Back to Archived Journals » Advances in Genomics and Genetics » Volume 4
K-type human endogenous retroviral elements in human melanoma
Authors Rincon L, Sedrak M, Sun H, Garza G, Kelly B, Dong J
Received 6 June 2014
Accepted for publication 21 July 2014
Published 4 October 2014 Volume 2014:4 Pages 153—159
DOI https://doi.org/10.2147/AGG.S68926
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Liliana Rincon,1 Michael P Sedrak,1 Huijian Sun,2 Gregorio Garza,3 Brent Kelly,4 Jianli Dong1
1Department of Pathology, 2School of Medicine, University of Texas Medical Branch, TX, USA; 3Ciencias de la Salud, Universidad de Monterrey, Nuevo León, Mexico; 4Department of Dermatology, University of Texas Medical Branch, TX, USA
Abstract: Human endogenous retroviral elements (HERVs) are thought to be germline-integrated genetic remnants of exogenous retroviral infections. HERVs comprise approximately 5%–8% of the human genome. Although all HERV genomes are highly defective, some, especially the K type (HERV-K), have the potential to be expressed and have biological activities. HERV-K expression has been detected in human melanomas. There are also reports on the regulation and potential activities of HERV-K in melanoma cells. Although a causal link between the activation of HERV-K and melanoma development has yet to be determined, existing data support the further research efforts in this area. In this review, we summarize the published studies on the expression, regulation, and activity of HERV-K in human melanoma.
Keywords: HERV, HERV-K, melanoma development
Introduction
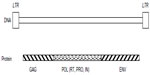
Human endogenous retroviral elements (HERVs) comprise approximately 5%–8% of the human genome.1–3 It is hypothesized that HERVs were once exogenous retroviruses that became incorporated into the human germline, which resulted in a vertical transfer of HERV DNA elements into the human genome, and thus HERVs became endogenous to the human genome.5 A complete HERV element has common sequences shared by other well-known retroviruses such as the human immunodeficiency virus, the human T lymphotropic virus, the mouse mammary tumor virus (MMTV), and the murine leukemia virus.6 These well-studied viruses can exist as proviruses in the host genome, and commonly contain open reading frames of group-specific antigen (gag), polymerase (pol), and envelope (env) that are flanked by non-coding regulatory sequences, namely long terminal repeats (LTRs).1–3,5 Similarly, HERVs can have homologous sequences of GAG, POL, ENV, and LTR. With open reading frames, the GAG, POL, and ENV sequences may be transcribed and translated to the corresponding proteins, similar to retroviruses. Thus, GAG encodes core and structural proteins; POL encodes reverse transcriptase (RT), protease, and integrase; and ENV encodes envelop proteins (Figure 1).
HERVs are degenerated, defective, and non-infectious due to the accumulated mutations and deletions within their DNA sequences. Without an established function, they are often referred to as part of the “junk DNA” of the human genome. Although expression of HERVs is limited due to their degenerated genetic sequences, some HERVs retain relatively complete genetic sequences, capable of encoding functional proteins,3 while others play cis-regulatory roles in gene transcription.7,8 If expressed in active form, as in retroviruses, RT and integrase may generate new copies of HERV DNA from messenger RNA and insert them back into the human genome. Protease might cause proteolytic cleavages to make mature GAG and POL proteins, and ENV might mediate cellular entry/exit of viral-like particles.5,9
HERVs can be classified into more than 20 families based on the transfer RNA specificity of the primer binding site used to initiate reverse transcription; thus, HERV-K would bind lysine and HERV-W, tryptophan, if they were replicating viruses.10 HERVs have also been broadly grouped into three classes based on phylogenetic studies with exogenous retroviruses.11 Class I HERVs are closer to gamma retroviruses, which include HERV-H and HERV-W; class II are closer to beta retroviruses and alpha retroviruses, and include HERV-K; class III are closer to spuma retroviruses, which include HERV-L and HERV-S.11 Class I and class III HERVs are older and less evolved viruses, compared to class II HERV-K, which has the most complete sequence and, therefore, most likely to be biologically active in the human genome.12 An alternative name for HERV-K is MMTV-like HML, due to HERV-K’s sequence homology with MMTV.13,14 Based on sequence homology, HERV-Ks are further classified into ten families, HML-1 to HML-10.15 Unlike most HERVs that harbor defective mutations, HERV-Ks have retained intact genes, can code for functional proteins, and may form viral-like particles.6,15–17
