Back to Journals » Drug Design, Development and Therapy » Volume 14
KLF6 Induces Apoptosis in Human Lens Epithelial Cells Through the ATF4-ATF3-CHOP Axis
Authors Tian F, Zhao J, Bu S, Teng H , Yang J, Zhang X, Li X, Dong L
Received 5 June 2019
Accepted for publication 1 December 2019
Published 9 March 2020 Volume 2020:14 Pages 1041—1055
DOI https://doi.org/10.2147/DDDT.S218467
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Cristiana Tanase
Fang Tian,* Jinzhi Zhao,* Shaochong Bu, He Teng, Jun Yang, Xiaomin Zhang, Xiaorong Li,* Lijie Dong*
Tianjin Key Laboratory of Retinal Functions and Diseases, Tianjin Medical University Eye Hospital, Tianjin, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Xiaorong Li; Lijie Dong
Tianjin Key Laboratory of Retinal Functions and Diseases, Tianjin Medical University Eye Hospital, No. 251 Fu Kang Road Nankai District, Tianjin 300384, People’s Republic of China
Tel +86 22 23346430;
Tel +86 1405326-6077
Email [email protected]; [email protected]
Background: Many studies have confirmed that high myopia is related to the high prevalence of cataracts, which results from apoptosis of lens epithelial cells (LECs) due to endoplasmic reticulum stress. Krüppel-like factor 6 (KLF6) is a tumor suppressor that is involved in the regulation of cell proliferation and apoptosis.
Purpose: In this study, our purpose was to find the relationship between KLF6-induced apoptosis in LECs and ATF4 (activating transcription factor 4)-ATF3 (activating transcription factor 3)-CHOP (C/EBP homologous protein) signaling pathway.
Methods: KLF6, ATF4, ATF3, and CHOP were ectopically expressed using cDNAs subcloned into the pCDNA3.1+ vector. ATF4, ATF3, and CHOP knockdown were performed by small interfering RNA (siRNA). Expression of relative gene was tested using QT-PCR and western-blot. Then, accompanied by UVB stimulation, cell viability was measured by CCK-8 assay; The cell damage was examined by live & dead staining; The apoptotic markers Bax and Bcl-2 were detected by immunoblotting; Quantitative apoptotic levels were measured with the Apoptosis Detection Kit; The expression level of reactive oxygen-free radical (ROS) was analyzed by DCFH-DA` probe.
Results: Ectopically expressed ATF4, ATF3, and CHOP-induced apoptosis in cells, whereas ATF4, ATF3, and CHOP knockdown by small interfering RNA (siRNA) blocked KLF6-induced apoptosis. In addition, we determined that ATF4 regulates ATF3 and CHOP expression and that ATF3 silencing reduces CHOP upregulation without changing ATF4 levels; however, ATF4 and ATF3 expression was unaffected by blockade of CHOP, suggesting that KLF6 triggers endoplasmic reticulum stress in LECs by mediating the ATF4-ATF3/CHOP axis. Besides, KLF6 overexpression significantly induced LEC apoptosis under UV radiation, as demonstrated by the elevated Bax/Bcl-2 ratio.
Conclusion: The ATF4-ATF3-CHOP pathway plays an important role in KLF6-induced apoptosis in HLECs. Our results increase our understanding of the mechanisms that regulate LEC apoptosis and contribute to the development of a new preventative strategy for cataract.
Keywords: KLF6, ATF4, apoptosis, HLECs, endoplasmic reticulum stress
Introduction
Cataract is a clouding of the lens in the eye that affects vision. The process of cataract initiation remains unclear, even though more and more related mechanisms have been revealed. It was demonstrated that all age-related cataract patients had a remarkable increase in apoptotic epithelial cells compared to healthy people, indicating apoptosis in lens may lead to cataract. It is generally accepted that endoplasmic reticulum stress (ERS) induces lens epithelial cell apoptosis.1 Although several drugs that can block LEC apoptosis have been identified, none has shown a definitive effect on cataracts. Therefore, noninvasive means for preventing LEC apoptosis are being investigated.
Many studies have revealed a strong correlation between high myopia (myopia exceeding 6.00 diopter (D) or axial length longer than 26 mm) and cataracts.2–4 It is well documented that myopia is a substantial risk factor for several subtypes of cataract, such as cortical cataract and posterior subcapsular cataract.5,6 Currently, the only effective treatment for cataract is phacoemulsification. However, operating on highly myopic eyes is challenging because of zonular weakness or posterior capsular rupture; moreover, this procedure could increase the risk of retinal detachment and macular edema postoperatively. In addition, a retrospective study in Taiwan reviewed that high myopia might accelerate nuclear sclerotic cataract in younger patients. Our initial object was to understand the mechanism of high myopia cataracts and develop targeted treatment protocols. In previous work, we performed mRNA microarray to find the differentially expressed genes between high myopia patients versus non-myopia patients and perform microarray analysis. GO functional enrichment analysis suggested the functions of differentially expressed genes between the two groups can be mainly divided into three parts: biological behavior regulation, cell component formation and molecular function. Differentially expressed genes participate in molecular biological processes and affect cell energy metabolism and protein synthesis (paper has not been published yet). We also identified two differentially expressed genes: Krüppel-like factor 6 (KLF6) and ATF4. According to this information, we designed a series of experiments to verified specialized function and interactions between KLF6/ATF4.
Our previous results have confirmed that KLF6 suppresses the IGF-1-induced proliferation of human LECs,7,8 a finding that is consistent with the inhibition of rat LEC proliferation by KLF6 overexpression.9 We also found that high KLF6 expression can induce apoptosis, suggesting that KLF6 might be a potential factor in the regulation of apoptosis.10 To further characterize the mechanism by which KLF6 induce apoptosis in LECs, we explored the molecular mechanism of KLF6 on the induction of ER stress and demonstrate a novel mechanism of ATF4 as a key mediator in KLF6-induced apoptosis.11
KLF6 is a tumor suppressor gene that is involved in regulating cell proliferation and apoptosis.12 The overexpression of KLF6 upregulates the proapoptotic genes FAS, TNF, TNFSF12, and PYCARD and inhibits the expression of the antiapoptotic gene IL10. FAS protein expression is positively correlated with KLF6 expression in Schwann cells.13 ERS is caused by the dysfunction of ER homeostasis and correlates with apoptosis in LECs.14 ATF4 is an activating transcription factor and is a key molecule in the induction of apoptosis during ER stress.15,16 ATF4, an upstream regulator of CHOP and a proapoptotic factor in cellular stress,17 mediates ER stress-induced cell apoptosis in cataract lens epithelium.18,19 To study this further, we put insight in several ER-related genes, such as ATF3 and CHOP.
