Back to Journals » Drug Design, Development and Therapy » Volume 10
Isolation and characterization of cyclo-(tryptophanyl-prolyl) and chloramphenicol from Streptomyces sp. SUK 25 with antimethicillin-resistant Staphylococcus aureus activity
Authors Alshaibani M , Jalil J, M.Sidik N, Edrada-Ebel R, Zin NM
Received 24 November 2015
Accepted for publication 22 January 2016
Published 31 May 2016 Volume 2016:10 Pages 1817—1827
DOI https://doi.org/10.2147/DDDT.S101212
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Prof. Dr. Wei Duan
Muhanna M Alshaibani,1 Juriyati Jalil,2 Nik M Sidik,3 Ruangelie Edrada-Ebel,4 Noraziah M Zin1
1Programme of Biomedical Science, School of Diagnostic and Applied Health Sciences, Faculty of Health Sciences, 2Drug and Herbal Research Centre, Faculty of Pharmacy, Universiti Kebangsaan Malaysia, Kuala Lumpur, 3School of Bioscience and Biotechnology, Faculty of Science and Technology, Universiti Kebangsaan Malaysia, Bangi, Malaysia; 4Strathclyde Institute of Pharmacy and Biomedical Sciences, University of Strathclyde, Glasgow, Scotland
Background: Zingiber spectabile, commonly known as Beehive Ginger, is used as an ethnobotanical plant in many countries as an appetizer or to treat stomachache, toothache, muscle sprain, and as a cure for swelling, sores and cuts. This is the first report of isolation of Streptomyces strain from the root of this plant. Strain Universiti Kebangsaan 25 (SUK 25) has a very high activity to produce secondary metabolites against methicillin-resistant Staphylococcus aureus (MRSA), which is associated with high morbidity and mortality rates due to acquired multidrug resistance genes and causes medication failure in some clinical cases worldwide. Phylogenetic analysis based on the 16S ribosomal RNA gene sequence exhibited that the most closely related strain was Streptomyces omiyaensis NBRC 13449T (99.0% similarity).
Aim: This study was conducted to carry out the extraction, identification, and biological evaluation of active metabolites isolated from SUK 25 against three MRSA strains, namely, MRSA ATCC 43300, MRSA ATCC 33591, and MRSA ATCC 49476.
Materials and methods: The production of secondary metabolites by this strain was optimized through Thronton’s media. Isolation, purification, and identification of the bioactive compounds were carried out using reversed-phase high-performance liquid chromatography, high-resolution mass spectrometry, Fourier transform infrared, and one-dimensional and two-dimensional nuclear magnetic resonance.
Results: During screening procedure, SUK 25 exhibited good antimicrobial potential against several strains of MRSA. The best biological activity was shown from fraction number VII and its subfractions F2 and F3 with minimum inhibitory concentration values at 16 µg/mL and 8 µg/mL, respectively. These two subfractions were identified as diketopiperazine cyclo-(tryptophanyl-prolyl) and chloramphenicol.
Conclusion: On the basis of obtained results, SUK 25 isolated from Z. spectabile can be regarded as a new valuable source to produce secondary metabolites against bacteria, especially MRSA.
