Back to Journals » OncoTargets and Therapy » Volume 12
ITGBL1 promotes EMT, invasion and migration by activating NF-κB signaling pathway in prostate cancer
Authors Li W, Li S, Yang J, Cui C, Yu M, Zhang Y
Received 1 January 2019
Accepted for publication 1 April 2019
Published 16 May 2019 Volume 2019:12 Pages 3753—3763
DOI https://doi.org/10.2147/OTT.S200082
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Jianmin Xu
Wenze Li,1,* Shuren Li,1,* Jie Yang,1 Chunyan Cui,2 Miao Yu,3 Yadong Zhang4
1Department of Urinary Surgery, The First hospital of Xiangtan city, Xiangtan 411101, People’s Republic of China; 2Department of Radiology, Sun Yat-sen University Cancer Center, State Key Laboratory of Oncology in South China, Collaborative Innovation Center for Cancer Medicine, Guangzhou 510060, People’s Republic of China; 3Center for Private Medical Service and Healthcare, The First Hospital of Sun Yat-sen University, Guangzhou, Guangdong 510080, People’s Republic of China; 4Department of Andrology, The First Affiliated Hospital of Sun Yat-sen University, Guangzhou 510080, People’s Republic of China
*These authors contributed equally to this work
Background: Integrin beta-like 1 (ITGBL1) was extensively demonstrated to contribute the metastasis and progression in a variety of cancers. However, its role of ITGBL1 in prostate cancer (PCa) is still not reported.
Methods: Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) and western blot were performed to detect ITGBL1 expression in PCa tissues and cell lines. Immunohistochemical (IHC) staining of ITGBL1 in 174 PCa tissues was performed. The influence of ITGL1 expression in PCa cells epithelial-mesenchymal transition (EMT), migration and invasion was investigated. Notably, the possible mechanisms underlying the action of ITGBL1 in vivo and vitro assays were explored.
Results: We analyzed PCa dataset from The Cancer Genome Atlas (TCGA) and found that ITGBL1 was upregulated in PCa tissues. Overexpression of ITGBL1 is positively associated with the progression and lymph node metastasis in PCa patients. Furthermore, upregulating ITGBL1 enhanced the invasion, migration abilities and EMT in PCa cells. Conversely, downregulating ITGBL1 exhibited an opposite effect. Our findings further demonstrated that ITGBL1 promoted invasion and migration via activating NF-κB signaling in PCa cells.
Conclusion: Therefore, our results identify a novel metastasis-related gene in PCa, which will help to develop a novel therapeutic strategy in metastatic PCa.
Keywords: ITGBL1, EMT, NF-κB signaling, lymph node metastasis and prostate cancer
Introduction
Prostate cancer (PCa) is one of the prevalent malignancies in male worldwide and occupies approximately 20% of newly diagnosed cancers in males.1 In China, although substantial progress in the treatment of localized PCa, such as radical prostatectomy and androgen ablation, metastatic PCa remains incurable due to limited therapeutic methods. The prime causes for the PCa patients deaths are mostly attributed to invasion and metastasis.2,3 Thus, exploring the underlying mechanism in metastasis of PCa is crucial to develop therapeutic avenues to treat or prevent PCa metastasis.
Integrin beta-like 1 (ITGBL1) encodes a class of κ-integrin-associated protein with ten tandem integrin EGF-like repeats since discovered from an osteoblast cDNA library.4 Unlike the most family members of EGF-like protein, neither an RGD (Arg–Gly–Asp)-binding domain nor a transmembrane domain was found in ITGBL1, implying that ITGBL1 exerts its function differing from those of integrin.5 Recently, the functional roles of ITGBL1 in the development and metastasis in numerous cancers have been extensively studied. For example, transcriptional upregulation of ITGBL1 by Runx2 promoted the bone metastasis via TGF-β signaling pathway in breast cancer;6 in addition, ITGBL1 has been reported to promote the adhesion and migration of ovarian cancer cell via FAK/SRC and Wnt/PCP signaling;7 in gastrointestinal cancers, including colorectal cancer and gastric cancer, high level of ITGBL1 was closely associated with the invasive phenotypes of cancer cells, and predicted poor prognosis.8,9 Therefore, the existing literature supports the idea that ITGBL1 functions as an important pro-metastatic oncogene in multiple human cancer types. Currently, however, any information regarding the clinical significance and functional role of ITGBL1 remain scanty in the metastasis of PCa.