Studies have shown that HERV DNA can be expressed and may play important roles in the physiological and pathological processes of human cells. For example, through millions of years of evolution, HERVs have become indispensable parts of the human genome, as demonstrated by syncytin-1 (ERVW-1), encoded by the ENV gene of HERV-W. Syncytin-1 mediates intercellular fusion of trophoblast cells to form syncytiotrophoblasts and prevents maternal immune attack against the developing embryo, thereby facilitating embryo implantation.2,3 HERV LTR sequences have integrated into human coding sequences, for example, as parts of splicing variants of methylthioadenosine phosphorylase gene (MTAP). Mutations in the LTR sequences in MTAP cause diaphyseal medullary stenosis with malignant fibrous histiocytoma.18 HERV-K proteins and virus-like particles have been detected not only in the placenta,19 but also in cancer cells, including teratocarcinoma,20 breast cancer,21 and melanoma.22 Up-regulation of HERV expression has been implicated in the pathogenesis of malignancy, inflammation, and autoimmune diseases.3,23
Melanoma is one of the most prevalent malignancies and has a very bleak prognosis. The rates of melanoma have been rising for the past 30 years. The American Cancer Society estimate that about 76,100 new cases of melanomas will be diagnosed, and about 9,710 melanoma patients are expected to die in the United States in 2014.24 Melanoma arises from melanocytes. The majority of melanomas arise in the skin; other sites of origin include oral and anogenital mucosal surfaces, esophagus, meninges, and eye. The natural history of melanomas involve multiple stages, often start with precursor lesions, progress from the primary site to regional or sentinel lymph nodes, and metastasize to distant sites. Melanomagenesis is driven by multiple genetic and epigenetic changes and presents multiple cancer hallmarks.25–27 Melanoma is characterized by early metastasis and is resistant to therapies.28,29 A better understanding of melanoma biology is necessary for the diagnosis and treatment of this devastating disease. It has been described that the abnormal expression of HERV-K seems to precipitate the pathological process leading to melanoma and also contributes to the morphological and functional cellular modifications implicated in melanoma maintenance and progression. In this review, we summarize the published literature on HERV-K expression, regulation, and potential activity in human melanoma.
Expression of HERV-K elements in human melanoma
The expression of HERV-K in human melanoma was first discovered during the characterization of melanoma antigens, which are families of antigens specifically recognized on human melanoma cells by cytolytic T lymphocytes.30,31 A short reading frame of the HERV-K ENV gene, namely HERV-K-MEL, encodes a melanoma antigen peptide on the autologous melanoma tumor cells.32 It was subsequently shown that human melanoma cells can produce retrovirus-like particles that exhibit RT activity and comprise mature GAG and ENV proteins.22
HERV-Ks exist in the human genome in multiple copies, generated through reinfection or intracellular formation of new copies following initial germline invasion.1–3 A recent study analyzed more than 1,400 HML-2 cDNA sequences that are transcribed in the human melanoma cell lines and melanoma tissues, and identified 23 HERV-K loci distributed on at least eleven chromosomes.33 This study underscores the complexity of HERV-K sequences, which is challenging when trying to assign specific loci for expressed HERV-K sequences. We also found that careful DNase I digestion is necessary to remove “contaminating” cellular DNA in order to accurately evaluate HERV-K RNA expression.34 As retroviral elements, HERV-K sequences do not have introns which are otherwise present in nearly all eukaryotic genes. The human genome harbors tens if not hundreds of HERV-K loci with homologous sequences. Without removal of genomic DNA (gDNA), HERV-K gDNA copies may be amplified, detected, and interpreted as HERV-K cDNA. This represents a technical challenge for the detection and quantification of human gene transcripts when using primers and probes that cannot distinguish gDNA from cDNA.
