lines than in NHAs (Figure 1A). Meanwhile, the level of miR-128 was down-regulated greatly (Figure 1B). We also tested the expression of HCP5 and miR-128 in clinical samples. The level of HCP5 was significantly higher in glioma tissues than normal brain tissues (NBTs). In high-grade glioma, including grade III glioma and glioblastoma multiforme (GBM), the level of HCP5 is high compared to low-grade glioma (LGG) (Figure 1C), which suggested that HCP5 expression was positively correlated with the pathological grade of glioma tissues. The results also showed that the level of miR-128 in glioma tissues was lower than in NBTs and was negatively correlated with the pathological grade of glioma (Figure 1D). The typical pathological photographs of NBT and gliomas are shown in Figure 1E.
It has been reported that miR-128 expression was decreased in glioma tissues compared to adjacent NBTs.38 To verify the expression profile of lncRNA HCP5 in gliomas, we analyzed the data of glioma from the TCGA database by an online tool Gepia2. It showed that the level of HCP5 is higher in GBM tissues than in NBTs, but there is no significant difference between LGG and NBTs. Analyzing the subtypes of LGG, the level of HCP5 was higher significantly in astrocytoma tissues than in NBTs, and there is no difference between tumor and NBTs in either oligoastrocytoma or oligodendroglioma (Figure 2A). Through analyzing downloaded data of gliomas from TCGA (Table 1), it showed that the expression of HCP5 and miR-128 was correlated with age and IDH1 status. The expression of HCP5 was lower in subjects with age ≤50 years than those whose age >50 years. The subjects with IDH1 mutant had a lower HCP5 level than those without IDH mutant (p<0.0001). The expression of miR-128 was also associated with age and IDH1 status (p<0.0001). The gender and MGMT status were not correlated with HCP5 or miR-128 expression. Analyzing the data of LGG and GBM from the Oncolnc database (www.oncolnc.org), the subjects with low HCP5 expression had a longer overall survival (OS) than those with high HCP5 expression in glioma (p<0.0001), which indicates that the level of HCP5 was a poor prognostic factor in glioma. In LGG, the level of HCP5 was also a poor prognostic factor (p<0.0001). However, there is no difference in OS between the low and high HCP5 group in GBM (Figure 2B). The possible reason was most GBM subjects expressed a higher HCP5 level than the LGG subjects (p<0.0001, Figure 2C). The prognostic value of miR-128 was also evaluated. Similarly, a high expression of miR-128 indicated a longer OS in all gliomas (p=0.0026) or LGG (p<0.0001) (Figure 2D). There was no difference in OS between low and high miR-128 groups in GBM (Figure 2D) due to the lower level of miR-128 in GBM compared to LGG (Figure 2E).
 |
Table 1 Multivariate Analysis of HCP5 and MiR-128 Expression |
Knockdown of LncRNA HCP5 Increased Radiosensitivity of Glioma Cells
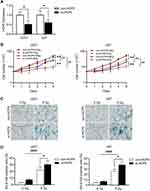
To investigate the role of HCP5 in the radiosensitivity of gliomas, si-HCP5 was transfected into U251 and U87 cells to knock down HCP5. Following 48h for transfection, the level of HCP5 was decreased greatly in both U251 and U87 (Figure 3A). After 8Gy X-ray radiation, the proliferation of U251 cells was slower than the 0Gy control group in both the con-HCP5 group and the si-HCP5 group (p<0.01). HCP knockdown promoted the inhibitory effect of X-ray on U251 cells (p<0.01). The same effect was observed in U87 cells (Figure 3B), which demonstrated that HCP5 knockdown was associated with increasing radiosensitivity in glioma cells. SA-β-gal staining showed that the knockdown of HCP5 did not change the percentage of cellular senescence in both U87 and U251. However, it increased the cellular senescence in glioma cells following X-ray radiation (Figure 3C and D). This indicated that HCP5 knockdown might increase the radiosensitivity of glioma cells by promoting cellular senescence.
