Back to Journals » Clinical, Cosmetic and Investigational Dermatology » Volume 15
ITGA9 Inhibits Proliferation and Migration of Dermal Microvascular Endothelial Cells in Psoriasis
Authors Hou H, Li J, Wang J, Zhou L, Li J, Liang J, Yin G, Li X, Cheng Y, Zhang K
Received 21 October 2022
Accepted for publication 9 December 2022
Published 20 December 2022 Volume 2022:15 Pages 2795—2806
DOI https://doi.org/10.2147/CCID.S394398
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Jeffrey Weinberg
Hui Hou, Jiao Li, Juanjuan Wang, Ling Zhou, Junqin Li, Jiannan Liang, Guohua Yin, Xinhua Li, Yueai Cheng, Kaiming Zhang
Shanxi Key Laboratory of Stem Cell for Immunological Dermatosis, Institute of Dermatology, Taiyuan Central Hospital of Shanxi Medical University, Taiyuan, People’s Republic of China
Correspondence: Kaiming Zhang, Shanxi Key Laboratory of Stem Cell for Immunological Dermatosis, Institute of Dermatology, Taiyuan Central Hospital of Shanxi Medical University, No. 5 Dong San Dao Xiang, Jiefang Road, Taiyuan, Shanxi Province, People’s Republic of China, Tel +86-351-5656080, Email [email protected]
Background: Cell proliferation, migration, and angiogenesis are aberrant in psoriatic human dermal microvascular endothelial cells (HDMECs), resulting in abnormal endothelial function and microvascular dilation in psoriasis.
Objective: To explore the role of Integrin subunit alpha 9 (ITGA9) in proliferation and migration of dermal microvascular endothelial cells.
Methods: HDMECs were isolated from the skin of 6 psoriatic patients and 6 healthy controls. Expression levels of ITGA9 mRNA and protein were assessed with qRT-PCR and Western blot, respectively, while miqRT-PCR was used to determine expression levels of miR-146a-3p. Cell proliferation and migration were assessed in human microvascular endothelial cell line (HMEC-1), following overexpression of either ITGA9 or miR-146a-3p, or co-transfection with miR-146a-3p-mimic and pLVX - ITGA9. Cell viability was detected by Cell Counting Kit-8 assay and 5-ethynyl-2′-deoxyuridine (EdU) cell proliferation assay. Cell apoptosis was assessed, using annexin V-FITC/PI apoptosis detection kit, while cell migration was detected by wound healing and transwell assay.
Results: Expression levels of ITGA9 were significantly decreased in psoriatic HDMECs compared to normal controls. Moreover, expression levels of miR-146a-3p were higher in psoriatic HDMECs than in normal controls. Overexpression of miR-146a-3p lowered expression levels of ITGA9, accompanied by increased proliferation and migration of HMEC-1 in vitro. In contrast, overexpression of ITGA9 inhibited proliferation and migration of HMEC-1, while increasing expression levels of cdc42, ki67, focal adhesion kinase (FAK), c-Src tyrosine kinase (Src), RAC1 and RhoA.
Conclusion: ITGA9 can repress the proliferation and migration of HMEC-1, suggesting utility of ITGA9 as a potential therapeutic intervention for psoriasis.
Keywords: ITGA9, miR-146a-3p, human dermal microvascular endothelial cells, psoriasis, proliferation, migration
Introduction
Psoriasis, a chronic immunoinflammatory skin disease, is characterized by epidermal hyperplasia, microvascular dilation and excessive angiogenesis, and affects approximately 2% of the global population.1 Altered endothelial cell function has been linked to the increased proliferation and migration of endothelial cells, and angiogenesis and vascular permeability in psoriasis.2 Our previous study showed that alterations in psoriatic human dermal microvascular endothelial cell (HDMECs) functions, including proliferation, migration, and angiogenesis, are associated with microvascular dilation in psoriasis.3
Recent studies have demonstrated alterations in the expression, localization, and function of several integrins in keratinocytes from psoriatic patients.4,5 A significant increase in endothelial integrin expression was also observed in dermal microvasculature of psoriatic lesion.6,7 Integrins are a family of cell surface heterodimeric transmembrane molecules composed of α and β subunits8,9 that transmit signals from the extracellular matrix to the interior of cells, regulating cell biological functions such as adhesion, proliferation, apoptosis, migration, and differentiation.10,11 Our previous studies showed that psoriatic dermal mesenchymal stem cells stimulate the adhesion and migration of human umbilical vein endothelial cells via upregulating the expression levels of integrins αvβ3 and α5β1.12 Basic fibroblast growth factor could also increase the expression of integrin αvβ3 in human dermal microvascular endothelial cells.13 Integrin subunit alpha 9 (ITGA9) regulates various signaling pathways involved in the tumor development by directly or indirectly affecting cell growth, adhesion, infiltration, proliferation, and migration through binding various extracellular matrix components.14–16
MicroRNAs (miRNAs or miRs) represent a class of evolutionarily conserved short noncoding RNA with about 20–25 nucleotides, which can partially bind to mRNA target genes and posttranscriptionally regulate gene expression.17,18 These miRNAs play important roles in pathogenesis of chronic inflammatory skin diseases, including psoriasis.19 Although their pathogenic mechanisms in psoriasis are not fully understood, the inhibitory effects are considered as a promising therapeutic strategy for psoriasis.20 Although studies have indicated the pathogenic role of miR-146a in psoriasis, the role of miR-146a in regulating psoriatic dermal microvascular endothelial cell (HDMECs) remains unexplored.