CHOP is a downstream component of endoplasmic reticulum (ER) stress pathways, which can be triggered by pancreatic ER kinase-like ER kinase-, activating downstream target and cause ER-related apoptosis. By now, CHOP-induced apoptosis in ER stress was related to various human diseases, such as neurodegenerative diseases, diabetes and tumor.20 ATF3, like ATF4, is also a stress responsive protein, which can be triggered by ER stress. It was previously reported that ATF3 played an integral part in the PERK/eIF2 signaling branch of the UPR.21 PERK/eIF2 cascade has been proved to be a remarkable pathway that could influence apoptosis.22
For the past few years, multiple researchers have reported a potential correlation in ATF4/ATF3/CHOP. Liu et al reported that ATF4-ATF3-CHOP could induce human pulmonary cancer cells apoptosis by mediating DR5.23 Joo et al demonstrate ATF4-ATF3-CHOP cascade act as an intrinsic pathway of the ER induced apoptosis.24 Those studies implying that there might be a connection between the three genes.
UV exposure is inevitable in daily life and is closely linked to eye diseases. There is growing evidence that oxidative stress under UV exposure plays a critical role in the HLECs apoptosis in cataract pathogenesis. UV radiation stimulate intracellular ROS production, a complicated event involving several potential pathways, such as p53 pathway or Nrf2/ARE pathway.25–27 Therefore, studying UV-induced apoptosis in human LECs may provide clues for the exploration of the causes of cataract formation and lead to the development of novel treatments. In our investigation, we used UV radiation to simulate the daily sunlight in natural environment, followed by comparison of cellular apoptosis level between KLF6 up-regulated HLECS and normal cells. Our data showed that KLF6 up-regulation was highly susceptible to UV-induced cell apoptosis cascade.
In our study, we examine the interaction between ATF4/ATF3/CHOP under HLECs apoptosis induced by KLF6. Our results represent a novel therapeutic target for ER stress-based cataract therapeutic approaches.
Materials and Methods
Cell Culture and Transient Transfection
Human lens epithelial cell line was kindly provided by Yang Chunbo (Tianjin Medical University, Eye Institute). The cells were grown in Eagle’s minimum essential medium that contained 10% fetal bovine serum. For transient transfection, plasmids were transfected into cells using Lipofectamine 2000 (Invitrogen) following the manufacturer’s protocol.8,28,29
Plasmid Construction: Human cDNAs encoding the full-length KLF6 gene, ATF4, ATF3, and CHOP were obtained by PCR. The resulting cDNAs were then subcloned into the pCDNA3.1+ vector to overexpress each protein, respectively.
siRNA design: To suppress KLF6, ATF3, CHOP and ATF4 expression, we used the following siRNAs: KLF6 sense (5’-GCCCGAGCUUUUGUUACAAtt-3’) and antisense (5’-UUGUAACAAAAGCUCGGGCtg-3’); ATF3 sense (5’-UCACAAAAGCCGAGGUAGCtt-3’) and antisense (5’-GCUACCUCGGCUUUUGUGAtg-3’); ATF4 sense (5’-ggagcgcauugauacuagad-3’) and antisense (5’-ucuaguaucaaugcgcuccd-3), and CHOP sense (5’-gccuaggucucuuagaugad-3’) and antisense (5’-ucaucuaagagaccuaggcd-3’), respectively. Nonsilencing siRNA (5’-uucuccgaacgugucacgudTdT-3’ and 5’-acgugacacguucggagaadTdT-3’) was used as a negative control. HLECs were transfected with KLF6, ATF3, ATF4, and CHOP using Lipofectamine RNAiMAX Transfection Reagent (Cat. # 13778150) in accordance with the manufacturer’s instructions. Protein samples were collected for Western blot analysis 48 h after transfection.
UVB Irradiation
At 80% to 90% confluence, HLECs were washed twice with PBS, and UVB irradiation was performed under a thin layer of PBS. After 24 hrs transfection, cells were exposed to UVB radiation (20 mJ/cm2) for 200 seconds, incubated for another 48 hrs, and collected for further use.30
Cell Viability Assay
Cell viability was measured using Cell Counting Kit-8 (Dojindo Laboratories, Kumamoto, Japan). Briefly, HLEC cells (5×104 cells/well) were seeded into 96-well plates and left to adhere overnight. After the specific treatments, the cells were incubated with plain DMEM basic (100 μL) that contained 10% CCK-8 solution (10μL) for 2 h at 37°C. The absorbance of the cell solution was measured at 450 nm on an Infinite 200 PRO Multimode Microplate Reader (Tecan Group Ltd., Switzerland). This experiment was repeated 3 times.
Hoechst Staining
Cell apoptosis was observed using Hoechst staining. HLECs were fixed in 4% formaldehyde for 15 min at room temperature. After being washed with PBS, the cells were stained with ready-to-use Hoechst staining buffer (Yeasan, China) at room temperature for 5 min. After absorbing the dye solution, the cells were washed with PBS twice and subsequently observed under a fluorescence microscope (Olympus) using a 350-nm excitation filter. Apoptotic cells were counted under 10 views in each condition and the average was calculated. This experiment was repeated 3 times.31
Live/Dead Staining Assay
Cell apoptosis was detected using LIVE/DEAD™ Viability/Cytotoxicity Kit for mammalian cells (L3224, Thermo Fisher). HLECs were seeded in a 12-well plate, and after 24 hrs of incubation, the cells were subjected to different treatments and washed with ice-cold PBS twice. Then, the cells were incubated in 500 μL tissue culture-grade D-PBS containing 1 μM calcein AM (Component A) and 2 μM ethidium homodimer-1 (EthD-1) (Component B) for 30 min at room temperature in the dark. Following the incubation, the live cells were observed using a 490-nm excitation filter, whereas the dead cells were observed using a 545-nm excitation filter under a fluorescence microscope (Olympus). Dead cells were counted under 10 views in each condition and the average was calculated. This experiment was repeated 3 times.
ROS Assay
DCFH-DA assay was employed to determine intracellular ROS (reactive oxygen species) levels. HLECs were seeded in a 24-well plate, and after 24 hrs of incubation, the cells were subjected to different treatments and washed with cold PBS twice. Then, the cells were treated with 30 µL PBS containing 10 μM DCFH-DA (Sigma–Aldrich, St. Louis, MO, USA) and 4 μM ER-tracker Red (Yeasan, China) for 30 mins at 37°C in the dark. ER-tracker Red staining was used to mark the endoplasmic reticulum. After the incubation, the cells were rinsed with PBS three times to remove excess DCFH-DA. Intracellular ROS levels were measured under a fluorescence microscope (Olympus). This experiment was repeated 3 times.29
TUNEL Assay
Terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling (TUNEL) was performed to confirm the existence of apoptotic cells. A total of 2 x 104 cells were seeded onto coverslips. After overnight incubation, the cells were treated or left untreated as indicated, fixed with 4% paraformaldehyde for 30 min, and permeabilized with 0.1% Triton X-100. Then, the cells were incubated with TUNEL reaction solution (Yeasan, China) for 1 h in the dark at 37°C. After the incubation, the cells were treated with a drop of 5 µg/mL DAPI (Thermo Fisher Scientific, Waltham, MA, USA) for 10 min and washed with PBS three times. Apoptotic cells were visualized, based on green fluorescence, under a fluorescence microscope. Nuclei were counterstained with DAPI (4’,6-diamidino-2-phenylindole). The positive control for apoptosis was pretreated with DNase I to enzymatically induce DNA strand breaks. This experiment was repeated 3 times.