HCP5 Bound to MiR-128 and Down-Regulated Its Expression
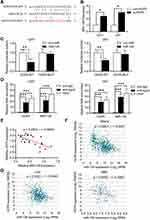
The online bioinformatics tool Starbase (http://starbase.sysu.edu.cn) was used to predict the possible miRNA binding site for LncRNA HCP5.35 The binding site of HCP5 and miR-128 is shown in Figure 4A. To verify whether HCP5 binds to miR-128 and regulates its expression, the miR-128 level was detected by qRT-PCR in U251 and U87 glioma cell lines following HCP5 knockdown. The result showed that knockdown of HCP5 increased the level of miR-128 in both U251 and U87 (Figure 4B). A cloned reporter plasmids containing the predicted miR-128 binding site (HCP5-WT or HCP5-MUT) was used to investigate whether HCP5 could bind functionally with miR-128 using a dual-luciferase reporter assay. As shown in Figure 4C, luciferase activity was significantly decreased while co-transfecting miR-128 with HCP5-WT but not with HCP5-MUT. This indicated that the miR-128 binding site within HCP5 is functional in glioma cells. Ago2 antibody was used with RIP assay to verify whether HCP5 and miR-128 were in the same RNA-induced silencing complex. Compared to the IgG group (control), HCP5 and miR-128 were captured greatly in the anti-Ago2 group (Figure 4D). In 25 clinical samples (including NBTs and glioma tissues), the level of HCP5 was negatively correlative with the level of miR-128 (Figure 4E). Besides, through analyzing the data from the TCGA database, it showed that HCP5 expression was also negatively correlated with the miR-128 expression in glioma tissues (Figure 4F). However, if analyzing the data of LGG or GBM respectively, there is no significant correlation between the expression of HCP5 and miR-128 (Figure 4G). All the above pieces of evidence supported that miR-128 was a target of HCP5 in gliomas.
HCP5 Knockdown Inhibited Cell Proliferation and Increased Radiosensitivity in Glioma Cells by Targeting MiR-128
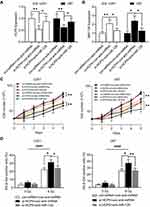
Our previous study has shown that over-expression of miR-128 could inhibit the cellular proliferation and increase the radiosensitivity of glioma cells through promoting cellular senescence.32 To further study whether the effect of HCP5 on the radiosensitivity of gliomas was associated with miR-128, U251 and U87 cells were co-transfected with si-HCP5 and anti-miR-128. Following 48h for co-transfection of si-HCP5 and anti-miRNA control, the expression of HCP5 was decreased significantly. However, in the presence of anti-miR-128, the reduction of RNA HCP5 level induced by si-HCP5 transfection was rescued partially (Figure 5A). Similar changes happened to the expression of miR-128. Si-HCP5 increased the level of miR-128 by co-transfection with anti-miRNA control, and this effect was inhibited in the presence of anti-miR-128 (Figure 5B). As shown in the cellular proliferation curve (Figure 5C), the knockdown of HCP5 increased the cell proliferation and the radiosensitivity in both U251 and U87, similar to the previous result (Figure 3B). However, down-regulation of miR-128 inhibited the change of cell proliferation and radiosensitivity caused by HCP5 knockdown. Furthermore, si-HCP5 induced an increase in cellular senescence following X-ray radiation. Co-transfection of si-HCP5 and anti-miR-128 did not change the percentage of cellular senescence caused by radiation (Figure 5D). It concluded that lncRNA HCP5 knockdown affects cell proliferation and radio-sensitivity by up-regulating miR-128 in gliomas.
Discussion
GBM is the most malignant type of gliomas. The patients with GBM have a poor prognosis due to their resistance to radiotherapy or chemotherapy.2,39 In our previous study, we have demonstrated that glioblastoma cells such as U87 were resistant to radiation-induced apoptosis. Cellular senescence was a possible mechanism for radiation-mediated inhibition of glioma cell proliferation.34 This involved reactive oxygen species (ROS) and the interaction between miR-128 and Bmi-1.32,33 It provided a novel possible target for increasing radiosensitivity of glioma.