In the present study, our results showed that ITGBL1 was elevated in PCa tissues, which was positively associated with poor clinicopathological characteristics and lymph node metastasis, as well as predicted poor progression-free survival. Furthermore, upregulating ITGBL1 promoted, while silencing ITGBL1 inhibited the invasion, migration abilities and EMT in PCa cells. Our findings further revealed that ITGBL1 promoted the migration and invasion abilities of PCa cells by activating NF-κB signaling. Therefore, our results unravel a new mechanism responsible for ITGBL1-induced invasion and migration abilities in PCa cells, suggesting that ITGBL1 may be used as a potential therapeutic target in metastatic PCa.
Materials and methods
Cell lines and cell culture
Normal prostate epithelial cells RWPE-1, primary PCa cell lines 22RV1, bone metastatic PCa cells PC-3 and VCaP, brain metastatic PCa DU145, lymph node metastatic PCa LNCaP were obtained from Shanghai Chinese Academy of Sciences cell bank (China). All were cultured according to the previous study.10
Patients and tumor tissues
A total of 174 paraffin-embedded PCa tissues and 8 fresh PCa tissues were collected at The First hospital of Xiangtan city (Xiangtan, Hunan, China). The clinical and pathological evidence were required for diagnosis of PCa patients. Prior patients’ consents and approval from the Institutional Research Ethics Committee were obtained from The First hospital of Xiangtan city. Furthermore, the patient consent was written informed consent, and in accordance with the Declaration of Helsinki. Table 1 summarized the clinicopathological characteristics of PCa patients.
 | Table 1 The relationship between ITGBL1 and clinicopathological characteristicin in 174 patients with prostate cancer |
RNA extraction, transfection, and real-time PCR
The total RNA from tissues or cells was extracted using TRIzol (Life Technologies) according to the manufacturer’s instructions. mRNA was reverse transcribed using the Revert Aid First Strand cDNA Synthesis Kit (Thermo, USA). cDNA was quantified and amplified on ABI 7500HT system (Applied Biosystems, Foster City, CA, USA) using SYBR Green I (Applied Biosystems). Real-time PCR was carried out as described previously.11 The primer sequence of ITGBL1 was forward, 5′-GACTGCAAAGCAGGCTGGTATG-3′; reverse, 5′-GGAGGATAGCAGGTGCATTTGC-3′. The endogenous control for mRNA was GAPDH. Relative fold expressions were calculated with the comparative threshold cycle (2−ΔΔCt) method as previously described.12
Plasmid, small interfering RNA and transfection
The cDNA of ITGBL1 was purchased from (Vigene Biosciences, Shandong, China) and cloned into the pSin-EF2 plasmid (Cambridge, MA, USA). Silencing ITGBL1 was carried out by cloning two short hairpin RNA (shRNA) oligonucleotides into the pSUPER-puro-retro vector (OligoEngine, Seattle, WA, USA). The control and pNFκB-luc plasmids were purchased from Promega Corporation. shRNAs or plasmids were transfected using Lipofectamine 3000 (Life Technologies) as previously described.13
Western blotting
Western blotting was performed as described previously.14 The E-cadherin (Cat#. 3195), Vimentin (Cat#. 5741) and Fibronectin (Cat#. 4706) antibodies were purchased from Cell Signaling Technology, ITGBL1 (Cat#: Ag11521) and p65 (Cat#. 10745–1-AP) from Proteintech, p84 (Cat#. PA5–27816) from Invitrogen. After stripped, the membranes were reprobed with an α-tubulin antibody (Sigma-Aldrich, USA) as the loading control.
Invasion and migration assay
The migration and invasion assays were carried out using Transwell chamber consisting of 8-mm membrane filter inserts (Corning) without or with coated Matrigel (BD Biosciences) respectively as described previously.15 Briefly, cells were trypsinized and resuspended in serum-free medium after serum starvation for 24 h. Thereafter, 1.5×105 cells were added to the upper chamber, while the lower chamber was filled with medium containing 10% FBS. After incubation for 48 h, cells that had invaded through the coated membrane to the lower surface were fixed and stained. The cell count was performed under a microscope (100×), and the random 10 fields were captured. The total cell number in these 10 fields were used to compare the effect of ITGBL1 on the invasion/migration ability of different groups of PCa cells.