Keywords: Zingiber spectabile, MRSA, SUK 25, cyclo-(tryptophanyl-prolyl), chloramphenicol, HR-MS, NMR
Introduction
Natural products are naturally derived metabolites and/or by-products from plants, animals, or microorganisms.1,2 Natural products, in particular from microbes, are important sources of novel compounds for pharmaceuticals. Approximately 300,000 plant species growing in unexplored area on the earth are host to one or more endophytes.3,4 One of these plants is Zingiber spectabile (common name Beehive Ginger), a member of the ginger family, which is used as an ethnobotanical plant in many countries, especially in South-East Asia. Ginger is used as a remedy ingredient to treat a wide range of diseases including cancer5 and arthritis,6,7 and it is also used as an antioxidant. It is used as an appetizer or to treat stomachache, toothache, muscle sprain, and as a cure for swelling, sores and cuts.8
The Streptomyces species is a Gram-positive, aerobic, and spore-forming bacteria belonging to the order Actinomycetales that belongs to the phylum Actinobacteria.9 It has a high DNA G-C content around 52%–70%.10–13 It is well known for its ability to produce secondary metabolites with diverse chemical structures and different activity against numerous pathogenic microorganisms. Several studies have reported that ~23,000 biologically active secondary metabolite compounds were produced by microorganisms, among which >10,000 of these compounds were produced by actinobacteria. This represents 45% of all bioactive microbial metabolites discovered. Streptomyces species alone produces around two-thirds of these compounds.14,15
Endophytic Streptomyces sp. Strain Universiti Kebangsaan (SUK 25) was isolated from the root of Z. spectabile plant.16 It appeared to have a good inhibitory activity against methicillin-resistant Staphylococcus aureus (MRSA).17 This bacterium is the major cause of hospital-associated infections (HAI-MRSA), and the resistance is the result of a supplemental penicillin-binding protein (PBP2a) encoded by the chromosomal mecA gene.18,19 The development of pathogenic bacteria resistance against antibiotic has become a serious problem worldwide. From this view, attempts have been made to extract, identify, and evaluate the active metabolites from SUK 25 against MRSA, and two compounds, namely, cyclo-(tryptophanyl-prolyl), which is a type of cyclic dipeptides diketopiperazines (DKPs), and chloramphenicol (CAP) have been isolated from this strain. Cyclic dipeptides DKPs or 2,5-DKPs, also known as dipeptide anhydrides (DPKs), are relatively simple compounds. Therefore, they are among the most common peptide derivatives found in nature.20 DPKs have been known since the beginning of the 20th century. They are found endogenously in many organisms and in large amounts in some foods and beverages.21 DPKs are extensively gained by extraction from natural sources such as plants, animals, and some microorganisms or by means of synthetic methods.22 CAP (previously called chloromycetin) is an antibiotic belonging to the family of nitroaromatic compounds; it was reported as a broad-spectrum antibiotic produced by Streptomyces species.23 The aim of this study was to screen the antagonistic activity of Streptomyces SUK 25 isolated from the root of Z. spectabile against multidrug-resistant pathogen, MRSA.
Materials and methods
Plant specimen and extract preparation
This study carried out the isolation, identification, and purification of active secondary metabolites from SUK 25, which were isolated from the root of the plant Z. spectabile. Root samples of Z. spectabile were collected in 2009 from Bukit Panchor Country Park, Penang Island, Northern Peninsular Malaysia (5.14°N latitude and 100.54°E). This plant was selected based on its ethnobotanical property. Specimen voucher number was given, and the plant sample was submitted to Universiti Kebangsaan Malaysia Herbarium collection. The pure colonies of endophytic actinomycetes obtained were subcultures onto International Streptomyces Project 2 (ISP2) agar and incubated at 28°C.16 The production of secondary metabolites by SUK 25 was optimized through Thronton’s media with some modification.17,24
Chemicals and media
All the chemicals used for extraction, column chromatography (CC), high-performance liquid chromatography (HPLC), and liquid chromatography–mass spectrometry (LC–MS) were HPLC grade purchased from EMD Millipore, Billerica, MA, USA; thin-layer chromatography (TLC) was performed on percolated plates (20×20 cm) silica gel 60-F254 purchased from EMD Millipore; and Sephadex LH20 was purchased from GE Healthcare Bio-Sciences AB, Uppsala, Sweden. Mueller Hinton Broth (MHB) and Mueller Hinton Agar (MHA) were purchased from Difco, France.