In this study, we determined the regulatory role of ITGA9 and miR-146a-3p in HDMECs function in vitro. Our results showed that ITGA9 inhibited HMEC-1 cell proliferation and migration, accompanied with downregulation of cdc42, ki67, focal adhesion kinase (FAK), c-Src tyrosine kinase (Src), RAC1 and RhoA expression. On contrary, miR-146a-3p stimulated HMEC-1 cell proliferation and migration. These results demonstrate the crucial role of ITGA9 in the pathogenesis of psoriasis.
Materials and Methods
Subjects
Six Han Chinese volunteers with psoriasis vulgaris (Table 1) and six healthy volunteers without a history of psoriasis were enrolled from Taiyuan Central Hospital. PASI=PASI(head)+PASI(upper limb)+PASI(trunk)+PASI(lower limbs). All subjects provided informed consent, and had not used immunosuppressants, corticosteroids, and retinoids three months prior to sampling. This study was approved by the Ethics Committee of Taiyuan Central Hospital, and was conducted in accordance with the principles expressed in the Declaration of Helsinki.
 |
Table 1 Information of Volunteers with Psoriasis Vulgaris |
Cell Culture
Skin samples were collected from foreskin of healthy subjects and lesional skin of psoriatic patients. After sequentially washing with Phosphate Buffer Saline (PBS), and a mixture of 100 U/mL penicillin and 100 μg/mL streptomycin, skin was cut into 2–5 mm3, and digested with 0.25% Dispase II (Gibco, USA) at 4°C for 12–16 h, followed by separation of the dermis and the epidermis. The dermis was incubated with 4 mg/mL Collagenase Type I (Sigma, USA) for 1–2 h at 37°C. The undigested tissue was removed by a 70 μm nylon filter, and the filtrate was centrifuged at 300 × g for 5 min. Afterwards, the cell pellet was re-suspended in endothelial cell growth medium (EGM) (Endothelial Cell Basal Medium (EBM-2, Lonza, Switzerland) supplemented with Microvascular Endothelial Cell Growth Medium SingleQuots (EGM-2 MV, Lonza, Switzerland)), and seeded into a T-25 culture flask for culturing at 37°C in an incubator with 5% CO2 and >95% humidity. The EGM was changed every 3–4 days. Human dermal microvascular endothelial cells (HDMECs) were then purified by CD31 MicroBead Kit (Miltenyi, Germany), using MACS Separator (Miltenyi, Germany). The adherent HDMECs were digested with 0.25% trypsin and passaged at 90% confluence. Cell morphology was observed under an inverted phase contrast microscope. HDMECs at passage 3 were used for all experiments. Human microvascular endothelial cell line (HMEC-1) was purchased from American Type Culture Collection (ATCC, USA), and cultured in MCDB131 medium (Thermo Fisher, USA) supplemented with 10% fetal bovine serum, 10 ng/mL EGF (Thermo Fisher, USA), 1ug/mL hydrocortisone (Sigma, USA), 10mM glutamine (ATCC 30–2214, USA) at 37 °C in a humidified incubator with 5% CO2.