Luciferase assay
Cells (4×104) were plated into the triplicate in 24-well plates and carried out as previously described.16 Briefly, the indicated number of cells were transfected with 100 ng the control or pNFκB reporter luciferase plasmid, plus the pRL-TK Renilla plasmid with concentration of 5 ng (Promega) as manufacturer’s protocol. A Dual-Luciferase Reporter Assay Kit (Promega) was used to measure Luciferase and Renilla signals 36 h after transfection. Briefly, 5 copies of an NF-κB response element (NF-κB-RE) were transferred into the pGL3-basic by restriction endonuclease, which is followed by the luciferase reporter gene. Cells were plated in 24-well plates, proliferating to 60–80% confluence after 24 h of culture, and the pGL3(NF-κB/luc) or pGL3, plus pRL-TK Renilla plasmid (Promega) were transfected into cells using Lipofectamine 3000 (Life Technologies). Forty-eight hours after transfection, the transfection medium was replaced with fresh RPMI-1640 medium; cells were harvested and washed with PBS, and lysed with passive lysis buffer (Promega). The cell lysates were analyzed immediately using Synergy™ 2 microplate system (BioTek, Winooski, VT, US). Luciferase and Renilla luciferase were measured using a Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer’s instructions. The luciferase activity of each lysate was normalized to Renilla luciferase activity.
Immunohistochemistry
The immunohistochemistry (IHC) was carried out as previously described.17 The IHC scores were obtained from two independent investigators to further comparatively evaluating ITGBL1 expression in PCa tissues. The IHC score was calculated as the product of the staining intensity score and the proportion of positive tumor cells, which were evaluated as previously described.18
Tumor xenografts
The twenty BALB/c-nu mice (6-week-old) were randomly divided into four groups. 1×106 22Rv1 tumor cells were injected subcutaneously into the right flanks of each mouse in 100 μl of RPMI mixed with Matrigel. In overexpression of ITGBL1 with JSH-23 treatment group, the ITGBL1 overexpression 22RV1cells were treated with JSH-23 (20 μmol/L)19 for 24h and then implanted subcutaneously in the right flanks of each mouse. In overexpression of ITGBL1with Y2409881 treatment group, the Y2409881 (50 mg/kg)20 was intraperitoneally given on every three days for 4 weeks. An external caliper and the equation (4/3) πr3 were used to calculate the tumor volume, where r=(length + width)/4. On day 28, the tumors tissues were collected and weighed. In addition to the approval from the Institutional Animal Care and Use Committee of Sun Yat-sen University, laboratory animal-guideline for ethical review of animal welfare based on the State Standard of the People’s Republic of China was followed for the welfare of the animals.
Statistical analysis
Statistical significance was determined by GraphPad 5.0 software (USA), and P values ≤0.05 considered significant. Statistical significance between two groups or multiple groups was determined by student’s ttest (unpaired two sample) or One-way ANOVA respectively. The association between ITGBL1 expression and clinicopathological characteristics of PCa patients was analyzed via the chi-square test. The Kaplan Meier method was used to plot survival curves and compared by the log-rank test. repetition of at least 3 times was applied to achieve reproducibility.
Results
ITGBL1 is upregulated in PCa tissues
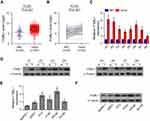
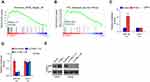
To determine the expression levels of ITGBL1 in PCa, the PCa dataset from The Cancer Genome Atlas (TCGA) was first analyzed. As shown in Figure 1A, ITGBL1 was found to be upregulated in 497 PCa tissues compared to the 52 adjacent normal tissues (ANT) (Figure 1A). Consistently, ITGBL1 expression level was upregulated in PCa tissues compared to those in the matched ANT (Figure 1B). The expression of ITGBL1 in our PCa tissues was further examined, and the results showed that ITGBL1 expression was elevated in PCa tissues compared to those in the matched ANT (Figure 1C and D). We further evaluated ITGBL1 expression in PCa cells and found that the expression levels of ITGBL1 were differentially upregulated in PCa cells compared with those in one normal prostate epithelial cells RWPE-1, particularly in metastatic PCa cells, including LNCaP and PC-3 (Figure 1E and F). Taken together, these results suggest that overexpression ITGBL1 may contribute to the development and metastasis of PCa.