Regulation of HERV-K expression in melanoma cells
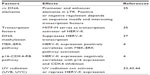
In general, HERV-K expression is suppressed. When expressed, there are differences in expression between cell and tissue types (Figure 2), which indicates that the expression is somehow regulated. However, the precise mechanism leading to the expression of HERV-K has yet to be clearly elucidated. Both endogenous and exogenous factors that can regulate the expression of HERV-K in melanoma cells have been identified (Table 1). Like other human genes, HERV-K transcription is regulated by cis-acting promoter and enhancer elements and trans-acting transcription factors.35 It has been shown that HERV-K LTR sequences contain promoter and enhancer elements that interact with melanocytic lineage-specific transcription factor microphthalmia-associated transcription factor (MITF),35 which may be necessary for HERV-K expression in melanoma cells. MITF is essential for the development of retinal pigmented epithelium and neural crest-derived melanocytes. MITF also induces genes that are essential for melanin synthesis and melanosome formation, and genes that support cell cycle progression and cell survival. MITF gene is classified as a lineage-specific oncogene based on its gene amplification and growth promoting function in melanoma. MITF gives rise to various isoforms, MITF-A, -B, -C, -H, and -M, depending of the N-terminal sequences. Only the MITF-M isoform undergoes melanocyte/melanoma-specific transcription in response to the alpha-melanocyte-stimulating hormone signaling.36 Katoh et al35 studied the HERV-K transcription controlling mechanism by determining the initiator (Inr) sites and enhancer sequences in the LTR, and found that an arrangement of (MITF motifs)-(TATA box)-(Inr) functions as the core enhancer/promoter inducible by MITF-M. The study suggests that MITF-M may be a prerequisite for the pigmented-cell-lineage-specific function of HERV-K LTR, leading to a high level of expression in malignant melanomas.35 Like other human genes, HERV-K promoter and enhancer elements are regulated by CpG DNA methylation.37
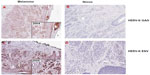  | Figure 2 Expression of HERV-K GAG and ENV in melanoma. |
Li et al4 have recently reported that the expression of HERV-K GAG and ENV correlates with p16 loss and ERK activation in melanoma specimens, and that CDK4 and MEK inhibitors can suppress HERV-K expression in cultured melanoma cells. This study suggests that HERV-K is downstream of p16-CDK4-RB and MEK-ERK pathways, which is significant given the critical importance of these pathways in melanoma biology and as proven and potential targets for melanoma treatment.38–40
Exposure to ultraviolet (UV) radiation (UVR) is a major environmental risk factor contributing to skin cancers, including melanoma.25,41 Sources of UV include natural sunlight and artificial lights such as tanning and germicidal lamps and lasers. UV radiation is divided into three categories, based on its wavelength: UVA (315–400 nm), UVB (280–315 nm), and UVC (100–280 nm).41 Cutaneous absorption of both UVA and UVB can reach the dermis, whereas UVC is limited to the epidermis. Therefore, all the three wavelengths of UVR can reach melanocytes that are located in the basal layers of the epidermis.41,42 UV radiation can cause mutations by direct interaction with DNA or indirectly through the generation of reactive oxygen species.25,41 Expression of HERV-K sequences in melanocytes and melanoma cells can be regulated by UVR.33,43,44 Reiche et al43 showed that after exposure to UVC, the expression of HERV-K was up-regulated in normal human epidermal melanocytes, but down-regulated or remained unchanged in melanoma cell lines. This study is consistent with a role, if any, of UVC and HERV-K in the initiation stage of melanoma development. Schanab et al44 reported that irradiation with UVB resulted in a significant transcriptional activation of HERV-K POL and ENV genes, and induced the production of retroviral-like particles. Since the positive effects were not observed in benign melanocytes, this study supports a role of UVB in the malignant progression of melanoma cells. On the other hand, Schmitt et al33 found that the overall HERV-K GAG gene transcripts were reduced both in melanocytes and melanoma cell lines after UVB radiation, and there were comparable responses in cultured melanoma and melanocytes. Interestingly, the study demonstrated that different HERV-K loci do not necessarily have comparable responses to UVB radiation, pointing to locus-specific regulation of HERV-K. These studies underscore the complex regulation of HERV-K by UV radiation; and the fact that HERV-K expression is dependent on factors such as the types and strength of UV radiation, melanocyte versus melanoma cells, and the presence of specific HERV-K loci and HERV-K genes.