Accumulating evidence showed that lncRNAs play important roles in a variety of malignant tumors and provide novel potential therapeutic targets.40–43 It is known that dozens of lncRNAs were up-regulated or down-regulated in gliomas, including lncRNA HCP5.44 LncRNA HCP5, located on chromosome 6p21.3, is known that associated with many types of malignant tumors (Figure 6A). For example, HCP5 was up-regulated in human prostate cancer,20 bladder cancer,21 gastric cancer,45 cervical cancer,19 including glioma.27 The fold change of HCP5 expression in glioma tissues was about six times of paired normal tissue, which is greater than other tumors (Figure 6B). That might be a reason for the poor prognosis of GBM. The malignant processes associated with HCP5 involve proliferation, invasion, migration, and epithelial-mesenchymal transition. However, the effect of HCP5 on radiosensitivity of glioma remains unclear.
In the present study, we showed that expression of the lncRNA HCP5 was increased in glioma tissues and cell lines, whereas miR-128 was decreased. The HCP5 expression was positively correlated with the histopathological grade in human glioma tissues, which suggested that HCP5 may have a predicted value as an oncogene in glioma. Basing on the analysis of the public database, high-level HCP5 or low-level miR128 was the poor prognostic factor respectively in low-grade glioma. HCP5 or miR-128 did not have the prognostic value in GBM, which might be because most GBM subjects expressed much higher HCP5 as well as much lower miR-128 than LGG or NBTs subjects. Considering all types of glioma, the level of HCP5 or miR-128 is very valuable in predicting the prognosis. Knockdown of HCP5 inhibited cellular proliferation, similar to the previous study.27 However, the trend increase about cellular senescence did not meet the significant difference, indicating the inhibition of cell proliferation might involve other pathways such as HCP5-miR-139-RUNX1.27 Our results showed that HCP5 knockdown increased cell radio-sensitivity and promoted cellular senescence in glioma cells.
It is well known that lncRNA act as a competitor for endogenous RNA (ceRNA) through binding to microRNA response elements competitively to intervene in the functions of miRNAs and their downstream genes.46–48 The studies have demonstrated that HCP549 act as a ceRNA for miR-17-5p, miR-140-5p, miR-219a-5p, miR-4656, miR-214-3p, miR-186-3p in some malignant tumors.20,50–54 By the bioinformatics tools, we predicted that HCP might be a ceRNA for miR-128 in gliomas. To explore the mechanisms about the increase of radiosensitivity medicated by HCP5 knockdown, we identified miR-128 as a target of HCP5 in glioma cells. HCP5 executed its effect on glioma by binding to miR-128. It has been confirmed previously that it did functionally down-regulated Bmi-1 to promote cellular senescence induced by X-ray radiation.32 MiR-128 has been reported to be down-regulated significantly in glioma38,55 and was associated with the proliferation and self-renewal of glioma stem-like cells.56 Our previous study also determined that miR-128 targeted Bmi-1 to regulate radiosensitivity of glioblastoma cells.32,33 Consistent with our results, recent reports demonstrated that miR-128 inhibited the proliferation of glioma cells by targeting E2F3a,57 NEK2,58 GRM1,10 NPTX1,38 RhoE.59 Another study showed that miR-128 enhances the chemosensitivity of temozolomide in glioblastoma.60 All the evidence supported that HCP5/miR-128/Bmi-1 pathway plays an important role in regulating the proliferation and radiosensitivity of glioma. Our results revealed the role of the interaction between HCP5 and miR-128 in glioma, and HCP5 knockdown increased radiosensitivity of glioma cells by up-regulating miR-128. The studies have reported that miR-128/Bmi-1 is associated with proliferation and self-renewal of glioma stem cells.61–63 Further experiments will be performed to explore whether HCP5 affects the stemness of glioma cells through miR-128/Bmi-1 pathway to regulate the proliferation and radiosensitivity in glioma.