Overexpression of ITGBL1 correlates with lymph node metastasis of PCa
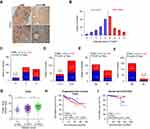
We further analyzed ITGBL1 expression via immunohistochemical (IHC) in 174 PCa tissues, and found that ITGBL1 expression in PCa tissues with lymph node metastasis was elevated compared to that in PCa tissues without lymph node metastasis (Figure 2A and B). Furthermore, ITGBL1 expression levels were positively associated with Gleason grade, serum PSA levels, tumor size, distant metastasis and lymph node metastasis in PCa patients (Figure 2C–F and Table 1). The association of ITGBL1 expression with clinicopathological features in PCa dataset from TCGA was further analyzed, and the results showed that high expression of ITGBL1 correlated with Gleason grades (Figure 2G) and poor progression-free survival (Figure 2H), but not with overall survival in PCa patients (Figure 2I). The results above indicate that overexpression of ITGBL1 correlated with progression and lymph node metastasis in PCa patients.
Upregulation of ITGBL1 enhances EMT, invasion and migration
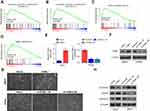
Gene Set Enrichment Analysis (GSEA) was performed to investigate the biological role of ITGBL1 in PCa. As shown in Figure 3A–D, the expression level of ITGBL1 was positively and strongly related with epithelial-mesenchymal transition (EMT)-associated gene signatures and metastatic phenotypes of cancer cells. Next, we constructed ITGBL1-stably expressing PCa cells by ectopically overexpressing ITGBL1 in 22RV1 cells, as well as endogenously knocking down ITGBL1 in LNCaP cells, respectively (Figure 3E and F). Upregulating ITGBL1 converted the epithelial profile to a mesenchymal phenotype in 22RV1 cells (Figure 3G). Conversely, silencing ITGBL1 converted the mesenchymal phenotype to the epithelial profile in LNCaP cells (Figure 3G). Western blot analysis revealed that upregulating ITGBL1 increased the expression of mesenchymal marker vimentin and fibronectin in PCa cells and decreased the expression of epithelial marker E-cadherin, and silencing ITGBL1 displayed the opposite effect in these PC cells (Figure 3H). Furthermore, upregulating ITGBL1 promoted, while downregulating ITGBL1 repressed the invasion and migration ability of PCa cells (Figure 4A and B). These findings demonstrated that ITGBL1 promotes the migration, invasion, and EMT in PCa cells.
ITGBL1 activates NF-κb signaling pathway
To determine the mechanism underlying the metastasis-stimulating role of ITGBL1 in PCa cells, GSEA based on ITGBL1 expression was performed, and our results revealed that overexpression of ITGBL1 was positively associated with hyperactivity of NF-κB signaling (Figure 5A and B) that has been identified to promote the tumorigenesis and metastasis in several human cancers.21–23 Our results showed that upregulating ITGBL1 increased, while downregulating ITGBL1decreased NF-κB-dependent luciferase activity in PCa cells via luciferase assay (Figure 5C and D). Moreover, upregulating ITGBL1 increased, while silencing ITGBL1 decreased nuclear translocation of NF-κB/p65 via cellular fractionation and western blotting analysis (Figure 5E). These results indicate that ITGBL1 activates NF-κB signaling in PCa cells.
The activity of NF-κb signaling is essential for ITGBL1-induced invasion and migration, and tumorigenesis of PCa cells
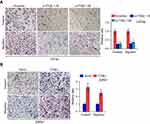
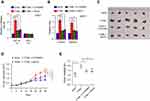
The function of NF-κB signaling in the metastasis-stimulating effect of ITGBL1 on PCa cells was further investigated using NF-κB signaling inhibitors JSH-23 and LY2409881. As shown in Figure 6A, JSH-23 or LY2409881 abolished the effect of ITGBL1 on the activity of NF-κB signaling. Furthermore, JSH-23 and LY2409881 dramatically reduced the migration and invasion abilities in ITGBL1-overexpressing PCa cells (Figure 6B). Importantly, the pro-tumorigenic role of ITGBL1-overexpressing PCa cells was partially blocked by LY2409881 and JSH-23 respectively (Figure 6C–E). Therefore, these results demonstrate that ITGBL1 activates NF-kB signaling pathway, which further promotes invasion, migration and tumorigenesis of PCa cells.