In vitro antibacterial activity
Primary screening (cross-streak plate method)
The antibacterial activity was carried out by cross-streak plate method as described by Williston et al25 and also by Alexander.26 A single streak line of the SUK 25 was made on nutrient agar and then incubated at 28°C for 7 days. The cultures of MRSA ATCC 43300, ATCC 33591, and ATCC 49476 were standardized to 0.5 McFarland standards using sterile MHB, corresponding to the reading of 0.08–0.1 at A625 and then streaked at right angles to the original streak of SUK 25 and incubated at 37°C. The diameter of inhibition zone (IZ, mm) was measured after 24 hours. The control plate was also maintained without inoculating SUK 25.
Secondary screening (disk diffusion assay method)
The crude extract from SUK 25 was subjected to secondary screening using disk diffusion assay method, in accordance with the Clinical and Laboratory Standards Institute (CLSI) guidelines. MRSA ATCC 43300 was inoculated into 10 mL MHB and incubated at 37°C for 2–6 hours. Turbidity was standardized to 0.5 McFarland (CLSI, 2011). The crude extract (1 mg/mL) of SUK 25 was dissolved in 10% methanol, and 30 μg/disk was loaded on a blank disk (6 mm diameter; Whatman™, Gred AA; Sigma–Aldrich Co., St Louis, MO, USA) and then dried in the hood. After that, the disk was placed on the MHA already lawned with MRSA. This culture was incubated overnight at 37°C. After overnight culture, the IZ in millimeters was measured for each plate, and vancomycin (30 μg/disk) (Thermo Fisher Scientific, Waltham, MA, USA) was used as a positive control.
Fermentation, production, and extraction of secondary metabolites
For isolation and identification of bioactive metabolites, the Streptomyces sp. SUK 25 was cultured on ISP2 agar media, followed by incubation at 28°C for 14 days. Then a few blocks of ISP2 media containing pure and good growth of SUK 25 were then transferred into 250 mL of modified Thronton’s medium is composed of (g/L) K2HPO4 1.0, KNO3 0.5, MgSO4·2H20 0.2, CaCl2·H2O 0.1, NaCl 0.1, FeCl3 0.01, asparagine 0.5, and glucose 1.5 instead of mannitol at pH 7.4.24 Then it was incubated at 28°C for 3 days with a gentle shake at 140 rpm using an orbital shaker. An aliquot of 60 mL of spore suspension of SUK 25 was aseptically transferred into 600 mL of Thronton’s broth as seeded media. After 12 days of incubation at 28°C, fermentation filtrates were extracted with three half-volumes of ethyl acetate (EtOAC), and the crude extracts were separated and dried using rotary evaporator at 40°C and 240 mbar.27
Phylogenetic tree full of 16S rDNA gene sequencing
DNA extraction
Genomic DNA was isolated according to Kieser et al,28 and the purity and concentration were determined using NanoDrop spectrophotometer. Polymerase chain reaction (PCR) was performed according to Coombs and Franco29 using two sets of gene primer, 27f (5′-AGAGTTTGATCMTGGCTCAG-3′) with 765r (5′-CTGTTTGCTCCCCACGCTTTC-3′) and 704f (5′-CTGGCGGTGAAATGCGTAGA-3′) with 1492r (5′-AAGGAGGTGWTCCARCC-3′), and the PCR product was sent to 1st BASE for sequencing. Sequencing data were analyzed using software BioEdit Version 7 and compared to the GenBank databases (BLAST) and EzTaxon Biocloud. Phylogenetic tree was built using the software Molecular Evolutionary Genetics Analysis (MEGA4) using the neighbor-joining (NJ) method.30
Chemotaxonomy analysis of ll-diaminopimelic acid cell wall of SUK 25
Chemotaxonomy features were analyzed through a type of ll-diaminopimelic acid (DAP) cell wall as described by Staneck and Roberts.31 Fifteen milligrams of mycelia dry weight of SUK 25 grown in ISP2 for 14 days was added to 1 mL of 6N hydrochloric acid, and vortexed and autoclaved for 15 minutes at 121°C. After cooling, the hydrolysate was filtered using a filter paper (Whatman no 1) before being taken to dryness in a heating block at 95°C. Then, 1 mL of dH2O was added, and these procedures were repeated for two times. The filtrate was evaporated to dryness in an oven overnight. After evaporation, 1 μL of hydrolysate and 0.01 M DAP standard (Sigma-Aldrich Co.) in 0.2 M NaOH, which contained both the meso- and L isomers, was spotted on cellulose aluminum plate (EMD Millipore). Descending chromatography was performed in the solvent system containing methanol:dH2O:6N HCl:piridin (80:26:4:10), respectively, sprayed with ninhydrin (0.2% in acetone), and heated at 100°C for 3 minutes to reveal the spot.