Conclusions
In conclusion, this study highlights the importance of the interactions among lncRNA HCP5 and microRNA-128 in regulating the radiosensitivity of glioma cells. HCP5 down-regulated miR-128 to affect a series of downstream target genic effects initiating by miR-128, promoting cellular senescence following exposure of X-ray radiation in glioma cells. Thus, HCP5/miR-128 may be used as potential radiosensitization targets for the treatment of glioma.
Funding
This work was supported by the Shandong Province Key Research and Development Plan [grant numbers 2019GSF108117]; the Young Taishan Scholars Program, Shandong, China [grant number: tsqn201909178]; Shandong Province Nature Science Foundation [grant number: ZR2020MH208]; the National Nature Science Foundation of China [grant number: 81902920]; and the Major Science and Technology Project of Shandong Province [grant number: 2014ZZCX02104].
Disclosure
The authors declare no conflicts of interest.
References
1. Chen R, Smith-Cohn M, Cohen AL, Colman H. Glioma subclassifications and their clinical significance. Neurotherapeutics. 2017;14(2):284–297. doi:10.1007/s13311-017-0519-x
2. Cancer Genome Atlas Research N. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–1068. doi:10.1038/nature07385
3. Homma T, Fukushima T, Vaccarella S, et al. Correlation among pathology, genotype, and patient outcomes in glioblastoma. J Neuropathol Exp Neurol. 2006;65(9):846–854. doi:10.1097/01.jnen.0000235118.75182.94
4. Khasraw M, Ameratunga MS, Grant R, Wheeler H, Pavlakis N, Antiangiogenic therapy for high-grade glioma. Cochrane Database Syst Rev. 2014;9:CD008218. doi:10.1002/14651858.CD008218.pub3
5. Deng G, Sui G. Noncoding RNA in oncogenesis: a new era of identifying key players. Int J Mol Sci. 2013;14(9):18319–18349. doi:10.3390/ijms140918319
6. Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136(4):629–641. doi:10.1016/j.cell.2009.02.006
7. Teppan J, Barth DA, Prinz F, Jonas K, Pichler M, Klec C. Involvement of long non-coding RNAs (lncRNAs) in tumor angiogenesis. Noncoding RNA. 2020;6(4):52.
8. Egranov SD, Hu Q, Lin C, Yang L. LncRNAs as tumor cell intrinsic factors that affect cancer immunotherapy. RNA Biol. 2020;17(11):1625–1627. doi:10.1080/15476286.2020.1767455
9. Lu W, Cao F, Wang S, Sheng X, Ma J. LncRNAs: the regulator of glucose and lipid metabolism in tumor cells. Front Oncol. 2019;9:1099. doi:10.3389/fonc.2019.01099
10. Fu C, Li D, Zhang X, Liu N, Chi G, Jin X. LncRNA PVT1 facilitates tumorigenesis and progression of glioma via regulation of MiR-128-3p/GREM1 axis and BMP signaling pathway. Neurotherapeutics. 2018;15(4):1139–1157. doi:10.1007/s13311-018-0649-9
11. Li XT, Li JC, Feng M, Zhou YX, Du ZW. Novel lncRNA-ZNF281 regulates cell growth, stemness and invasion of glioma stem-like U251s cells. Neoplasma. 2019;66(1):118–127. doi:10.4149/neo_2018_180613N391
12. Wang P, Liu YH, Yao YL, et al. Long non-coding RNA CASC2 suppresses malignancy in human gliomas by miR-21. Cell Signal. 2015;27(2):275–282. doi:10.1016/j.cellsig.2014.11.011
13. Cai H, Xue Y, Wang P, et al. The long noncoding RNA TUG1 regulates blood-tumor barrier permeability by targeting miR-144. Oncotarget. 2015;6(23):19759–19779. doi:10.18632/oncotarget.4331
14. Hu YW, Kang CM, Zhao JJ, et al. LncRNA PLAC2 down-regulates RPL36 expression and blocks cell cycle progression in glioma through a mechanism involving STAT1. J Cell Mol Med. 2018;22(1):497–510. doi:10.1111/jcmm.13338
15. Yao Y, Ma J, Xue Y, et al. Knockdown of long non-coding RNA XIST exerts tumor-suppressive functions in human glioblastoma stem cells by up-regulating miR-152. Cancer Lett. 2015;359(1):75–86. doi:10.1016/j.canlet.2014.12.051