Potential activities of HERV-K in melanoma
Although the activity of HERV-K in melanoma is not completely understood, clues have been revealed through a number of studies (Table 2). HERV-K has been reported to encode antigen epitope HERV-K-MEL, which can activate cell-mediated immunity, and serves as a melanoma vaccine candidate.32,45 Conversely, in immunocompetent mice, the inhibition of murine melanoma-associated retrovirus (MelARV) resulted in a reduction of melanoma aggressiveness and rejection of melanoma cells inoculated in nude mice, suggesting that MelARV acts to evade the immune system.46–48 It has also been reported that, compared to healthy individuals, sera from patients with melanoma were much more likely to harbor HERV-K, ENV, and GAG antibodies.49–51 The antibody concentration varied from 16%–22% depending on the study, but was negligible in healthy controls and correlated with worse prognosis.49–51 These studies show that HERV-K may have a positive or negative effect on cell-mediated and humoral immune systems.
Retrotransposition-mediated mutagenesis has been reported in murine melanoma cells, which promotes tumor cell mobility, metastasis, and in vivo growth.48 Another function of HERV-K in melanoma, proposed by Huang et al,52 is the blockage of HERV-K by RNA interference and monoclonal antibodies in melanoma cells, resulting in a marked reduction in colonic growth of melanoma cells in vitro.52 The down-regulation of the HERV-K reverse transcriptase by RNA interference or pharmacological inhibitors resulted in a lower mitotic rate and induced morphological differentiation,52–54 and slowed down melanoma growth and formation of metastasis in vivo.46–48
Conclusion
In summary, the human genome projects have identified approximately 20,000 protein-coding genes and 55,000 non-coding loci with evidence of gene transcription.55–57 Significant numbers of both protein-coding and non-coding loci contain mobile genetic elements consisting of DNA transposons and retro-elements.58–60 HERVs are a type of retro-element, with HERV-K being the more complete with open reading frames that have been expressed and have biological activities in melanoma and other human cells. Expression of HERV-K has been detected in melanoma cells and is increasingly implicated in melanoma pathogenesis. Recently, HERV-K has been studied as a molecular biomarker for melanoma and other malignancies, and as a vaccine candidate for cancer treatment.61–63 Further studies are warranted to examine and validate the activation and function of HERV-K in melanoma and other cancers, which should help improve the diagnosis and treatment of melanoma and other human diseases.
Acknowledgment
We would like to thank Dr Animesh Chandra for critical review and editing of the manuscript.
Disclosure
The authors report no conflicts of interest in this work.