Separation and purification of antimicrobial compounds by CC and TLC
Fractionations and purifications of the dry crude extract were carried out using reversed-phase open CC and TLC. A 700 mg dry crude extract was dissolved in 10 mL methanol. This mixture was then subjected to separation through size-exclusion CC containing 150 mg Sephadex LH20, using column size 2×40 cm. Gradient elution started with 100% chloroform, and the polarity of methanol was increased until 100% (100:0, 95:5, 90:10, 85:15, 80:20, 70:30, 60:40, 50:50, and 0:100 v:v). Further purification using silica gel column was carried out using column size 2×40 cm. After CC, the fractions were applied to TLC plate and developed using mixture of hexane, ethyl acetate, and methanol (4:4:2 v:v) as mobile phase. The separated compounds were visualized under short ultraviolet (UV) at λ254 nm (absorbance) and long wavelength UV at λ365 nm (fluorescence), and then sprayed with 10% H2SO4 in ethanol and heated at 80°C–100°C for 10 minutes. The compounds that had similar Rf in TLC were mixed together.
Bioautography on disk diffusion
Disk diffusion assay method for all fractions that were purified from open CC was carried out in order to test the activity of each fraction separately against MRSA ATCC 43300.
Determination of the minimum inhibitory concentration and minimum bactericidal concentration
The minimum inhibitory concentrations (MICs) of the compounds were determined according to the method described by the CLSI.32,33 Twofold serial dilutions of each fraction dissolved in 10% MeOH were made with MHB in a 96-well microtiter plate to give concentrations ranging from 0.12 μg/mL to 1,000 μg/mL. Fifty microliters of test bacterial suspension was inoculated in each well to give a final concentration of 1×105 CFU/mL. Bacteria inoculum in MHB was used as a positive control, whereas the tested compounds in MHB were used as a negative control. Determination of minimum bactericidal concentrations (MBCs) was performed by inoculating 20 μL of each dilution with apparent no bacterial growth in MIC wells and streak on MHA plates. Then, the plates were incubated for 24 hours at 37°C. The presence of colonies was considered an evidence of bacteriostatic action, while the absence of colonies indicated bactericidal activity.
Purification of compounds using reversed-phase high-performance liquid chromatography
The crude extract of each fraction isolated from open CC was analyzed using an Agilent 1200 HPLC system equipped with a C-18 column (4.6×250 mm, 5 μm). The mobile phase consisted of H2O:Acetonitrile (ACN) with 0.1% trifluoroacetic acid (TFA) added to both solvents, and a gradient elution step was applied as shown in Table S1. The flow rate was 1 mL/min with an injection volume of 30 μL from 1 mg/mL, and the elution was monitored by the UV absorption at 210 nm, 240 nm, and 360 nm. The preparative HPLC protocol was carried out as same as analytical HPLC, only the quantity of injection and flow rate was injected to C-18 separation column (size 22×150 mm, 5 μm), where the flow rate was 10 mL/min with an injection volume of 100 μL each time from 7 mg/mL. Fractions were collected from a fraction collector.
Structure determination of antimicrobial compounds
FT-IR spectrophotometer, optical rotation, and melting point
Fourier transform infrared (FT-IR) spectrophotometer (Spectrum one FTIR spectrometer, PerkinElmer Inc., Waltham, MA, USA) using potassium bromide (KBr) disk method was carried out to determine the absorption bands of compounds. Optical rotation was measured on an Autopol VI automatic polarimeter (Rudolph Research analysis, USA). Melting point was measured on Stuart (Bibby Scientific, UK).