16. Wang C, Chen Y, Wang Y, et al. Inhibition of COX-2, mPGES-1 and CYP4A by isoliquiritigenin blocks the angiogenic Akt signaling in glioma through ceRNA effect of miR-194-5p and lncRNA NEAT1. J Exp Clin Cancer Res. 2019;38(1):371. doi:10.1186/s13046-019-1361-2
17. Liu Y, Helms C, Liao W, et al. A genome-wide association study of psoriasis and psoriatic arthritis identifies new disease loci. PLoS Genet. 2008;4(3):e1000041. doi:10.1371/journal.pgen.1000041
18. Kulski JK. Long noncoding RNA HCP5, a hybrid HLA Class I endogenous retroviral gene: structure, expression, and disease associations. Cells. 2019;8(5). doi:10.3390/cells8050480
19. Yu Y, Shen HM, Fang DM, Meng QJ, Xin YH. LncRNA HCP5 promotes the development of cervical cancer by regulating MACC1 via suppression of microRNA-15a. Eur Rev Med Pharmacol Sci. 2018;22(15):4812–4819. doi:10.26355/eurrev_201808_15616
20. Hu R, Lu Z. Long noncoding RNA HCP5 promotes prostate cancer cell proliferation by acting as the sponge of miR4656 to modulate CEMIP expression. Oncol Rep. 2020;43(1):328–336. doi:10.3892/or.2019.7404
21. Zhao C, Li Y, Hu X, et al. LncRNA HCP5 promotes cell invasion and migration by sponging miR-29b-3p in human bladder cancer. Onco Targets Ther. 2020;13:11827–11838. doi:10.2147/OTT.S249770
22. Liang L, Xu J, Wang M, et al. LncRNA HCP5 promotes follicular thyroid carcinoma progression via miRNAs sponge. Cell Death Dis. 2018;9(3):372. doi:10.1038/s41419-018-0382-7
23. Chen J, Zhao D, Meng Q. Knockdown of HCP5 exerts tumor-suppressive functions by up-regulating tumor suppressor miR-128-3p in anaplastic thyroid cancer. Biomed Pharmacother. 2019;116:108966. doi:10.1016/j.biopha.2019.108966
24. Wu J, Chen H, Ye M, et al. Corrigendum to “Downregulation of long noncoding RNA HCP5 contributes to cisplatin resistance in human triple-negative breast cancer via regulation of PTEN expression” [Biomed. Pharmacother. 115 (2019) 108869]. Biomed Pharmacother. 2020;122:109789. doi:10.1016/j.biopha.2019.109789
25. Liu N, Zhang R, Zhao X, et al. A potential diagnostic marker for ovarian cancer: involvement of the histone acetyltransferase, human males absent on the first. Oncol Lett. 2013;6(2):393–400. doi:10.3892/ol.2013.1380
26. Lange CM, Bibert S, Dufour JF, et al. Comparative genetic analyses point to HCP5 as susceptibility locus for HCV-associated hepatocellular carcinoma. J Hepatol. 2013;59(3):504–509. doi:10.1016/j.jhep.2013.04.032
27. Teng H, Wang P, Xue Y, et al. Role of HCP5-miR-139-RUNX1 feedback loop in regulating malignant behavior of glioma cells. Mol Ther. 2016;24(10):1806–1822. doi:10.1038/mt.2016.103
28. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi:10.1016/S0092-8674(04)00045-5
29. Tumilson CA, Lea RW, Alder JE, Shaw L. Circulating microRNA biomarkers for glioma and predicting response to therapy. Mol Neurobiol. 2014;50(2):545–558. doi:10.1007/s12035-014-8679-8
30. Smirnova L, Grafe A, Seiler A, Schumacher S, Nitsch R, Wulczyn FG. Regulation of miRNA expression during neural cell specification. Eur J Neurosci. 2005;21(6):1469–1477. doi:10.1111/j.1460-9568.2005.03978.x
31. Persengiev SP, Kondova II, Bontrop RE. The impact of MicroRNAs on brain aging and neurodegeneration. Curr Gerontol Geriatr Res. 2012;2012:359369. doi:10.1155/2012/359369
32. Sun J, Ye L, Wang C, Li N, Wang D, Li X. MicroRNA-128 increases glioma cell radio-sensitivity by suppressing senescent evasion through oncogene Bmi-1. Int J Clin Exp Pathol. 2018;11(3):1423–1430.