Transfection
Both the miR-146a-3p-mimic-negative control (miR-146a-3p-mimic-NC) and miR-146a-3p-mimic were from RIBOBIO (Guangzhou, China), and were used to transfect human microvascular endothelial cell line (HMEC-1) with lipofectamine 3000 reagent (Invitrogen, Carlsbad, CA, USA) at a dose of 50nM. ITGA9 control plasmid (pLVX-vector) and ITGA9 overexpressing plasmid (pLVX-ITGA9) were purchased from PPL (Shanghai, China), and were used to transfect HMEC-1 with lipofectamine 3000 reagent at a dose of 2.2ug/mL. miR-146a-3p-mimic and ITGA9 overexpressing plasmid (pLVX-ITGA9) were co-transfected into HMEC-1 with lipofectamine 3000 reagent at doses of 50nM and 2.2ug/mL, respectively. The expression levels of miR-146a-3p and ITGA9 were analyzed to identify the transfection efficiency. The medium was changed 6 hours after transfection, and further experiments were conducted three days after transfection.
miRNA Isolation and Quantitative Real-Time Polymerase Chain Reaction (miqRT-PCR)
Total miRNA was extracted and purified from HDMECs and transfected HMEC-1, using miRNeasy Mini Kit (Qiagen, Germany), and reverse transcribed by miScript II RT Kit (Qiagen, Germany). Specifically, 1 ug of total miRNA, 4 ul 5*miScript HiFlex Buffer, 2 ul 10*Nucleics Mix, 2 ul miScript Reverse Transcriptase Mix and variable RNase-free dH2O were mixed in a total volume of 20 ul. The reaction was performed at 37°C for 60 min and terminated by heating at 95°C for 5 min. U6 (HmiRQP9001) and miR-146a-3p primers (HmiRQP0195) were purchased from GeneCopoeia (Guangzhou, China). miqRT-PCR reactions were performed using miScript SYBR Green PCR Kit (Qiagen) in an Applied BiosystemsTM StepOneTM Real-Time PCR Systems (Thermo Fisher, USA). U6 was used as internal controls.
RNA Isolation and Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
Total RNA was extracted from HDMECs and transfected HMEC-1 with the TRIzol reagent (Invitrogen, USA), and first-strand of cDNA was synthesized by reverse transcription. All qRT-PCR reactions were performed using SYBR Green Mix (Takara, Japan) in an Applied BiosystemsTM StepOneTM Real-Time PCR Systems (Thermo Fisher, USA). β-actin was used as internal controls. Primer sequences designed by primer 5.0 are shown in Table 2.
 |
Table 2 Primer Information |
Western Blot
Total proteins were extracted from HDMECs and transfected HMEC-1 with RIPA lysis buffer (Solarbio, China), containing protease inhibitors, and protein concentrations were determined by the BCA kit (Solarbio, China). Protein lysates were carried out with ProteinSimple Wes (ProteinSimple, USA), an automatic protein expression quantitative analysis system for ultramicro samples, according to the manufacturer’s protocol. The following primary antibodies were used: rabbit anti-human ITGA9 (1:100, Abcam, UK) and rabbit anti-human β-actin (1:100, Abcam, UK). HRP-conjugated secondary antibody was goat anti-rabbit IgG (ProteinSimple, USA). β-actin was used as an internal standard, and expression levels of protein were normalized to β-actin.
Cell Counting Kit-8 Assay
HMEC-1 cell proliferation was assessed, using Cell Counting Kit-8 (CCK-8) (Boster, China) assay, following the manufacturer’s instructions. Briefly, 3×103 cells containing 100 µL of MCDB131 complete medium were seeded into 96-well plate, while the wells without cells served as the blank control. Cells in each well were incubated daily with 10 µL of CCK-8 solution for 2 h at 37 °C for 4 consecutive days. Afterwards, the absorbance was read at a wavelength of 450 nm, using a microplate reader. Absorbance intensity is positively correlated with the number of survival cells.
5-Ethynyl-2′-Deoxyuridine (EdU) Cell Proliferation Assay
HMEC-1 cell proliferation was detected with the EdU Cell Proliferation Assay Kit (RiboBio, Guangzhou, China). After incubation in 50 mM EdU for 2 hours, cells were fixed in 4% paraformaldehyde and stained with Apollo 567. Cell nuclei were stained with Hoechst 33342 (5 mg/mL) for 30 minutes, and EdU-positive cells were counted in five random fields under a laser scanning confocal microscopy (Olympus, Japan).