Discussion
A number of studies have shown that ITGBL1 was upregulated in various types of cancers, including colorectal cancer,8 breast cancer6 and gastric cancer,9,24 which contributed to the metastasis and progression of cancers via the varying mechanism. Strikingly, ITGBL1 expression was downregulated in non-small cell lung cancer (NSCLC), and upregulating ITGBL1 inhibited invasion and migration abilities of NSCLC cells,25 indicating that ITGBL1 functions it role as a tumor suppressor in NSCLC. These findings imply that the anti- or pro-tumor roles of ITGBL1 vary based on tumor type. Notably, ITGBL1 was found to be overexpressed in PCa tissues by transcriptomic analysis.26 However, the biological function and clinical significance of ITGBL1 in PCa is still unclear. In the current study, our results revealed that ITGBL1 expression was upregulated in PCa tissues, which was positively associated with progression and metastasis in PCa patients. Functional experiments in vitro showed that upregulating ITGBL1 enhanced, while silencing ITGBL1 inhibited the EMT, invasion and migration in PCa cells. Therefore, our results indicate that ITGBL1 plays an oncogenic role in PCa cells.
Since been identified, the vast majority of studies have reported that ITGBL1 functions as a metastasis-related gene in several cancers, which promotes the migration, invasion, and EMT of cancer cells, resulting in the distant metastasis of cancer.6–9 Consistent with these observations, our results showed that ITGBL1 in PCa tissues with lymph node metastasis was upregulated compared to that in PCa tissues without lymph node metastasis. Furthermore, upregulating ITGBL1 promoted the development of EMT, inducing the epithelial profile to a mesenchymal phenotype, enhancing Vimentin and Fibronectin expression and reduced E-cadherin expression, as well as aggravated the migration and invasion ability in PCa cells. Conversely, silencing ITGBL1 yielded an opposite effect on PCa cells. Collectively, our results determine a metastasis-stimulating role of ITGBL1 in PCa.
As mentioned above, our study demonstrated that upregulating ITGBL1 promoted, while silencing ITGBL1 repressed the EMT in PCa cells. In fact, accumulating evidence have reported that EMT is identified to be one of the critical steps in tumor metastasis, including PCa, by allowing cancer cells acquire mesenchymal features that permit escape from the primary tumor.27–30 Given the crucial role of EMT in metastasis of PCa and our findings in this study, we infer that ITGBL1 promote lymph node metastasis of PCa via inducing EMT of PCa cells.
Accumulating evidence has shown that ITGBL1 can be used as a potential marker to predict prognosis in cancer patients. For example, overexpression of ITGBL1 was demonstrated to predict poor prognosis in gastric cancer or colorectal cancer patients;8,9,24 moreover, ITGBL1 has been identified to be a prognostic gene to be able to predict progression-free survival in multiple myeloma patients.31 Here, our results indicated that overexpression of ITGBL1 was positively associated with Gleason grade, tumor size, distant metastasis and lymph node metastasis in PCa patients. Consistently, our results further revealed that high expression of ITGBL1 predicted poor progression-free survival in PCa patients through analyzing the ITGBL1 expression in PCa dataset from TCGA. These findings indicate that ITGBL1 harbors great promise as a candidate biomarker for the progression-free and metastasis-free survivals in PCa patients. However, a solid conclusion requires further investigation in the following work.
In summary, our findings reveal that ITGBL1 promotes the metastasis, invasion, and EMT by activating NF-kB signaling in PCa cells, providing the evidence that ITGBL1 plays a pro-metastatic role in PCa. Therefore, better identification of the biological function of ITGBL1 in PCa will facilitate the development of novel anti-metastasis therapeutic measures in PCa.