33. Ye L, Yu G, Wang C, et al. MicroRNA128a, BMI1 polycomb ring finger oncogene, and reactive oxygen species inhibit the growth of U87 MG glioblastoma cells following exposure to Xray radiation. Mol Med Rep. 2015;12(4):6247–6254. doi:10.3892/mmr.2015.4175
34. Ye L, Wang C, Yu G, et al. Bmi-1 induces radioresistance by suppressing senescence in human U87 glioma cells. Oncol Lett. 2014;8(6):2601–2606. doi:10.3892/ol.2014.2606
35. Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42(Database issue):D92–97. doi:10.1093/nar/gkt1248
36. Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98–W102. doi:10.1093/nar/gkx247
37. Dimri GP, Lee X, Basile G, et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci U S A. 1995;92(20):9363–9367. doi:10.1073/pnas.92.20.9363
38. Huo L, Wang B, Zheng M, et al. miR-128-3p inhibits glioma cell proliferation and differentiation by targeting NPTX1 through IRS-1/PI3K/AKT signaling pathway. Exp Ther Med. 2019;17(4):2921–2930. doi:10.3892/etm.2019.7284
39. Van Meir EG, Hadjipanayis CG, Norden AD, Shu HK, Wen PY, Olson JJ. Exciting new advances in neuro-oncology: the avenue to a cure for malignant glioma. CA Cancer J Clin. 2010;60(3):166–193. doi:10.3322/caac.20069
40. Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011;1(5):391–407. doi:10.1158/2159-8290.CD-11-0209
41. Renganathan A, Felley-Bosco E. Long noncoding RNAs in cancer and therapeutic potential. Adv Exp Med Biol. 2017;1008:199–222.
42. Fu D, Shi Y, Liu JB, et al. Targeting long non-coding RNA to therapeutically regulate gene expression in cancer. Mol Ther Nucleic Acids. 2020;21:712–724. doi:10.1016/j.omtn.2020.07.005
43. Choudhari R, Sedano MJ, Harrison AL, et al. Long noncoding RNAs in cancer: from discovery to therapeutic targets. Adv Clin Chem. 2020;95:105–147.
44. Peng Z, Liu C, Wu M. New insights into long noncoding RNAs and their roles in glioma. Mol Cancer. 2018;17(1):61. doi:10.1186/s12943-018-0812-2
45. Chen S, Ren C, Zheng H, Sun X, Dai J. The effect of long non-coding RNA (lncRNA) HCP5 on regulating epithelial-mesenchymal transition (EMT)-related markers in gastric carcinoma is partially reversed by miR-27b-3p. Med Sci Monit. 2020;26:e921383.
46. Qi X, Zhang DH, Wu N, Xiao JH, Wang X, Ma W. ceRNA in cancer: possible functions and clinical implications. J Med Genet. 2015;52(10):710–718. doi:10.1136/jmedgenet-2015-103334
47. Ergun S, Oztuzcu S. Oncocers: ceRNA-mediated cross-talk by sponging miRNAs in oncogenic pathways. Tumour Biol. 2015;36(5):3129–3136. doi:10.1007/s13277-015-3346-x
48. Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505(7483):344–352. doi:10.1038/nature12986
49. Wu Y, Qian Z. Long non-coding RNAs (lncRNAs) and microRNAs regulatory pathways in the tumorigenesis and pathogenesis of glioma. Discov Med. 2019;28(153):129–138.