Cell Apoptosis Assay
Annexin V-FITC/PI apoptosis detection kit (KeyGEN BioTECH, China) was used to detect the percentage of apoptotic cells. HMEC-1 cells were digested by the addition of 0.25% trypsin without EDTA, and neutralized by 10% fetal bovine serum. Following centrifugation at 800 × g for 5 min and washing twice with cold phosphate-buffered saline (PBS), cells were resuspended in 500 μL 1 × binding buffer, and incubated with 5 μL annexin V-FITC and 5 μL propidium iodide (PI) for 15 minutes in dark. Flow cytometry (Miltenyi, Germany) was used to determine apoptosis of HMEC-1 cells.
Wound Healing Assay
HMEC-1 cells were seeded in a 6-well plate with marker on the outside of the bottom used as reference points during image acquisition, and cultured in MCDB131 complete medium until complete confluence. The culture medium was then removed, and a scratch wound was performed using a 200-µL pipette tip. The exfoliated cells were gently washed off with PBS. HDMECs were cultured in MCDB131 medium supplemented with 1% FBS. During wound healing, images in the same area were recorded by an inverted phase contrast microscope (Olympus, Japan) at the 0h and 24h of incubation. The wound closures were measured using ImageJ software.
Transwell Assay
Migration assay was performed in 8-µm Transwell with polycarbonate membrane inserts (Corning, USA). A total of 600 µL MCDB131 complete medium was added into the bottom compartment, while transfected HMEC-1 were loaded into the upper chamber at a density of 5×104 cells per chamber in MCDB131 medium supplemented with 1% fetal bovine serum (FBS), and allowed to migrate for 24 h at 37 °C. Afterwards, non-migrating cells on the upper side of the insert were removed with a cotton swab, and the migrated cells on the lower part were fixed with 4% paraformaldehyde for 20 min and stained with 0.5% crystal violet (Solarbio, China) for 25 min. The number of migrated cells was determined and averaged by counting 5 random fields under an inverted phase contrast microscope (Olympus, Japan).
Statistics Analysis
Data were analyzed using the SPSS 17.0 software (SPSS Inc, USA) and OriginPro 8.5 (OriginLab, Northampton, MA, USA) and expressed as the mean ± SEM. The significances between two groups were determined using an unpaired Student’s t-test. P < 0.05 was considered statistically significant.
Results
ITGA9 is Downregulated in Psoriatic HDMECs
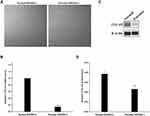
After 14-day culture, both normal HDMECs and psoriatic HDMECs arranged in a “paving stone” pattern and appeared the endothelium-like morphology under an inverted phase contrast microscope (Figure 1A). Moreover, the expression levels of both ITGA9 mRNA and protein were significantly lower in psoriatic HDMECs than in the normal HDMECs (P < 0.001 for mRNA, P < 0.01 for protein) (Figure 1B–D).
Overexpression of ITGA9 Inhibits HMEC-1 Proliferation and Migration
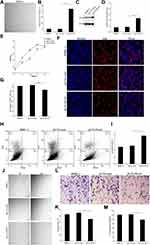
We next determined the functional significance of decreased ITGA9 expression in psoriasis. We assessed HMEC-1 proliferation, migration and apoptosis following cultured with or without overexpression of ITGA9. As seen in Figure 2, human microvascular endothelial cell line (HMEC-1) appeared the endothelium-like morphology under an inverted phase contrast microscope (Figure 2A). Transfection of HMEC-1 with pLVX-ITGA9 markedly increased expression levels of both ITGA9 mRNA and protein (Figure 2B–D, P < 0.001), indicating successful transfection of ITGA9 into HMEC-1. Both cell viability assessed by CCK-8 assay (Figure 2E) and percentage of EdU-positive cells (Figure 2F and G), P < 0.01) were significantly decreased over a 96-h period after transfection with ITGA9. In contrast, transfection with pLVX-ITGA9 increased the number of apoptotic cells (Figure 2H and I, p<0.05 vs controls). In parallel, transfection of HMEC-1 with ITGA9 delayed wound closure (Figure 2J and K, P < 0.01 vs controls), and decreased the number of migrated HMEC-1 (Figure 2L and M, P < 0.01 vs controls) at 24 hours. Taken together, these results indicate that ITGA9 inhibits cell proliferation and migration, while increasing cell apoptosis, in human microvascular endothelial cell cultures.