Acknowledgments
This work is supported by a grant from the Medical Association of Xiangtan city (C2016114), Guangdong Provincial Science and Technology Project (2017A020211021) and Guangdong Medical Research Fund Project (No. A2016320).
Ethical approval
For the use of these clinical materials for research purposes, prior patients’ consents and approval from the Institutional Research Ethics Committee were obtained. All mouse experiments were approved by The Institutional Animal Care and Use Committee of Sun Yat-sen University and the approval-No.was L102012018120P.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10–29. doi:10.3322/caac.20138
2. Sabbatini P, Larson SM, Kremer A, et al. Prognostic significance of extent of disease in bone in patients with androgen-independent prostate cancer. J Clin Oncol. 1999;17:948–957. doi:10.1200/JCO.1999.17.3.948
3. Norgaard M, Jensen AO, Jacobsen JB, Cetin K, Fryzek JP, Sorensen HT. Skeletal related events, bone metastasis and survival of prostate cancer: a population based cohort study in Denmark (1999 to 2007). J Urol. 2010;184:162–167. doi:10.1016/j.juro.2010.03.034
4. Berg RW, Leung E, Gough S, et al. Cloning and characterization of a novel beta integrin-related cDNA coding for the protein TIED ("ten beta integrin EGF-like repeat domains”) that maps to chromosome band 13q33: a divergent stand-alone integrin stalk structure. Genomics. 1999;56:169–178. doi:10.1006/geno.1998.5707
5. Takagi J, Beglova N, Yalamanchili P, Blacklow SC, Springer TA. Definition of EGF-like, closely interacting modules that bear activation epitopes in integrin beta subunits. Proc Natl Acad Sci USA. 2001;98:11175–11180. doi:10.1073/pnas.201420198
6. Li XQ, Du X, Li DM, et al. ITGBL1 is a Runx2 transcriptional target and promotes breast cancer bone metastasis by activating the TGFbeta signaling pathway. Cancer Res. 2015;75:3302–3313. doi:10.1158/0008-5472.CAN-15-0240
7. Sun L, Wang D, Li X, Zhang L, Zhang H, Zhang Y. Extracellular matrix protein ITGBL1 promotes ovarian cancer cell migration and adhesion through Wnt/PCP signaling and FAK/SRC pathway. Biomed Pharmacother. 2016;81:145–151. doi:10.1016/j.biopha.2016.03.053
8. Qiu X, Feng JR, Qiu J, et al. ITGBL1 promotes migration, invasion and predicts a poor prognosis in colorectal cancer. Biomed Pharmacother. 2018;104:172–180. doi:10.1016/j.biopha.2018.05.033
9. Li R, Zhuang C, Jiang S, et al. ITGBL1 predicts a poor prognosis and correlates EMT phenotype in gastric cancer. J Cancer. 2017;8:3764–3773. doi:10.7150/jca.20900
10. Dai Y, Ren D, Yang Q, et al. The TGF-beta signalling negative regulator PICK1 represses prostate cancer metastasis to bone. Br J Cancer. 2017. doi:10.1038/bjc.2017.212
11. Ren D, Lin B, Zhang X, et al. Maintenance of cancer stemness by miR-196b-5p contributes to chemoresistance of colorectal cancer cells via activating STAT3 signaling pathway. Oncotarget. 2017. doi:10.18632/oncotarget.17971
12. Zhang X, Ren D, Wu X, et al. miR-1266 contributes to pancreatic cancer progression and chemoresistance by the STAT3 and NF-kappaB signaling pathways. Mol Ther Nucleic Acids. 2018;11:142–158. doi:10.1016/j.omtn.2018.01.004
13. Wu N, Ren D, Li S, et al. RCC2 over-expression in tumor cells alters apoptosis and drug sensitivity by regulating Rac1 activation. BMC Cancer. 2018;18:67. doi:10.1186/s12885-017-3908-y
14. Zhang X, Zhang L, Lin B, et al. Phospholipid phosphatase 4 promotes proliferation and tumorigenesis, and activates Ca2+-permeable cationic channel in lung carcinoma cells. Mol Cancer. 2017;16:147. doi:10.1186/s12943-017-0717-5