Nuclear magnetic resonance
The structure of compounds was determined using nuclear magnetic resonance (NMR) spectroscopy FT–NMR spectroscopy 600 MHz with Cryoprobe AVANCE III (Berker, USA). The probe temperature was maintained at 298 K, and a standard 5 mm NMR tube was used. Four milligrams of purified solid compound was dissolved in 200 μL MeOH-d4 (99.8 atom% D; Sigma-Aldrich Co.). Experiments that included one-dimensional (1D) 1H, 13C and two-dimensional (2D) NMR as sequences used in structural elucidation involving correlation spectroscopy (COSY), nuclear overhauser effect spectroscopy, and heteronuclear multiple-bond correlation spectroscopy (HMBC) were carried out to Berker standard pulse sequences. 1H and 13C chemical shifts (δ) are expressed in parts per minute (ppm) relative to solvent peak (MeOH-d4: 1H δ 5.18 ppm and 13C δ 47.7 ppm).
High-resolution mass liquid chromatography–mass spectrometry (HR-MS)
HR-MS was performed on an Agilent 6410 triple quadrupole mass spectrometer (Agilent Technologies, Santa Clara, CA, USA) equipped with an electrospray ionization interface negative mode coupled to an Agilent 1200 HPLC (Agilent Technologies). Column Agilent Zorbax Eclipse Plus (150×2.1 mm, 5 μm) HR-MS (Agilent Technologies) was used with column temperature 30°C, flow rate was 0.3 mL/min, injection volume was 20 μL, and mobile phase: pH 8.5 H2O (A) and acetonitrile (B). The retention time (RT) was measured in minutes. The flow rate was as shown in Table S1.
Statistical analysis
Statistical analysis of the data was performed using SPSS. One-way analysis of variance complemented by Tukey’s post hoc test was performed, and all data were reported as the mean ± standard deviation of the mean. All values were considered significant at P<0.05.
Results
The present work describes a comprehensive methodology for the screening, purification, and characterization of bioactive secondary metabolites isolated from endophytic Streptomyces SUK 25.
Screening for antibacterial activity
Primary screening
Primary screening of SUK 25 was carried out to determine the potential of this strain to production of antimicrobial secondary metabolites against MRSA. SUK 25 showed good activity against MRSA ATCC 43300, MRSA ATCC 33591, and MRSA ATCC 49476 as a primary screening. The distance between the edge of the tested bacterial growth and the SUK 25 bacterial growth was measured and recorded in millimeters. The IZ was 12 mm from the streaked right angle to the original streak of SUK 25 as shown in Figure S1.
Secondary screening by disk diffusion assays
SUK 25 was subjected to secondary screening using a sterile disk 6 mm in diameter; 30 μg/disk of either compounds isolated from SUK 25 or vancomycin was used as a positive control. The plates were incubated at 37°C overnight. The mean diameters of triplicate samples of IZ were 20±1.2 mm and 18±1.3 mm for crude extract and vancomycin, respectively. The effect of this fraction was larger than vancomycin as shown in Figure S2.
Bioautography
The results of bioautography using the disk diffusion method of purified fractions from CC against MRSA are listed in Table 1. Thirteen fractions were isolated. Only fraction numbers I, II, and VII using chloroform (CHCl3) as an eluting solvent revealed activity against MRSA ATCC 43300 with a diameter of IZ at 14 mm, 15 mm, and 22 mm, respectively. However, only fraction numbers IX and X showed activity when MeOH was used as an elution solvent; the diameter of IZ was at 13 mm and 14 mm, respectively, as shown in Table 1 and Figure S3.
Determination of the MIC and MBC
MIC is defined as the lowest concentration of an antimicrobial compound that inhibited the visible growth of a sensitive stain after the appropriate incubation period. MICs of all fractions are presented in Table 1. MICs of fraction numbers VII, F2, F3, and vancomycin against MRSA ATCC 43300 were 8 μg/mL, 16 μg/mL, 8 μg/mL and 2 μg/mL, respectively.