Apoptosis Assays
Apoptosis was quantified using the Cell Death Detection ELISA PLUS kit (Roche Applied Science, Catalog No. 11774425001, Mannheim, Germany) following the manufacturer’s protocol. This assay evaluates apoptosis by measuring the contents of mono- and oligonucleosomes in the lysates of apoptotic cells. The cell lysates were placed in a streptavidin-coated microplate and incubated with a mixture of anti-histone/biotin and anti-DNA/peroxidase. The amount of peroxidase that was retained in the immunocomplex was determined photometrically with 2,2-azino-bis(3-ethylbenzthiazoline-6-sulfonic acid as the substrate. Absorbance was measured at 405 nm. This experiment was repeated 3 times.
RNA Extraction, Reverse Transcription, and Quantitative Reverse-Transcription Polymerase Chain Reaction (qRT-PCR)
Total RNA was extracted from LECs using the GeneJET RNA Purification Kit (Thermo Fisher Scientific, Beijing, China) as previously described.32,33 RNA concentration and purity were determined on a Nanodrop 2000 (Thermo Fisher Scientific, Waltham, MA, USA). Total RNA (6μg) was digested with DNase I to eliminate residual DNA, and then, 1 μg of total RNA was subjected to reverse transcription in a 20-μL reaction mixture using the RevertAid cDNA Synthesis Kit (Thermo Fisher Scientific, USA) according to the manufacturer’s instructions. Real-time PCR was performed on an HT7900 Real-time PCR System (Applied Biosystems, Foster City, CA, USA). The reaction mixture contained SYBR Green FastStart 2X Master Mix (Roche, Beijing, China), cDNA template, and gene-specific primers. The primers are listed in Table 1. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) served as the internal control. The cDNA content of related genes in each LEC sample was normalized to that of GAPDH. The relative expression levels of genes were quantified using the comparative threshold cycle (2−ΔΔCt) method. This experiment was repeated 3 times.34,35,36
 |
Table 1 Primers Used for qRT-PCR |
Western Blot Analysis
Lysates of LECs were prepared in RIPA buffer (1% Nonidet P-40, 0.5% sodium deoxycholate, and 0.1% sodium dodecyl sulfate [SDS] in PBS) following previously described protocols.14 A complete protease inhibitor mixture (catalog no. 04693159001, Roche Applied Science, Mannheim, Germany) was added to the lysis buffer prior to analysis. Protein concentration was determined by BCA protein assay (Catalog No. 23228, Pierce, Thermo Scientific, Rockford, IL, USA). Equal amounts of proteins were separated on 10% SDS polyacrylamide gels (100 V for 120 min) and transferred to polyvinylidene fluoride (PVDF) membranes (Millipore, MA) on a semi-dry transfer apparatus (Bio-Rad) for 40 min at 20 V. Binding of nonspecific proteins to the membrane was blocked with blocking buffer (5% nonfat milk, 200 mM NaCl, 50 mM Tris, 0.05% Tween 20) at room temperature (RT) for 1 h. The blocked membrane was then incubated with the following primary antibodies: anti-KLF6, C-terminal (Abcam, ab135783); anti-ATF4 [EPR18111] (Abcam, ab184909); anti-ATF3 [44C3a] (Abcam, ab58668), anti-DDIT3 [9C8] (Abcam, ab11419), anti-Bax [E63] (Abcam, ab32503); and anti-Bcl-2 [E17] (Abcam, ab32124), with GAPDH as a reference at 4°C for overnight. After the membrane was washed three times with TBST (20 mM Tris, 500 mM NaCl, 0.1% Tween 20) for 10 min each time at RT, the membrane was incubated with Goat Anti-Rabbit IgG H&L (HRP) (ab6721) in a dark place for 1 h at RT. Then, the membrane was washed three times with TBST for 10 min each time at RT in the dark. Detection was performed using an ECL chemiluminescence kit (GE Healthcare, Freiburg, USA), and the WB film was scanned on a MultiSpectral Imaging System (EC3 410, UVP, Upland, CA, USA) and analyzed with Image J software to quantify protein band intensities. This experiment was repeated 3 times.37,38,39
Statistics
All data were expressed as mean±SD. All experiments were performed at least three times. Statistical analyses were performed using two-tailed Student t test and one-way ANOVA followed by Tukey post-test. A p-value of less than 0.05 was considered to be significant.40
Results
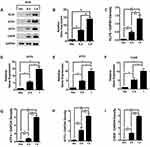
KLF6 Upregulates ATF4/ATF3/CHOP Expression
To confirm the relationship between KLF6 and ATF4/ATF3/CHOP, pCDNA3.1+-KLF6 plasmid was employed to increase KLF6 levels in HLECs, and the effects were measured by Western blot and qRT-PCR. As shown in Figure 1A–C, p 3.1+-KLF6 transfection upregulated KLF6 mRNA and protein compared with the empty vector (Vec) group. Upregulation of ATF4/ATF3/CHOP by pCDNA3.1+-KLF6 transfection was detected by Western blot and qRT-PCR. pCDNA3.1+-KLF6 transfection can significantly increase ATF4/ATF3/CHOP expression in HLECs (Figure 1A and D–I).
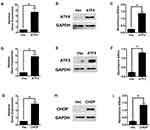
ATF4, ATF3, and CHOP are Involved in the Regulation of HLEC Apoptosis
To determine the role of ATF4 in the apoptosis of HLECs, ATF4 was overexpressed in HLECs by transient transfection of pCDNA3.1+-ATF4, and the upregulation of ATF4 was verified by real-time PCR and Western blot. As shown in Figure 2A and B, overexpression of ATF4 by transient transfection significantly increased ATF4 mRNA and protein, compared with cells that were transfected with empty vector. Furthermore, ATF4 expression significantly induced the apoptosis of HLECs (Figure 2C). Based on these results, we also determine the effects of ATF3 and CHOP overexpression on HLECs apoptosis. Figure 2D and E show that pCDNA3.1+-ATF3/CHOP upregulated ATF3/CHOP mRNA and protein in HLECs (Figure 2D, E and G, H). Moreover, the increase in ATF3/CHOP was positively related to HLECs apoptosis (Figure 2F and I). These data indicate that ATF4, ATF3, and CHOP play a role in the regulation of cell apoptosis.