15. Ren D, Wang M, Guo W, et al. Double-negative feedback loop between ZEB2 and miR-145 regulates epithelial-mesenchymal transition and stem cell properties in prostate cancer cells. Cell Tissue Res. 2014;358:763–778. doi:10.1007/s00441-014-2001-y
16. Li X, Liu F, Lin B, et al. miR150 inhibits proliferation and tumorigenicity via retarding G1/S phase transition in nasopharyngeal carcinoma. Int J Oncol. 2017. doi:10.3892/ijo.2017.3909
17. Zhang X, Ren D, Guo L, et al. Thymosin beta 10 is a key regulator of tumorigenesis and metastasis and a novel serum marker in breast cancer. Breast Cancer Res. 2017;19:15. doi:10.1186/s13058-016-0785-2
18. Ren D, Dai Y, Yang Q, et al. Wnt5a induces and maintains prostate cancer cells dormancy in bone. J Exp Med. 2018. doi:10.1084/jem.20180661
19. Ahmed KM, Zhang H, Park CC. NF-kappaB regulates radioresistance mediated by beta1-integrin in three-dimensional culture of breast cancer cells. Cancer Res. 2013;73:3737–3748. doi:10.1158/0008-5472.CAN-12-3537
20. Deng C, Lipstein M, Rodriguez R, et al. The novel IKK2 inhibitor LY2409881 potently synergizes with histone deacetylase inhibitors in preclinical models of lymphoma through the downregulation of NF-kappaB. Clin Cancer Res. 2015;21:134–145. doi:10.1158/1078-0432.CCR-14-0384
21. Boldin MP, Baltimore D. MicroRNAs, new effectors and regulators of NF-kappaB. Immunol Rev. 2012;246:205–220. doi:10.1111/j.1600-065X.2011.01089.x
22. Ren D, Yang Q, Dai Y, et al. Oncogenic miR-210-3p promotes prostate cancer cell EMT and bone metastasis via NF-kappaB signaling pathway. Mol Cancer. 2017;16:117. doi:10.1186/s12943-017-0688-6
23. Huber MA, Azoitei N, Baumann B, et al. NF-kappaB is essential for epithelial-mesenchymal transition and metastasis in a model of breast cancer progression. J Clin Invest. 2004;114:569–581. doi:10.1172/JCI21358
24. Liu X, Wu J, Zhang D, et al. Identification of potential key genes associated with the pathogenesis and prognosis of gastric cancer based on integrated bioinformatics analysis. Front Genet. 2018;9:265. doi:10.3389/fgene.2018.00265
25. Gan X, Liu Z, Tong B, Zhou J. Epigenetic downregulated ITGBL1 promotes non-small cell lung cancer cell invasion through Wnt/PCP signaling. Tumour Biol. 2016;37:1663–1669. doi:10.1007/s13277-015-3919-8
26. Shan M, Xia Q, Yan D, et al. Molecular analyses of prostate tumors for diagnosis of malignancy on fine-needle aspiration biopsies. Oncotarget. 2017;8:104761–104771. doi:10.18632/oncotarget.22289
27. Montanari M, Rossetti S, Cavaliere C, et al. Epithelial-mesenchymal transition in prostate cancer: an overview. Oncotarget. 2017;8:35376–35389. doi:10.18632/oncotarget.15686
28. Guo W, Ren D, Chen X, et al. HEF1 promotes epithelial mesenchymal transition and bone invasion in prostate cancer under the regulation of microRNA-145. J Cell Biochem. 2013;114:1606–1615. doi:10.1002/jcb.24502
29. Ren D, Wang M, Guo W, et al. Wild-type p53 suppresses the epithelial-mesenchymal transition and stemness in PC-3 prostate cancer cells by modulating miR145. Int J Oncol. 2013;42:1473–1481. doi:10.3892/ijo.2013.1825
30. Wang M, Ren D, Guo W, et al. N-cadherin promotes epithelial-mesenchymal transition and cancer stem cell-like traits via ErbB signaling in prostate cancer cells. Int J Oncol. 2016;48:595–606. doi:10.3892/ijo.2015.3270
31. Schinke C, Qu P, Mehdi SJ, et al. The pattern of mesenchymal stem cell expression is an independent marker of outcome in multiple myeloma. Clin Cancer Res. 2018;24:2913–2919. doi:10.1158/1078-0432.CCR-17-2627
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.