50. Zhu K, Wang L, Zhang X, et al. LncRNA HCP5 promotes neuroblastoma proliferation by regulating miR-186-5p/MAP3K2 signal axis. J Pediatr Surg. 2020. doi:10.1016/j.jpedsurg.2020.10.011
51. Zhao J, Bai X, Feng C, Shang X, Xi Y. Long non-coding RNA HCP5 facilitates cell invasion and epithelial-mesenchymal transition in oral squamous cell carcinoma by miR-140-5p/SOX4 axis. Cancer Manag Res. 2019;11:10455–10462. doi:10.2147/CMAR.S230324
52. Wang L, Luan T, Zhou S, et al. LncRNA HCP5 promotes triple negative breast cancer progression as a ceRNA to regulate BIRC3 by sponging miR-219a-5p. Cancer Med. 2019;8(9):4389–4403. doi:10.1002/cam4.2335
53. Liu Y, Wang J, Dong L, et al. Long noncoding RNA HCP5 regulates pancreatic cancer gemcitabine (GEM) resistance by sponging Hsa-miR-214-3p to target HDGF. Onco Targets Ther. 2019;12:8207–8216. doi:10.2147/OTT.S222703
54. Chen R, Xin G, Zhang X. Long non-coding RNA HCP5 serves as a ceRNA sponging miR-17-5p and miR-27a/b to regulate the pathogenesis of childhood obesity via the MAPK signaling pathway. J Pediatr Endocrinol Metab. 2019;32(12):1327–1339. doi:10.1515/jpem-2018-0432
55. Ciafre SA, Galardi S, Mangiola A, et al. Extensive modulation of a set of microRNAs in primary glioblastoma. Biochem Biophys Res Commun. 2005;334(4):1351–1358. doi:10.1016/j.bbrc.2005.07.030
56. Godlewski J, Nowicki MO, Bronisz A, et al. Targeting of the Bmi-1 oncogene/stem cell renewal factor by microRNA-128 inhibits glioma proliferation and self-renewal. Cancer Res. 2008;68(22):9125–9130. doi:10.1158/0008-5472.CAN-08-2629
57. Zhang Y, Chao T, Li R, et al. MicroRNA-128 inhibits glioma cells proliferation by targeting transcription factor E2F3a. J Mol Med. 2009;87(1):43–51. doi:10.1007/s00109-008-0403-6
58. Ye Y, Zhi F, Peng Y, Yang CC. MiR-128 promotes the apoptosis of glioma cells via binding to NEK2. Eur Rev Med Pharmacol Sci. 2018;22(24):8781–8788. doi:10.26355/eurrev_201812_16645
59. Shang C, Hong Y, Guo Y, Liu YH, Xue YX. miR-128 regulates the apoptosis and proliferation of glioma cells by targeting RhoE. Oncol Lett. 2016;11(1):904–908. doi:10.3892/ol.2015.3927
60. She X, Yu Z, Cui Y, et al. miR-128 and miR-149 enhance the chemosensitivity of temozolomide by Rap1B-mediated cytoskeletal remodeling in glioblastoma. Oncol Rep. 2014;32(3):957–964. doi:10.3892/or.2014.3318
61. Shan ZN, Tian R, Zhang M, et al. miR128-1 inhibits the growth of glioblastoma multiforme and glioma stem-like cells via targeting BMI1 and E2F3. Oncotarget. 2016;7(48):78813–78826. doi:10.18632/oncotarget.12385
62. Tu Y, Gao X, Li G, et al. MicroRNA-218 inhibits glioma invasion, migration, proliferation, and cancer stem-like cell self-renewal by targeting the polycomb group gene Bmi1. Cancer Res. 2013;73(19):6046–6055. doi:10.1158/0008-5472.CAN-13-0358
63. Peruzzi P, Bronisz A, Nowicki MO, et al. MicroRNA-128 coordinately targets polycomb repressor complexes in glioma stem cells. Neuro Oncol. 2013;15(9):1212–1224. doi:10.1093/neuonc/not055
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.