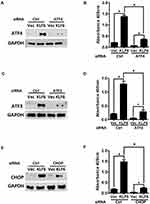
ATF4, ATF3, and CHOP Mediates KLF6-Induced Apoptosis
To determine the importance of ATF4 in KLF6-induced apoptosis in HLECs, we employed a siRNA that targeted ATF4 (Figure 3A) to specifically knock down the expression of ATF4 in HLECs. As shown in Figure 3A, transfection with ATF4 siRNA significantly downregulated KLF6-induced ATF4 expression. Although KLF6-induced apoptosis in HLECs that were transfected with the negative control siRNA, it failed to do so in cells in which ATF4 was knocked down by siRNA (Figure 3B). Furthermore, similar results were observed with ATF3 (Figure 3C) and CHOP (Figure 3E) siRNA. Knockdown of ATF4, ATF3, or CHOP reduced that ratio of apoptotic cells that were induced by KLF6 (Figure 3D and F). These data indicate that in addition to ATF4, ATF3 and CHOP regulate KLF6-induced apoptosis.
The Relationship Between ATF4, ATF3, and CHOP Under KLF6 Induction
Knocking down each protein in turn reduced the level of apoptosis (Figure 3). Following this experiment, we validated the relationship between ATF4, ATF3, and CHOP in KLF6-dependent cell apoptosis. Cells were transfected with ATF4, ATF3, or CHOP siRNA, respectively. Transfection with ATF4 siRNA decreased the expression of ATF3 and CHOP (Figure 4A). Furthermore, by inhibiting ATF3 expression using siRNA, we found that the induction of ATF4 was unaffected, whereas the expression of CHOP declined (Figure 4B). In cells that were transfected with CHOP siRNA, KLF6-induced expression including ATF4 and ATF3 was still unchanged (Figure 4C). These results suggest that KLF6 mediate induction of the ATF4-ATF3/CHOP axis, which leads to KLF6-dependent apoptosis.
KLF6 Aggravates Apoptosis in LECs Under UV Treatment
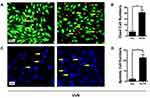
UV radiation was used as a classic oxidative stressor to induce cell apoptosis. To determine the effect of KLF6 on the survival of HLECs under UV radiation, pCDNA3.1+-KLF6 plasmid was employed to increase KLF6 levels in HLECs. Viability and cytotoxicity were measured to assess viability under UV treatment with or without KLF6. As shown in Figure 5A and B, living cells showed green signals, whereas dead cells generated red signals. There were only a few red cells in the normal group by fluorescence microscopy; however, the decrease in green signals was accompanied by a dramatic increase in red signals and KLF6 levels.
We performed Hoechst staining to determine the effect of KLF6 on UV-induced HLEC apoptosis. UV treatment significantly increased the number of apoptotic cells, as shown by the increase in apoptotic nuclei (condensed, crescent-shaped, and fragmented nuclei), which KLF6 exacerbated. Taken together, these results suggest that KLF6 aggravates HLECs death during UV-induced injury in vitro (Figure 5C and D).
To confirm the role of KLF6 in UV-induced HLEC apoptosis, cells were pretreated with or without KLF6 overexpression and subjected to TUNEL assay. As shown in Figure 6A, the increase in TUNEL-positive cells was accompanied by a rise in KLF6.
On this basis, we also checked the expression of Bax and Bcl-2 in HLECs. Consistent with the increased apoptosis in HLECs, by Western blot, KLF6 upregulated the pro-apoptosis protein Bax, whereas the active form of the pro-viability protein Bcl-2 decreased (Figure 6B). Considering that the ratio of Bax to Bcl-2 protein is a decisive factor in transmitting apoptosis signals, we compared the gray values of the two corresponding bands and make clear the ratio of Bax/Bcl-2 to be augmented with KLF6 upregulation (Figure 6C–E). HLECs died through apoptosis, as confirmed by ELISA of mono- and oligonucleosome content in the lysates of apoptotic cells (Figure 6F). These data confirm that KLF6 increased the susceptibility of UV-induced apoptosis in HLECs.
Discussion
Cataract is the leading cause of blindness worldwide, and its prevalence rises in high myopia patients with much younger age. As the relationship between LEC apoptosis and cataract is studied in greater detail, scientists are inclined to believe that the apoptosis of lens epithelial cells is the common pathogenesis of all types of cataract, except congenital cataract. UV radiation, aging, hypoxia, and other stimuli could cause excess apoptosis of lens epithelial cells, thus causing cytoskeletal degradation, accumulation of crystal protein, and ultimately cataract.37 It has been approved that the apoptosis of lens epithelial cells is much more different in high myopia cataract and ordinary age-related cataract patients. We analyzed lens capsule gene microarray data in high myopia patients and identified two differentially expressed genes: Krüppel-like factor 6 (KLF6) and ATF4. We supposed that the two difference may play a pivotal role in the apoptosis of lens epithelial cells so that the stimuli above can cause more damage and protein degradation followed by subsequent cataract formation in lens of high myopia. Therefore, first of all, understanding the mechanisms by which KLF6 induces apoptosis in the lens is important for developing the novel pathogenic hypothesis of cataracts.
KLF6 is a member of the specificity protein 1/Krüppel-like transcription factor family (Sp1/KLF) and was initially cloned from leukocytes.41 KLF6 is a tumor suppressor protein that is downregulated or mutated in several types of cancers, including prostate cancer.42–44 KLF6 suppresses tumor growth by activating p21WAF1/Cip1, an inhibitor of cyclin-dependent kinases, in cultured cells and a transgenic mouse model.45,46 KLF6 also interacts directly with cyclin D1 to suppress cyclin-dependent kinase 4 and causes cell-cycle arrest.47 In addition, the overexpression of KLF6 increases the susceptibility of hepatic stellate cells to apoptotic stress, as evidenced by PARP cleavage in the liver.48 Moreover, KLF6 overexpression induces apoptosis in several human cancers, including prostate cancer, non-small-cell lung cancer, and osteosarcoma.49–52 However, the mechanism of KLF6-induced apoptosis remains unknown. In previous study, we found that KLF6 induces apoptosis via ATF4 activation and that the induction of apoptosis likely has a key role in cataract formation.
In the presence of disequilibrium in the endoplasmic reticulum, such as hypoxia and oxidative stress, endoplasmic reticulum stress that is induced by the accumulation of misfolded endoplasmic reticulum proteins is called the unfolded protein response (UPR).53,54 Three pathways are known to mediate the UPR according to different transmembrane receptors: inositol-requiring enzyme 1 (IRE1), protein kinase R-like ER kinase (PERK), and activating transcription factor 6 (ATF6), the foremost of which is the PERK-mediated PERK-eIF2α-ATF4 pathway.55 PERK phosphorylation is activated during endoplasmic reticulum stress, leading to eIF2α phosphorylation, which subsequently inhibits protein translation and reduces protein synthesis, thereby reducing false protein folding and maintaining endoplasmic reticulum homeostasis. This procedure can improve ATF4 mRNA translation, and ATF4 combined with amino acid regulatory element, AARE, increased the expression of CHOP, and further induces oxidative damage through the endoplasmic reticulum stress pathway. Initially, activation of PERK is a protective measure under mild stress. However, activation of PERK also leads to the induction of CHOP, which is an important factor that switches the survival signal to a pro-death signal.54,56–58 ATF4 plays an important role in anti-tumor responses and anti-oxidation and promotes renal interstitial fibrosis through the endoplasmic reticulum stress pathway (Figure 7).59–61 Previously, our study has revealed that the direct activation of ATF4 mRNA expression can elevate apoptosis in HLECs. In addition, ATF4 knockdown by siRNA blocks KLF6-induced apoptosis, indicating that ATF4 is a key component in HLECs apoptosis.