References
Belshaw R, Katzourakis A, Paces J, Burt A, Tristem M. High copy number in human endogenous retrovirus families is associated with copying mechanisms in addition to reinfection. Mol Biol Evol. 2005;22(4):814–817. | |
de Parseval N, Heidmann T. Human endogenous retroviruses: from infectious elements to human genes. Cytogenet Genome Res. 2005; 110(1–4):318–332. | |
Kurth R, Bannert N. Beneficial and detrimental effects of human endogenous retroviruses. Int J Cancer. 2010;126(2):306–314. | |
Li Z, Sheng T, Wan X, Liu T, Wu H, Dong J. Expression of HERV-K correlates with status of MEK-ERK and p16INK4A-CDK4 pathways in melanoma cells. Cancer Invest. 2010;28(10):1031–1037. | |
Löwer R, Löwer J, Kurth R. The viruses in all of us: characteristics and biological significance of human endogenous retrovirus sequences. Proc Natl Acad Sci U S A. 1996;93(11):5177–5184. | |
Turner G, Barbulescu M, Su M, Jensen-Seaman MI, Kidd KK, Lenz J. Insertional polymorphisms of full-length endogenous retroviruses in humans. Curr Biol. 2001;11(19):1531–1535. | |
Lee E, Iskow R, Yang L, et al; Cancer Genome Atlas Research Network. Landscape of somatic retrotransposition in human cancers. Science. 2012;337(6097):967–971. | |
Macfarlan TS, Gifford WD, Driscoll S, et al. Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature. 2012;487(7405):57–63. | |
Bannert N, Kurth R. Retroelements and the human genome: new perspectives on an old relation. Proc Natl Acad Sci U S A. 2004; 101(Suppl 2):14572–14579. | |
Blomberg J, Benachenhou F, Blikstad V, Sperber G, Mayer J. Classification and nomenclature of endogenous retroviral sequences (ERVs): problems and recommendations. Gene. 2009;448(2):115–123. | |
Griffiths DJ. Endogenous retroviruses in the human genome sequence. Genome Biol. 2001;2(6):REVIEWS1017. | |
Medstrand P, Mager DL. Human-specific integrations of the HERV-K endogenous retrovirus family. J Virol. Dec 1998;72(12):9782–9787. | |
Andersson ML, Lindeskog M, Medstrand P, Westley B, May F, Blomberg J. Diversity of human endogenous retrovirus class II-like sequences. J Gen Virol. 1999;80(Pt 1):255–260. | |
Mayer J, Sauter M, Rácz A, Scherer D, Mueller-Lantzsch N, Meese E. An almost-intact human endogenous retrovirus K on human chromosome 7. Nat Genet. 1999;21(3):257–258. | |
Hohn O, Hanke K, Bannert N. HERV-K(HML-2), the best preserved family of HERVs: endogenization, expression, and implications in health and disease. Front Oncol. 2013;3:246. | |
Beimforde N, Hanke K, Ammar I, Kurth R, Bannert N. Molecular cloning and functional characterization of the human endogenous retrovirus K113. Virology. 2008;371(1):216–225. | |
Magiorkinis G, Belshaw R, Katzourakis A. ‘There and back again’: revisiting the pathophysiological roles of human endogenous retroviruses in the post-genomic era. Philos Trans R Soc Lond B Biol Sci. 2013;368(1626):20120504. | |
Camacho-Vanegas O, Camacho SC, Till J, et al. Primate genome gain and loss: a bone dysplasia, muscular dystrophy, and bone cancer syndrome resulting from mutated retroviral-derived MTAP transcripts. Am J Hum Genet. 2012;90(4):614–627. | |
Kalter SS, Helmke RJ, Heberling RL, et al. Brief communication: C-type particles in normal human placentas. J Natl Cancer Inst. 1973;50(4):1081–1084. | |
Löwer R, Boller K, Hasenmaier B, et al. Identification of human endogenous retroviruses with complex mRNA expression and particle formation. Proc Natl Acad Sci U S A. 1993;90(10):4480–4484. | |
Seifarth W, Frank O, Zeilfelder U, et al. Comprehensive analysis of human endogenous retrovirus transcriptional activity in human tissues with a retrovirus-specific microarray. J Virol. 2005;79(1):341–352. | |