miR-146a-3p Stimulates Cell Proliferation and Migration via Targeting ITGA9 in HMEC-1 Cultures
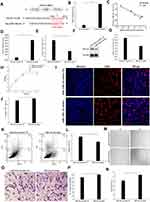
Using the TargetScan software, we predicted that the seed sequence of miR-146a-3p matches the ITGA9 3’-UTR from 3256 to 3263 nucleotides, and identified ITGA9 gene as the potential target of miR-146a-3p (Figure 3A). The expression levels of miR-146a-3p in psoriatic HDMECs were significantly higher than that in the normal HDMECs (Figure 3B, P < 0.001). Correlation analysis showed a negative correlation of miR-146a-3p with ITGA9 expression in HDMECs (Figure 3C, R2=0.9120, P < 0.01), implying that ITGA9 is a potential target of miR-146a-3p. To validate this speculation, expression levels of ITGA9 were measured following transfection of HMEC-1 with either miR-146a-3p-mimic-NC or miR-146a-3p-mimic (Figure 3D, P < 0.001). As expected, transfection of HMEC-1 with miR-146a-3p-mimic decreased expression levels of both ITGA9 mRNA and protein (Figure 3E, P < 0.05; 3F and G, P < 0.001), indicating that ITGA9 is the target of miR-146a-3p in HMEC-1. To determine whether miR-146a-3p-mimic transfection affects proliferation of HMEC-1, we assessed the cell viability and the percentage of EdU-positive cells daily over a 96-h period. Indeed, miR-146a-3p-mimic significantly increased both cell viability (Figure 3H, P < 0.001) and EdU-positive cells (Figure 3I and J, P < 0.05). Moreover, the results of cell apoptosis assay showed that miR-146a-3p-mimic decreased the number of apoptotic cells compared to the controls (Figure 3K and L, P < 0.05). Additionally, miR-146a-3p-mimic increased both wound closure rate (Figure 3M and N, P < 0.01) and the number of migrated HMEC-1 (Figure 3O and P, P < 0.01) 24 hours after transfection. These results demonstrate that miR-146a-3p increases the HMEC-1 proliferation and migration, while decreasing cell apoptosis by targeting ITGA9.
Overexpression of ITGA9 Reverses the Effects of miR-146a-3p on HMEC-1 Proliferation and Migration
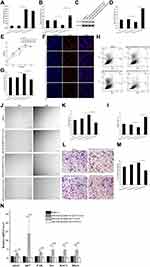
To further confirm that miR-146a-3p targets ITGA9 to regulate HMEC-1 function, HMEC-1 cells were co-transfected with ITGA9 and miR-146a-3p-mimic, followed by assessment of HMEC-1 functions. As shown in Figure 4A, expression levels of miR-146a-3p were significantly higher in HMEC-1 transfected with miR-146a-3p-mimic +pLVX-vector than in that transfected with miR-146a-3p-mimic-NC+pLVX-vector group (P < 0.001). But expression levels of miR-146a-3p were comparable between miR-146a-3p-mimic+pLVX-ITGA9 group and miR-146a-3p-mimic+pLVX-vector group (Figure 4A). Co-transfection with 146a-3p-mimic+pLVX-vector decreased, while co-transfection with miR-146a-3p-mimic+pLVX-ITGA9 increased expression levels of both ITGA9 mRNA and protein (Figure 4B–D). In comparison to co-transfection of HMEC-1 with 146a-3p-mimic+pLVX-vector, co-transfection of HMEC-1 with miR-146a-3p-mimic+pLVX-ITGA9 decreased cell viability (Figure 4E, P < 0.001), EdU-positive cells (Figure 4F and G, P < 0.001) and cell migration (Figure 4J and K, Figure 4L and M, P < 0.001), while increasing cell apoptosis (Figure 4H and I, P < 0.001). Correspondingly, co-transfection of HMEC-1 with miR-146a-3p-mimic+pLVX-ITGA9 lowered expression levels of cell proliferation markers such as cdc42 and ki67 (Figure 4N, P < 0.05), and cell migration markers, including FAK, Src, RAC1 and RhoA (Figure 4N, P < 0.01 for FAK and Src; P < 0.05 for RAC1 and RhoA). These results indicate that overexpression of ITGA9 reverses the effects of miR-146a-3p on HMEC-1 proliferation and migration.