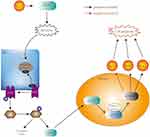 |
Figure 7 A model of KLF6-induced cell apoptosis via ATF4-ATF3/CHOP pathway under UV. |
KLF6 could induce endoplasmic reticulum stress under UV radiation, large amount of unfolded protein can bind to BIP (immunoglobulin binding protein) and inhibit the bound of BIP to PERK, thereby activate the PERK pathway. PERK activation leads to phosphorylation of the subunit of eukaryotic initiation factor 2 (eIF2a), which inhibits protein synthesis and subsequently inhibits protein translation, thereby reducing false protein folding and maintaining endoplasmic reticulum homeostasis. This procedure can improve ATF4 mRNA translation, and ATF4 is known to activate the ATF3 gene by binding to a ATF/cAMP response element binding (ATF/CREB). ATF3 controls CHOP expression and CHOP leads to decrease in Bcl-2 but increase in DOCs, TRB3, directly induce cell apoptosis.
ATF3 and ATF4 belong to the ATF/cAMP response element binding (ATF/CREB) family of transcription factors.21,62 Members of the ATF/CREB family sense environmental stimulation signals and participate in cellular processes. Liu reported that the UPR increases ATF3 expression via p-eIF2-ATF4 signaling.21 Studies have shown that ATF3 is an adaptive response gene that has dual functions in cell apoptosis and the cell cycle.63 ATF3 can induce glomerular mesangial cell apoptosis under stress conditions via KLF6 upregulation.64 It also triggers HDACI-induced proapoptotic activity through the ATF4/ATF3/CHOP pathway.65 Conversely, ATF3 might have anti-apoptotic effects. Nakagomi found that ATF3 protects PC12 cells and superior ganglion neurons from JNK-induced apoptosis.66 Several genes have been found to be its targets, including p53, CHOP, DR5, and Nrf2.23,67,68 CHOP is a target gene of ATF3. ATF3 can promote the transcription of CHOP and induce apoptosis of tumor cells.69 Liu has indicated that salermide promotes the expression of CHOP by upregulating ATF3, which contributes to overexpression of DR5 and promotes salermide-induced apoptosis.23 The mainly accepted mechanism of how CHOP lead to cell apoptosis is CHOP worked as a transcriptional factor mediating multiple apoptotic/anti-apoptotic genes such as DOCs (for down-stream of CHOP), Bcl-2 (B-cell lymphoma-2), TRB3 (tribbles-related protein3).20 DOCs TRB3 is an intracellular pseudokinase that modulates the activity of several signal transduction cascades. During ER stress, TRB3 is up-regulated by an ER stress-inducible transcriptional factor, over-expression of TRB3 would work as feedback to downregulates its own expression via the repression of ATF4/CHOP transcriptional activity;70 Bcl-2 family contains both pro-apoptotic (BAX, Bak, Bok) and antiapoptotic (Bcl-2, Bcl-X, Bcl-W). Under ER stress, CHOP down-regulates the expression of Bcl-2 but not Bcl-X, leading cells to apoptosis.71 DOCs are a family composed of three members: DOC1, DOC4, and DOC6. However, the mechanism of DOCs effect either survival or death fell far behind the pharmacological and biochemical research.40
In this study, we assumed that KLF6 induces HLECs apoptosis through ATF4 and ATF3. We employed siRNA to block ATF4, ATF3, and CHOP expression separately. The results showed that ATF4 regulates ATF3 and CHOP expression and that ATF3 regulates CHOP expression; however, when blocking CHOP, ATF4 and ATF3 expression remained unchanged. Our further work would be focusing on how the three molecules interact with each other. Wang speculates that ATF4 promotes ATF3 expression; thus, ATF4 and ATF3 could form a complex and subsequently enhance the transcription of CHOP,72 Some earlier studies have found that ATF3 can suppress CHOP gene transcription, whereas CHOP inhibits ATF3 protein function.73 However, our data show that ATF3 can promote CHOP expression. Anyway, how CHOP is regulated by ATF3 and ATF4 remains unknown and needs to be studied further.
In our research, we stressed HLECs with UV, which is a physiological stressor in vivo and relevant for study of oxidative stress-induced ocular disorders. KLF6 overexpression significantly accelerated UV-induced HLECs apoptosis, as verified by cell death detection and cell staining, such as Hoechst, and live and dead staining. We proved that KLF6 upregulation can activate the proapoptotic protein Bax, inactivate the anti-apoptotic protein Bcl-2, and simultaneously elevate the Bax/Bcl-2 ratio in HLECs that are transfected with KLF6. Bax/Bcl-2 ratio is a crucial “molecular switch” in cell apoptosis reflecting the apoptotic state of HLECs.74,75
In summary, KLF6 is a novel transcriptional activator that induces apoptosis in HLECs via the ATF4-ATF3-CHOP pathway. Moreover, our data provide evidence that KLF6 could make HLECs prone to ER stress and elevated apoptosis level in HLECs under UV radiation. Our results increase our understanding of the regulatory mechanisms of HLECs apoptosis for the development of novel approaches to prevent and treat high myopia cataract.
Abbreviations
AARE, amino acid regulatory element; ATF3, activating transcription factor 4; ATF4, activating transcription factor 3; ATF6, activating transcription factor 6; ATF/CREB, ATF/cAMP response element binding; Bax, Bcl-2-associated X protein; Bcl-2, B-cell lymphoma 2; CHOP, C/EBP homologous protein; DAPI, 4’,6-diamidino-2-phenylindole; DR5, death receptor 5; ERS, endoplasmic reticulum stress; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; IRE1, inositol-requiring enzyme 1; JNK, c-Jun N-terminal kinase; KLF6, Krüppel-like factor 6; LECs, lens epithelial cells; Nrf2, nuclear factor (erythroid-derived 2)-like 2; PERK, protein kinase R-like ER kinase; ROS, reactive oxygen species; TUNEL, terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling; UPR, unfolded protein response; UVB, ultraviolet radiation B.
Data Sharing Statement
The data used to support the findings of this study are included within the article.
Acknowledgments
This work was supported by a grant from the National Natural Science Foundation of China (NSFC:81570872), Tianjin Municipal Science and Technology Commission Grants (15JCYBJC24900), The Science & Technology Development Fund of Tianjin Education Commission for Higher Education (2016YD08), Tianjin Medical University Natural Science Foundation (2016KYZM14, 2015KYZM10), Tianjin Medical University Eye Institute Foundation (15YKYJS002).