Muster T, Waltenberger A, Grassauer A, et al. An endogenous retrovirus derived from human melanoma cells. Cancer Res. 2003;63(24):8735–8741. | |
Voisset C, Weiss RA, Griffiths DJ. Human RNA “rumor” viruses: the search for novel human retroviruses in chronic disease. Microbiol Mol Biol Rev. 2008;72(1):157–196, table of contents. | |
DeSantis CE, Lin CC, Mariotto AB, et al. Cancer treatment and survivorship statistics, 2014. CA Cancer J Clin. 2014;64(4):252–271. | |
Bastian BC. The molecular pathology of melanoma: an integrated taxonomy of melanocytic neoplasia. Annu Rev Pathol. 2014;9:239–271. | |
Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. | |
Leong SP, Mihm MC Jr, Murphy GF, et al. Progression of cutaneous melanoma: implications for treatment. Clin Exp Metastasis. 2012;29(7):775–796. | |
Coit DG, Olszanski AJ. Progress in the management of melanoma in 2013. J Natl Compr Canc Netw. 2013;11(Suppl 5):645–648. | |
Shah DJ, Dronca RS. Latest advances in chemotherapeutic, targeted, and immune approaches in the treatment of metastatic melanoma. Mayo Clin Proc. 2014;89(4):504–519. | |
Chomez P, De Backer O, Bertrand M, De Plaen E, Boon T, Lucas S. An overview of the MAGE gene family with the identification of all human members of the family. Cancer Res. 2001;61(14):5544–5551. | |
Parmiani G, Anichini A, Castelli C. New tumour-restricted melanoma antigens as defined by cytotoxic T-cell responses. Melanoma Res. 1997;7(Suppl 2):S95–S98. | |
Schiavetti F, Thonnard J, Colau D, Boon T, Coulie PG. A human endogenous retroviral sequence encoding an antigen recognized on melanoma by cytolytic T lymphocytes. Cancer Res. 2002;62(19):5510–5516. | |
Schmitt K, Reichrath J, Roesch A, Meese E, Mayer J. Transcriptional profiling of human endogenous retrovirus group HERV-K(HML-2) loci in melanoma. Genome Biol Evol. 2013;5(2):307–328. | |
Esqueda D, Xu F, Moore Y, et al. Lack of correlation between HERV-K expression and HIV-1 viral load in plasma specimens. Ann Clin Lab Sci. 2013;43(2):122–125. | |
Katoh I, Mirova A, Kurata S, et al. Activation of the long terminal repeat of human endogenous retrovirus K by melanoma-specific transcription factor MITF-M. Neoplasia. 2011;13(11):1081–1092. | |
Shibahara S, Takeda K, Yasumoto K, et al. Microphthalmia-associated transcription factor (MITF): multiplicity in structure, function, and regulation. J Investig Dermatol Symp Proc. 2001;6(1):99–104. | |
Stengel S, Fiebig U, Kurth R, Denner J. Regulation of human endogenous retrovirus-K expression in melanomas by CpG methylation. Genes Chromosomes Cancer. 2010;49(5):401–411. | |
Li J, Xu M, Yang Z, Li A, Dong J. Simultaneous inhibition of MEK and CDK4 leads to potent apoptosis in human melanoma cells. Cancer Invest. 2010;28(4):350–356. | |
Zhao Y, Zhang Y, Yang Z, Li A, Dong J. Simultaneous knockdown of BRAF and expression of INK4A in melanoma cells leads to potent growth inhibition and apoptosis. Biochem Biophys Res Commun. 2008;370(3):509–513. | |
Rotolo S, Diotti R, Gordon RE, et al. Effects on proliferation and melanogenesis by inhibition of mutant BRAF and expression of wild-type INK4A in melanoma cells. Int J Cancer. 2005; 115(1):164–169. | |
Ayala F, Palla M, Di Trolio R, Mozzillo N, Ascierto PA. The role of optical radiations in skin cancer. ISRN Dermatol. 2013;2013:842359. | |
Campbell C, Quinn AG, Angus B, Farr PM, Rees JL. Wavelength specific patterns of p53 induction in human skin following exposure to UV radiation. Cancer Res. 1993;53(12):2697–2699. | |
Reiche J, Pauli G, Ellerbrok H. Differential expression of human endogenous retrovirus K transcripts in primary human melanocytes and melanoma cell lines after UV irradiation. Melanoma Res. 2010;20(5):435–440. | |