Discussion
Psoriasis is featured by angiogenesis of the superficial skin microvascular system. The capillaries of dermal papilla are dilated, more permeable, and marked elongated. These morphological changes occur before visible epidermal hyperplasia.21 In addition to morphological changes, papillary dermal microvessels in psoriatic lesions exhibit increased expression of inflammation-related adhesion molecules such as E-selectin, intercellular cell adhesion molecule-1 (ICAM-1), and vascular cell adhesion molecule-1 (VCAM-1), along with increases in endothelial cell proliferation, dilation, and capillary permeability.22 A large number of miRNAs are involved in the pathogenesis of psoriasis such as angiogenesis, keratinocyte differentiation, and cutaneous inflammation.23,24 More than 250 miRNAs have been found aberrantly expressed in psoriatic skin.25 Compared to normal skin, hematopoietic-specific miRNAs such as miR-142-3p and miR-223/223*, and angiogenic miRNAs, including miR-21, miR-378, miR-100 and miR-31, are upregulated in psoriatic skin.26 Sonkoly et al reported that miR-21, miR-125b, miR-146a and miR-203 are psoriatic specific miRNAs.27 By comparison of miRNA and mRNA expression profile among involved and noninvolved psoriatic skin, and normal healthy skin, Zibert et al identified 10 upregulated miRNAs in psoriatic skin, including miR-17, miR-20a, miR-21, miR-31, miR-141, miR-142-3p, miR-146b-5p, miR-146a, miR-200a, and miR-203.28 Expression levels of miRNA and mRNA differ between psoriasis-noninvolved and healthy skin. Studies have demonstrated that miR-146a, one of the most highly upregulated miRNAs in psoriatic skin, promotes immune response by negatively regulating nuclear factor kappa-light-chain enhancer of activated B cells (NF-kB) - dependent inflammation by targeting of TNF receptor associated factor 6 (TRAF6) and IL-1 receptor associated kinase 1 (IRAK1).29,30 MiR-146a expression is also increased in psoriatic peripheral blood mononuclear cells, and its expression levels correlate positively with IL-17, an important cytokine in the pathogenesis of psoriasis.31,32
Integrins play important roles in angiogenesis by mediating adhesion to neighboring cells and extracellular matrix, and initiating intracellular signal transduction.33 ITGA9, a member of integrin family, regulates cell proliferation and migration. ITGA9 is highly expressed in the fetal lung and in lung cancer, the invasion and migration of breast cancer cells are significantly inhibited by anti-ITGA9 antibodies and ITGA9-siRNA. The changes in ITGA9 expression affects the interaction between extracellular matrix and tumor cells, and invasion and metastasis of cancer cells.34–36 Elucidating the molecular mechanisms that mediate the proliferation and migration of psoriatic dermal microvascular endothelial cells could contribute to the identification of novel biomarkers and therapeutic targets for psoriasis. Here, we showed that ITGA9 is down-regulated in psoriatic dermal microvascular endothelial cells, leading to increased endothelial cell proliferation and migration, and decreased apoptosis, possibly contributing to the development of psoriasis. We further identified ITGA9 as a target of miR-146a-3p in human microvascular endothelial cell line. Importantly, miR-146a-3p overexpression represses ITGA9 mRNA and protein expression, while increasing the expression of ITGA9 decreases the proliferation and migration of HMEC-1, and expression levels of genes related to cell proliferation (cdc42, ki67) and migration (FAK, Src, RAC1, RhoA). These results suggest that upregulation of ITGA9 expression could alleviate psoriasis.
Conclusions
We determined the regulatory role of ITGA9 and miR-146a-3p in HDMEC function in vitro. Our results showed that ITGA9 inhibited HMEC-1 cell proliferation and migration. On contrary, miR-146a-3p stimulated HMEC-1 cell proliferation and migration. These results demonstrate the role of ITGA9 as a therapeutic intervention for psoriasis.
Abbreviations
HDMECs, Human dermal microvascular endothelial cells; HMEC-1, Human microvascular endothelial cell line; ITGA9, Integrin subunit alpha 9; FAK, Focal adhesion kinase; Src, c-Src tyrosine kinase; RAC1, Rac family small GTPase 1; RhoA, ras homolog family member A; EGM, Endothelial cell growth medium; PBS, Phosphate Buffer Saline; MACS, Magnetic Activated Cell Sorting; CCK-8, Cell Counting Kit-8; EDU, 5-Ethynyl-2’-deoxyuridine; qRT-PCR, Quantitative real-time reverse transcription PCR; FBS, Fetal bovine serum; EGF, Epidermal growth factor; NF-kB, Nuclear factor kappa-light-chain enhancer of activated B cells; TRAF6, TNF receptor associated factor 6; IRAK1, IL-1 receptor associated kinase 1; ICAM-1, Intercellular cell adhesion molecule-1; VCAM-1, Vascular cell adhesion molecule-1; IL-17, Interleukin-17.