MBC was recorded as the lowest extract concentration, killing 99.9% of the bacterial inoculum after 24 hours incubation at 37°C. The MBCs of all active fractions were in the range of 32–128 μg/mL. MBCs of fraction numbers VII, F2, and F3, standard CAP, and vancomycin were 64 μg/mL, 32 μg/mL, 64 μg/mL, 64 μg/mL, and 4 μg/mL, respectively. The MICs and MBCs of all active fractions are shown in Table 1. From the bioautography, MICs, MBCs, and preparative HPLC results, fraction number VII (including subfractions F2 and F3) was selected for further analysis, where these subfractions exhibited strong anti-MRSA biological activity.
Phylogenetic tree of 16S rDNA sequences of SUK 25
Genomic DNA was extracted, and the 16S ribosomal RNA (rRNA) gene sequence was amplified by PCR using universal bacterial 16S rRNA gene primers as described.29 SUK 25 isolate sequences were compared with isolates in GenBank with Blast N software through the website www.ncbi.nlm.nih.gov. Sequences were aligned and edited using Bioedit software Version 7. Phylogenetic analysis was performed using MEGA 4.0 software to generate phylogenetic trees. The results showed that SUK 25 (1,450 nucleotides) is closely related to Streptomyces omiyaensis NBRC 13449T with 99% similarities (Figure 1).
Chemotaxonomy analysis of DAP cell wall of SUK 25
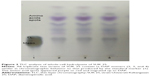
For TLC analysis, all the triplicate samples of SUK 25 strains were found to support the identification they obtained in the DAP isomer, chromatographically similar to the standard from Sigma-Aldrich Co. The DAP spots were seen as gray-green fading to yellow, with the L-DAP isomer moving ahead of the meso isomer with hydrolysates; amino acid spots appeared purple or red and migrated ahead of the DAP spot31 as shown in Figure 2.
Chemical characterization of isolated compounds
Analytical and preparative HPLC, melting point, optical rotations, and HR-MS
Different separation techniques such as TLC, CC, analytical and reversed-phase high-performance liquid chromatography (RP-HPLC), melting point, optical rotations, FT-IR, and HR-MS were carried out to obtain and identify the fraction FVII, which yielded two subfractions, namely, compound (F2) and compound (F3). HPLC purification peak is shown in Figures S3 and S4.
Compound F2: cyclo-(L-tryptophanyl-L-prolyl) (3S, 8aR)-3-(1H-indol-3ylmethyl) hexahydropyrrolo [1, 2-a] pyrazine-1, 4-dione
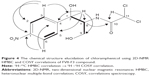
Analytical HPLC of this fraction showed RT at 11.64 minutes with a UV absorbing band at 254 nm, by using RP-HPLC method as shown in Figure S1. The F2 was isolated as a colorless solid, gave a dark red coloration with anisaldehyde/sulfuric acid, and became pink with Ehrlich’s reagent and blue with chlorine/o-anisidine as indication of a peptide with Rf =0.47 (CHCl3:MeOH; 95:5, v/v). The melting point of this fraction was between 165°C–168°C. The optical rotations of this compound [α]25 C (0.001 g) was (−46.819°) using methanol as a solvent. The m/z value of this compound was 283.1321 [M+] at RT 6.63 minutes. The mass spectrum confirmed the structure based on the proposed fragmentation mechanism pathway. The molecular formula is C16H17N3O2. The 1H and 13C signals were assigned on the basis of chemical shifts, spin–spin coupling constants, splitting patterns, and signal intensities. The 1H NMR spectrum of F2 showed a broad 1H singlet at δ 8.20 characteristic for an NH of an indole moiety, five aromatic protons at δ 7.59, 7.40, 7.24, 7.14 (m, 2H) confirming the indole skeleton substituted at 3-position. In