We thank Liwen Bianji, Edanz Editing China, for editing the English text of a draft of this manuscript.
The use of gifted HLECs was approved by Tianjin Medical University Eye Hospital ethics committee.
Disclosure
The authors declare that they have no conflicts of interest.
References
1. Zheng XY, Xu J, Chen XI, Li W, Wang TY. Attenuation of oxygen fluctuation-induced endoplasmic reticulum stress in human lens epithelial cells. Exp Ther Med. 2015;10:1883–1887. doi:10.3892/etm.2015.2725
2. Ikuno Y. Overview of the complications of high myopia. Retina. 2017;37:2347–2351. doi:10.1097/IAE.0000000000001489
3. Pan CW, Boey PY, Cheng C-Y, et al. Myopia, axial length, and age-related cataract: the Singapore Malay eye study. Invest Ophthalmol Vis Sci. 2013;54:4498. doi:10.1167/iovs.13-12271
4. Pan CW, Cheung CY, Aung T, et al. Differential associations of myopia with major age-related eye diseases: the Singapore Indian Eye Study. Ophthalmology. 2013;120(2):284–291.
5. Pan CW, Cheng CY, Saw S-M, Wang JJ, Wong TY. Myopia and age-related cataract: a systematic review and meta-analysis. Am J Ophthalmol. 2013;156:1021–1033.e1. doi:10.1016/j.ajo.2013.06.005
6. Mehdizadeh M, Ashraf H. Prevalence of cataract type in relation to axial length in subjects with high myopia and emmetropia in an Indian population. Am J Ophthalmol. 2008;146:329–330. doi:10.1016/j.ajo.2008.04.004
7. Yu Z, Lijie D, Hong Z, Fang T. Pilot study on suppression of Krüppel-like factor 6 for proliferation of human lens epithelial cells. Chin J Exp Ophthalmol. 2014;32(4):325–330.
8. Dong L, Nian H, Shao Y, et al. PTB-associated splicing factor inhibits IGF-1-induced VEGF upregulation in a mouse model of oxygen-induced retinopathy. Cell Tissue Res. 2015;360:233–243. doi:10.1007/s00441-014-2104-5
9. Su Y, Wang F, Zhou D, et al. Inhibition of proliferation of rat lens epithelial cell by overexpession ofKLF6. Mol Vis. 2011;17:1080–1084.
10. Tian F, Zhao J, Huang L, et al. Effects of Kriippel·like factor 6 overexpression towards apoptosis of human lens epithelial cells with ultraviolet radiation B treatment. Chin J Exp Ophthalmol. 2019;37:257–262.
11. Tian F, Zhao J, Teng H, et al. Regulation of Krüppel-like factor 6 via activating transcription factor 4 pathway to apoptosis of human lens epithelial cells. Chin J Exp Ophthalmol. 2018;36:181–186.
12. Andreoli V, Gehrau RC, Bocco JL. Biology of Krüppel-like factor 6 transcriptional regulator in cell life and death. IUBMB Life. 2010;62:896–905. doi:10.1002/iub.v62.12
13. Gui T, Wang Y, Zhang L, Wang W, Zhu H, Ding W. Kruppel-like factor 6 rendered rat Schwann cell more sensitive to apoptosis via upregulating FAS expression. PLoS One. 2013;8:e82449. doi:10.1371/journal.pone.0082449
14. Li Z, Zhou A, Tang C. Molecular mechanism on endoplasmic reticulum stress responses. Chin J Biochem Mol Biol. 2004;20:283–288.
15. Fatma N, Singh P, Chhunchha B, et al. Deficiency of Prdx6 in lens epithelial cells induces ER stress response-mediated impaired homeostasis and apoptosis. Am J Physiol Cell Physiol. 2011;301:C954. doi:10.1152/ajpcell.00061.2011
16. Palsamy P, Bidasee KR, Shinohara T. Valproic acid suppresses Nrf2/Keap1 dependent antioxidant protection through induction of endoplasmic reticulum stress and Keap1 promoter DNA demethylation in human lens epithelial cells. Exp Eye Res. 2014;121:26–34. doi:10.1016/j.exer.2014.01.021
17. Qing G, Li B, Vu A, et al. ATF4 regulates MYC-mediated neuroblastoma cell death upon glutamine deprivation. Cancer Cell. 2012;22:631–644. doi:10.1016/j.ccr.2012.09.021
18. Ikesugi K, Yamamoto R, Mulhern ML, Shinohara T. Role of the unfolded protein response (UPR) in cataract formation. Exp Eye Res. 2006;83:508–516. doi:10.1016/j.exer.2006.01.033
19. Yang J, Zhou S, Gu J, Guo M, Xia H, Liu Y. UPR activation and the down–regulation of α-crystallin in human high myopia-related cataract lens epithelium. PLoS One. 2015;10:e0137582. doi:10.1371/journal.pone.0137582
20. Li Y, Guo Y, Tang J, Jiang J, Chen Z. New insights into the roles of CHOP-induced apoptosis in ER stress. http://Acta Biochim Biophys Sin (Shanghai). 2014;46(8):629–640.