Schanab O, Humer J, Gleiss A, et al. Expression of human endogenous retrovirus K is stimulated by ultraviolet radiation in melanoma. Pigment Cell Melanoma Res. 2011;24(4):656–665. | |
Krone B, Kölmel KF, Henz BM, Grange JM. Protection against melanoma by vaccination with Bacille Calmette-Guerin (BCG) and/or vaccinia: an epidemiology-based hypothesis on the nature of a melanoma risk factor and its immunological control. Eur J Cancer. 2005;41(1):104–117. | |
Mangeney M, Pothlichet J, Renard M, Ducos B, Heidmann T. Endogenous retrovirus expression is required for murine melanoma tumor growth in vivo. Cancer Res. 2005;65(7):2588–2591. | |
Oricchio E, Sciamanna I, Beraldi R, Tolstonog GV, Schumann GG, Spadafora C. Distinct roles for LINE-1 and HERV-K retroelements in cell proliferation, differentiation and tumor progression. Oncogene. 2007;26(29):4226–4233. | |
Pothlichet J, Mangeney M, Heidmann T. Mobility and integration sites of a murine C57BL/6 melanoma endogenous retrovirus involved in tumor progression in vivo. Int J Cancer. 2006;119(8):1869–1877. | |
Büscher K, Trefzer U, Hofmann M, Sterry W, Kurth R, Denner J. Expression of human endogenous retrovirus K in melanomas and melanoma cell lines. Cancer Res. 2005;65(10):4172–4180. | |
Humer J, Waltenberger A, Grassauer A, et al. Identification of a melanoma marker derived from melanoma-associated endogenous retroviruses. Cancer Res. 2006;66(3):1658–1663. | |
Hahn S, Ugurel S, Hanschmann KM, et al. Serological response to human endogenous retrovirus K in melanoma patients correlates with survival probability. AIDS Res Hum Retroviruses. 2008;24(5):717–723. | |
Huang G, Li Z, Wan X, Wang Y, Dong J. Human endogenous retroviral K element encodes fusogenic activity in melanoma cells. J Carcinog. 2013;12:5. | |
Sciamanna I, Landriscina M, Pittoggi C, et al. Inhibition of endogenous reverse transcriptase antagonizes human tumor growth. Oncogene. 2005;24(24):3923–3931. | |
Serafino A, Balestrieri E, Pierimarchi P, et al. The activation of human endogenous retrovirus K (HERV-K) is implicated in melanoma cell malignant transformation. Exp Cell Res. 2009;315(5):849–862. | |
Dunham I, Kundaje A, Aldred SF; ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. | |
Gerstein MB, Kundaje A, Hariharan M, et al. Architecture of the human regulatory network derived from ENCODE data. Nature. 2012;489(7414):91–100. | |
Qu H, Fang X. A brief review on the Human Encyclopedia of DNA Elements (ENCODE) project. Genomics Proteomics Bioinformatics. 2013;11(3):135–141. | |
Kim YJ, Lee J, Han K. Transposable elements: no more ‘junk DNA’. Genomics Inform. 2012;10(4):226–233. | |
Solyom S, Kazazian HH Jr. Mobile elements in the human genome: implications for disease. Genome Med. 2012;4(2):12. | |
Kim HS. Genomic impact, chromosomal distribution and transcriptional regulation of HERV elements. Mol Cells. 2012;33(6):539–544. | |
Kraus B, Fischer K, Büchner SM, et al. Vaccination directed against the human endogenous retrovirus-K envelope protein inhibits tumor growth in a murine model system. PloS One. 2013;8(8):e72756. | |
Kraus B, Fischer K, Sliva K, Schnierle BS. Vaccination directed against the human endogenous retrovirus-K (HERV-K) gag protein slows HERV-K gag expressing cell growth in a murine model system. Virol J. 2014;11:58. | |
Tran T, Burt D, Eapen L, Keller OR. Spontaneous regression of metastatic melanoma after inoculation with tetanus-diphtheria-pertussis vaccine. Curr Oncol. 2013;20(3):e270–e273. |
 © 2014 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2014 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.