Data Sharing Statement
All data generated or used during the study appear in the submitted article.
Ethical Statement
The study was approved by the institutional ethics committee of Taiyuan City Central Hospital. The study was conducted according to the Declaration of Helsinki principles.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (grant no. 81472888, 81371736 and 81803146). The funding was attributed to the design of the study and the collection, analysis, and interpretation of data.
Disclosure
The authors have no conflict of interest to declare.
References
1. Lowes MA, Bowcock AM, Krueger JG. Pathogenesis and therapy of psoriasis. Nature. 2007;445(7130):866–873. doi:10.1038/nature05663
2. Bhushan M, Mclaughlin B, Weiss JB, Griffiths CE. Levels of endothelial cell stimulating angiogenesis factor and vascular endothelial growth factor are elevated in psoriasis. Br J Dermatol. 1999;141(6):1054–1060. doi:10.1046/j.1365-2133.1999.03205.x
3. Li J, Hou H, Zhou L, et al. Increased angiogenesis and migration of dermal microvascular endothelial cells from patients with psoriasis. Exp Dermatol. 2021;30(7):973–981. doi:10.1111/exd.14329
4. Romero MR, Carroll JM, Watt FM. Analysis of cultured keratinocytes from a transgenic mouse model of psoriasis: effects of suprabasal integrin expression on keratinocyte adhesion, proliferation and terminal differentiation. Exp Dermatol. 1999;8(1):53–67. doi:10.1111/j.1600-0625.1999.tb00348.x
5. Teige I, Backlund A, Svensson L, Kvist PH, Petersen TK, Kemp K. Induced keratinocyte hyper-proliferation in alpha2beta1 integrin transgenic mice results in systemic immune cell activation. Int Immunopharmacol. 2010;10(1):107–114. doi:10.1016/j.intimp.2009.10.004
6. Creamer D, Allen M, Sousa A, Poston R, Barker J. Altered vascular endothelium integrin expression in psoriasis. Am J Pathol. 1995;147(6):1661–1667.
7. Gal B, Dulic S, Kiss M, et al. Increased circulating anti-α6-integrin autoantibodies in psoriasis and psoriatic arthritis but not in rheumatoid arthritis. J Dermatol. 2017;44(4):370–374. doi:10.1111/1346-8138.13667
8. Sun G, Guillon E, Holley SA. Integrin intra-heterodimer affinity inversely correlates with integrin activatability. Cell Rep. 2021;35(10):109230. doi:10.1016/j.celrep.2021.109230
9. Bhatwadekar AD, Kansara V, Luo Q, Ciulla T. Anti-Integrin therapy for retinovascular diseases. Expert Opin Investig Drugs. 2020;29(9):935–945. doi:10.1080/13543784.2020.1795639
10. Hynes RO. A reevaluation of integrins as regulators of angiogenesis. Nat Med. 2002;8(9):918–921. doi:10.1038/nm0902-918
11. Shimaoka M, Springer TA. Therapeutic antagonists and conformational regulation of integrin function. Nat Rev Drug Discov. 2003;2(9):703–716. doi:10.1038/nrd1174
12. Han QX, Niu XP, Hou RX, et al. Dermal mesenchymal stem cells promoted adhesion and migration of endothelial cells by integrin in psoriasis. Cell Biol Int. 2021;45(2):358–367. doi:10.1002/cbin.11492
13. Sepp NT, Li LJ, Lee KH, et al. Basic fibroblast growth factor increases expression of the alpha v beta 3 integrin complex on human microvascular endothelial cells. J Invest Dermatol. 1994;103(3):295–299. doi:10.1111/1523-1747.ep12394617
14. Khan Z, Marshall JF. The role of integrins in TGFβ activation in the tumour stroma. Cell Tissue Res. 2016;365(3):657–673. doi:10.1007/s00441-016-2474-y
15. Zheng W, Jiang C, Li R. Integrin and gene network analysis reveals that ITGA5 and ITGB1 are prognostic in non-small-cell lung cancer. Onco Targets Ther. 2016;9:2317–2327. doi:10.2147/OTT.S91796
16. Mostovich LA, Prudnikova TY, Kondratov AG, et al. Integrin alpha9 (ITGA9) expression and epigenetic silencing in human breast tumors. Cell Adh Migr. 2011;5(5):395–401. doi:10.4161/cam.5.5.17949
17. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi:10.1016/S0092-8674(04)00045-5
18. Farh KH, Grimson A, Jan C, et al. The widespread impact of mammalian microRNAs on mRNA repression and evolution. Science. 2005;310(5755):1817–1821. doi:10.1126/science.1121158
19. Lovendorf MB, Skov L. miRNAs in inflammatory skin diseases and their clinical implications. Expert Rev Clin Immunol. 2015;11(4):467–477. doi:10.1586/1744666X.2015.1020301
20. Moles R. MicroRNAs-based therapy: a novel and promising strategy for cancer treatment. Microrna. 2017;6(2):102–109. doi:10.2174/2211536606666170710183039
21. Archid R, Patzelt A, Asschenfeldt BL, et al. Confocal laser-scanning microscopy of capillaries in normal and psoriatic skin. J Biomed Opt. 2012;17(10):101511. doi:10.1117/1.JBO.17.10.101511
22. Boer OD, Wakelkamp IM, Pals ST, Claessen N, Bos JD, Daset PK. Increased expression of adhesion receptors in both lesional and non-lesional psoriatic skin. Arch Dermatol Res. 1994;286(6):304–311. doi:10.1007/BF00402220
23. Timis TL, Orasan R. Understanding psoriasis: role of miRNAs. Biomed Rep. 2018;9(5):367–374. doi:10.3892/br.2018.1146
24. Sileno S, Beji S, Agostino M, Carassiti A, Melillo G, Magenta A. microRNAs involved in psoriasis and cardiovascular diseases. Vasc Biol. 2021;3(1):49–68. doi:10.1530/VB-21-0007
25. Hawkes JE, Nguyen GH, Fujita M, et al. microRNAs in Psoriasis. J Invest Dermatol. 2016;136(2):365–371. doi:10.1038/JID.2015.409
26. Joyce CE, Zhou X, Xia J, et al. Deep sequencing of small RNAs from human skin reveals major alterations in the psoriasis miRNAome. Hum Mol Genet. 2011;20(20):4025–4040. doi:10.1093/hmg/ddr331
27. Sonkoly E, Wei T, Janson PC, et al. MicroRNAs: novel regulators involved in the pathogenesis of psoriasis? PLoS One. 2007;2(7):610. doi:10.1371/journal.pone.0000610
28. Zibert JR, Lovendorf MB, Litman T, Olsen J, Kaczkowski B, Skov L. MicroRNAs and potential target interactions in psoriasis. J Dermatol Sci. 2010;58(3):177–185. doi:10.1016/j.jdermsci.2010.03.004
29. Xia P, Fang X, Zhang ZH, et al. Dysregulation of miRNA146a versus IRAK1 induces IL-17 persistence in the psoriatic skin lesions. Immunol Lett. 2012;148(2):151–162. doi:10.1016/j.imlet.2012.09.004
30. Meisgen F, Landen NX, Wang A, et al. MiR-146a negatively regulates TLR2-induced inflammatory responses in keratinocytes. J Invest Dermatol. 2014;134(7):1931–1940. doi:10.1038/jid.2014.89
31. Lovendorf MB, Mitsui H, Zibert JR, et al. Laser capture microdissection followed by next-generation sequencing identifies disease-related microRNAs in psoriatic skin that reflect systemic microRNA changes in psoriasis. Exp Dermatol. 2015;24(3):187–193. doi:10.1111/exd.12604
32. O’Connell RM, Rao DS, Baltimore D. microRNA regulation of inflammatory responses. Annu Rev Immunol. 2012;30:295–312. doi:10.1146/annurev-immunol-020711-075013
33. Susini BG, Varner JA. Roles of integrins in tumor angiogenesis and lymphangiogenesis. Lymphat Res Biol. 2008;6(3–4):155–163. doi:10.1089/lrb.2008.1011
34. Zhang YL, Xin X, Cai LB, et al. Integrin α9 suppresses hepatocellular carcinoma metastasis by Rho GTPase signaling. J Immunol Res. 2018;2018:1–11.
35. Yang Y, Enis D, Zheng H, et al. Cell adhesion mediated by VCAM-ITGα9 interactions enables lymphatic development. Arterioscler Thromb Vasc Biol. 2015;35(5):1179–1189. doi:10.1161/ATVBAHA.114.304997
36. Bazigou E, Xie S, Chen C, et al. Integrin-alpha 9 is required for fibronectin matrix assembly during lymphatic valve morphogenesis. Dev Cell. 2009;17(2):175–186. doi:10.1016/j.devcel.2009.06.017
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.