21. Liu Z, Shi Q, Song X, et al. ATF4-ATF3-CHOP cascade activation play an important role in ER stress-induced sensitization of tetrachlorobenzoquinone-challenged PC12 cells to ROS-mediated apoptosis through DR5 signaling. Chem Res Toxicol. 2016;29:1510–1518. doi:10.1021/acs.chemrestox.6b00181
22. Atsushi S, Kimiko O, Shinichi K, et al. Endoplasmic reticulum stress response mediated by the PERK-eIF2(alpha)-ATF4 pathway is involved in osteoblast differentiation induced by BMP2. J Biol Chem. 2011;286:4809–4818. doi:10.1074/jbc.M110.152900
23. Liu G, Su L, Hao X, et al. Salermide up-regulates death receptor 5 expression through the ATF4-ATF3-CHOP axis and leads to apoptosis in human cancer cells. J Cell Mol Med. 2012;16:1618–1628. doi:10.1111/j.1582-4934.2011.01401.x
24. Joo JH, Eiichiro U, Bortner CD, Xiao-Ping Y, Grace L, AM J. Farnesol activates the intrinsic pathway of apoptosis and the ATF4-ATF3-CHOP cascade of ER stress in human T lymphoblastic leukemia Molt4 cells. Biochem Pharmacol. 2015;97:256–268. doi:10.1016/j.bcp.2015.08.086
25. Saha S, Bhattacharjee P, Mukherjee S, et al. Contribution of the ROS-p53 feedback loop in thuja-induced apoptosis of mammary epithelial carcinoma cells. Oncol Rep. 2014;31:1589. doi:10.3892/or.2014.2993
26. Kong L, Wang S, Wu X, Zuo F, Qin H, Wu J. Paeoniflorin attenuates ultraviolet B-induced apoptosis in human keratinocytes by inhibiting the ROS-p38-p53 pathway. Mol Med Rep. 2016;13:3553. doi:10.3892/mmr.2016.4953
27. Liu C, Dijana V, Kochevar IE, Jurkunas UV. UV-A irradiation activates Nrf2-regulated antioxidant defense and induces p53/Caspase3-dependent apoptosis in corneal endothelial cells. Invest Ophthalmol Vis Sci. 2016;57:2319–2327. doi:10.1167/iovs.16-19097
28. Li WB, Ma MW, Dong L-J, Wang F, Chen L-X, Li X-R. MicroRNA-34a targets notch1 and inhibits cell proliferation in glioblastoma multiforme. Cancer Biol Ther. 2011;12:477–483. doi:10.4161/cbt.12.6.16300
29. Shao Y, Chen J, Freeman W, et al. Canonical Wnt Signaling Promotes Neovascularization Through Determination of Endothelial Progenitor Cell Fate via Metabolic Profile Regulation. Stem cells. 2019;37(10):1331–1343. doi:10.1002/stem.3049
30. Teng H, Huang LY, Tian F, Dong LJ, Zhang H. [Effects of SMP-30 overexpression on apoptosis of human lens epithelial cells induced by ultraviolet B irradiation]. Zhonghua Yan Ke Za Zhi. 2017;11(53):835–841. doi:10.3760/cma.j.issn.0412-4081.2017.11.007
31. Xing X, Huang L, Lv Y, et al. DL-3-n-butylphthalide protected retinal muller cells dysfunction from oxidative stress. Curr Eye Res. 2019;44:1112–1120. doi:10.1080/02713683.2019.1624777
32. Liu X, Dong L, Zhang X, et al. Identification of p100 target promoters by chromatin immunoprecipitation-guided ligation and selection (ChIP-GLAS). Cell Mol Immunol. 2011;8(1):88–91. doi:10.1038/cmi.2010.47
33. Li W, Dong L, Ma M, et al. Preliminary in vitro and in vivo assessment of a new targeted inhibitor for choroidal neovascularization in age-related macular degeneration. Drug Des Devel Ther. 2016;10:3415–3423.
34. Dong L, Zhang X, Fu X, et al. PTB-associated splicing factor (PSF) functions as a repressor of STAT6-mediated Ig epsilon gene transcription by recruitment of HDAC1. J Biol Chem. 2011;286(5):3451–3459.
35. Shao Y, Dong LJ, Takahashi Y, et al. miRNA-451a regulates RPE function through promoting mitochondrial function in proliferative diabetic retinopathy. Am J Physiol Endocrinol Metab. 2019;316:E443–E452. doi:10.1152/ajpendo.00360.2018
36. Dong L, Chen X, Shao H, Bai L, Li X, Zhang X. Mesenchymal Stem Cells Inhibited Dendritic Cells Via the Regulation of STAT1 and STAT6 Phosphorylation in Experimental Autoimmune Uveitis. Curr Mol Med. 2018;17(7):478–487. doi:10.2174/1566524018666180207155614
37. Tian F, Dong L, Zhou Y, et al. Rapamycin-induced apoptosis in HGF-stimulated lens epithelial cells by AKT/mTOR, ERK and JAK2/STAT3 pathways. Int J Mol Sci. 2014;15:13833–13848. doi:10.3390/ijms150813833
38. Wei R, Dong L, Xiao Q, Sun D, Li X, Nian H. Engagement of Toll-like receptor 2 enhances interleukin (IL)-17(+) autoreactive T cell responses via p38 mitogen-activated protein kinase signalling in dendritic cells. Clin Exp Immunol. 2014;178:353–363. doi:10.1111/cei.2014.178.issue-2
39. Shao Y, Chen J, Dong LJ, et al. A Protective Effect of PPARα in Endothelial Progenitor Cells Through Regulating Metabolism. Diabetes. 2019;68(11):2131–2142. doi:10.2337/db18-1278.
40. Shao Y, Dong LJ, Zhang Y, et al. Surgical induced astigmatism correlated with corneal pachymetry and intraocular pressure: transconjunctival sutureless 23-gauge versus 20-gauge sutured vitrectomy in diabetes mellitus. Int J Ophthalmol. 2015;8(3):528.
41. El RS, Newcomb EW. Identification of Bcd, a novel proto-oncogene expressed in B-cells. Oncogene. 1996;13:2623–2630.
42. Selvanayagam ZE, Cheung TH, Wei N, et al. Prediction of chemotherapeutic response in ovarian cancer with DNA microarray expression profiling. Cancer Genet Cytogenet. 2004;154:63–66. doi:10.1016/j.cancergencyto.2004.01.024
43. Spentzos D, Levine DA, Kolia S, et al. Unique gene expression profile based on pathologic response in epithelial ovarian cancer. J Clin Oncol. 2005;23:7911. doi:10.1200/JCO.2005.02.9363
44. Brown GJ, John DS, Macrae FA, Aittomäki K. Cancer risk in young women at risk of hereditary nonpolyposis colorectal cancer: implications for gynecologic surveillance. Gynecol Oncol. 2001;80:346. doi:10.1006/gyno.2000.6065
45. Narla G, Heath KE, Reeves HL, et al. KLF6, a candidate tumor suppressor gene mutated in prostate cancer. Science. 2001;294:2563–2566. doi:10.1126/science.1066326
46. Narla G, Kremer-Tal S, Matsumoto N, et al. In vivo regulation of p21 by the Kruppel-like factor 6 tumor-suppressor gene in mouse liver and human hepatocellular carcinoma. Oncogene. 2007;26:4428. doi:10.1038/sj.onc.1210223
47. Benzeno S, Narla G, Allina J, et al. Cyclin-dependent kinase inhibition by the KLF6 tumor suppressor protein through interaction with cyclin D1. Cancer Res. 2004;64:3885–3891. doi:10.1158/0008-5472.CAN-03-2818
48. Ghiassi-Nejad Z, Hernandez-Gea V, Woodrell C, et al. Reduced hepatic stellate cell expression of kruppel‐like factor 6 tumor suppressor isoforms amplifies fibrosis during acute and chronic rodent liver injury. Hepatology. 2013;57:786. doi:10.1002/hep.26056
49. Banck MS, Beaven SW, Narla G, Walsh MJ, Friedman SL, Beutler AS. KLF6 degradation after apoptotic DNA damage. FEBS Lett. 2006;580:6981. doi:10.1016/j.febslet.2006.10.077
50. Huang X, Li X, Guo B. KLF6 induces apoptosis in prostate cancer cells through up-regulation of ATF3. J Biol Chem. 2008;283:29795. doi:10.1074/jbc.M802515200
51. Ito G, Uchiyama M, Kondo M, et al. Krüppel-like factor 6 is frequently down-regulated and induces apoptosis in non-small cell lung cancer cells. Cancer Res. 2004;64:3838–3843. doi:10.1158/0008-5472.CAN-04-0185
52. Jianwei Z, Enzhong B, Fan L, Jian L, Ning A. Effects of Kruppel-like factor 6 on osteosarcoma cell biological behavior. Tumour Biol. 2013;34:1097–1105. doi:10.1007/s13277-013-0651-0
53. Szegezdi E, Logue SE, Gorman AM, Samali A. Mediators of endoplasmic reticulum stress‐induced apoptosis. EMBO Rep. 2006;7:880–885. doi:10.1038/sj.embor.7400779
54. Oakes SA, Papa FR. The role of endoplasmic reticulum stress in human pathology. Annu Rev Pathol. 2014;10:173. doi:10.1146/annurev-pathol-012513-104649
55. Fels DR, Koumenis C. The PERK/eIF2alpha/ATF4 module of the UPR in hypoxia resistance and tumor growth. Cancer Biol Ther. 2006;5:723–728. doi:10.4161/cbt.5.7.2967
56. Cao J, Dai DL, Yao L, et al. Saturated fatty acid induction of endoplasmic reticulum stress and apoptosis in human liver cells via the PERK/ATF4/CHOP signaling pathway. Mol Cell Biochem. 2012;364:115–129. doi:10.1007/s11010-011-1211-9
57. Lai X, Ouyang J, Shi Y, Pharmacy DO. Role of endoplasmic reticulum stress PERK- ATF4- CHOP pathway in matrine induced retinoblastoma apoptosis. Chin Pharm. 2015.
58. Chen Y, Gui D, Chen J, He D, Luo Y, Wang N. Down-regulation of PERK-ATF4-CHOP pathway by astragaloside IV is associated with the inhibition of endoplasmic reticulum stress-induced podocyte apoptosis in diabetic rats. Cell Physiol Biochem. 2014;33:1975–1987. doi:10.1159/000362974
59. Rozpedek W, Pytel D, Mucha B, Leszczynska H, Diehl JA, Majsterek I. The role of the PERK/eIF2α/ATF4/CHOP signaling pathway in tumor progression during endoplasmic reticulum stress. Curr Mol Med. 2016;16:533–544. doi:10.2174/1566524016666160523143937
60. Afonyushkin T, Oskolkova OV, Philippova M, et al. Oxidized phospholipids regulate expression of ATF4 and VEGF in endothelial cells via NRF2-dependent mechanism: novel point of convergence between electrophilic and unfolded protein stress pathways. Arterioscler Thromb Vasc Biol. 2010;30:1007–1013. doi:10.1161/ATVBAHA.110.204354
61. Cui Z, Zhou Y, Zhou Y, Ying L, Wang D. Regulation of eIF2α expression and renal interstitial fibrosis by resveratrol in rat renal tissue after unilateral ureteral obstruction. Ren Fail. 2016;38:1.
62. Persengiev SP, Green MR. The role of ATF/CREB family members in cell growth, survival and apoptosis. Apoptosis. 2003;8:225. doi:10.1023/A:1023633704132
63. Zmuda EJ, Qi L, Zhu MX, Mirmira RG, Montminy MR, Hai T. The roles of ATF3, an adaptive-response gene, in high-fat-diet-induced diabetes and pancreatic beta-cell dysfunction. Mol Endocrinol. 2010;24:1423–1433. doi:10.1210/me.2009-0463
64. Xu K, Zhou Y, Qiu W, et al. Activating transcription factor 3 (ATF3) promotes sublytic C5b-9-induced glomerular mesangial cells apoptosis through up-regulation of Gadd45α and KLF6 gene expression. Immunobiology. 2011;216:871–881. doi:10.1016/j.imbio.2011.02.005
65. Jia L, Edagawa M, Goshima H, et al. Role of ATF3 in synergistic cancer cell killing by a combination of HDAC inhibitors and agonistic anti-DR5 antibody through ER stress in human colon cancer cells. Biochem Biophys Res Commun. 2014;445:320–326. doi:10.1016/j.bbrc.2014.01.184
66. Nakagomi S, Suzuki Y, Namikawa K, Kiryuseo S, Kiyama H. Expression of the activating transcription factor 3 prevents c-Jun N-terminal kinase-induced neuronal death by promoting heat shock protein 27 expression and Akt activation. J Neurosci. 2003;23:5187–5196. doi:10.1523/JNEUROSCI.23-12-05187.2003
67. Yan C, Lu D, Hai T, Boyd DD. Activating transcription factor 3, a stress sensor, activates p53 by blocking its ubiquitination. EMBO J. 2014;24:2425–2435. doi:10.1038/sj.emboj.7600712
68. Shan Y, Akram A, Amatullah H, et al. ATF3 protects pulmonary resident cells from acute and ventilator-induced lung injury by preventing Nrf2 degradation. Antioxid Redox Signal. 2015;22:651–668. doi:10.1089/ars.2014.5987
69. Weng S, Zhou L, Deng Q, et al. Niclosamide induced cell apoptosis via upregulation of ATF3 and activation of PERK in Hepatocellular carcinoma cells. BMC Gastroenterol. 2016;16:1–10. doi:10.1186/s12876-016-0442-3
70. Tsai DH, Chung CH, Lee KT. Antrodia cinnamomea induces autophagic cell death via the CHOP/TRB3/Akt/mTOR pathway in colorectal cancer cells. Sci Rep. 2018;8. doi:10.1038/s41598-018-35780-y
71. Zhou W, Fang H, Wu Q, et al. Ilamycin E, a natural product of marine actinomycete, inhibits triple-negative breast cancer partially through ER stress-CHOP-Bcl-2. Int J Biol Sci. 2019;15:1723–1732. doi:10.7150/ijbs.35284
72. Wang Q, Morajensen H, Weniger MA, et al. ERAD inhibitors integrate ER stress with an epigenetic mechanism to activate BH3-only protein NOXA in cancer cells. Proc Natl Acad Sci U S A. 2009;106:2200–2205. doi:10.1073/pnas.0807611106
73. Yong-Min C, Hey-Young C, Muhammad Ayaz A, Han-Kyul K, Ji-Woong K, Sangdun C. ATF3 attenuates cyclosporin A-induced nephrotoxicity by downregulating CHOP in HK-2 cells. Biochem Biophys Res Commun. 2014;448:182–188. doi:10.1016/j.bbrc.2014.04.083
74. Yang MU, Liang-Liang LI, Liu YY, Zhang C, Zhou EM. Bax and Bcl-2 gene expression in porcine alveolar macrophages infected by porcine reproductive and respiratory syndrome virus. Chin J Vet Sci. 2013;33:1313–1615. doi:10.1007/s11425-013-4582-4
75. Theron KE, Penny CB, Hosie MJ. The Bax/Bcl-2 apoptotic pathway is not responsible for the increase in apoptosis in the RU486-treated rat uterus during early pregnancy. Reprod Biol. 2013;13:290–297. doi:10.1016/j.repbio.2013.09.002
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.