the aliphatic region, another broad 1H singlet of an acidic proton at δ 5.75 due to NH of an amide, two oxygenated or aminated methines at δ 4.39 (dd) and 4.08 (t), three different resonances each of 1H, 2H, and 1H, respectively, at δ 3.76 (dd), 3.64 (m), and 2.97 (dd) were seen. In addition, three multiplets of 4H between δ 2.36 and 1.90 were representative for two methylene groups. The 13C NMR spectrum exhibited four signals in the region between 22.48 and 45.33 ppm attributed to four CH2. The rest of the spectrum displayed 12 signals, among them seven were CH (123.4–110.0) and five were quaternary carbons (169.5–111.6), two of them belong to amide carbonyls (169.5 and 165.7) as shown in Figure S3. The prediction data from NMR suggested that this subfractionation is similar with the NMR data of cyclo-(L-tryptophanyl-L-prolyl), also known as brevianamide F.34 1H and 13C NMR data are shown in Table 2 and Figure 3. A search in AntiBase revealed that F2 was known as L,L-cyclo-(tryptophanyl-prolyl) or cyclo-(L-Trp-L-Pro), which was further confirmed by comparison with the previous literature by Kobayashi and Palumbo35 and Amar et al.36
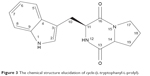  | Figure 3 The chemical structure elucidation of cyclo-(L-tryptophanyl-L-prolyl). |
Compound F3: CAP 2,2-dichloro-N-[(1R, 2R)-2-hydroxy-1-(hydroxymethyl)-2-(4-nitrophenyl) ethyl]-acetamide
Analytical HPLC of this fraction showed RT at 14.3 minutes with UV absorbing band at 273 nm as shown in Figures S1 and S2. By using RP-HPLC, the F3 was isolated as a grayish-white or yellowish-white crystalline powder, gave a dark red coloration with anisaldehyde/sulfuric acid, and became pink with Ehrlich’s reagent and blue with chlorine/o-anisidine as indication of a peptide with Rf =0.75 (CHCl3:MeOH; 95:5, v/v). The melting point of this fraction was between 150°C and 152°C. The optical rotation of this compound [α]25 C (0.001 g) was +31.493°, whereas the optical rotation of standard CAP [α]25 C (1 g) was +17.496° using ethyl acetate. F3 and standard CAP were characterized by HR-MS spectra using negative ionization mode due to the better response in relation to the positive ionization mode. The m/z value of this compound was 322 [M-H]– at RT 11.9 minutes. The mass spectrum confirmed the structure based on the proposed fragmentation mechanism pathway. The molecular formula is C11H12Cl2N2O5 and was supported by 1H and 13C NMR data as shown in Table 3 and Figures S4–S8. The 1H and 13C signals were assigned on the basis of chemical shifts, spin–spin coupling constants, splitting patterns, and signal intensities and by using 1H–1H COSY, 1H–13C heteronuclear single quantum coherence (HSQC), and 1H–13C HMBC experiments. The 1H and 13C chemical shifts of compound FVII-F3 are given in Table 3 and Figure 4. Chemical shifts were calibrated internally against the residual signal of the solvent in which the sample was dissolved in MeOH-d4 with δ H at 5.18 and C at 47.7. 1H NMR (600 MHz, MeOH-d4) of FVII-F3 showed 12 protons, at 8.3 (2H, m, aromatic-H), 7.67 (2H, m, aromatic-H), 6.3 (1H, s), 5.18 (1H, s), 4.16 (1H, s), and 3.62–3.8 (2H, dd, J=6.0 Hz; 12.0 Hz). The 13C and HSQC spectra show eleven carbon signal peaks at 165.2, 148.6, 126.9, 126, 150.2, 69.9, 57.10, and 60.80 (six aromatic carbons). The 2D 1H–1H and 1H–13C experiments, and especially the long-range 1H–13C couplings observed in the HSQC, COSY, and HMBC spectrum as shown in Figures S9–S11, permitted the connectivity between all the groups of the molecule to be established.
Fourier transform infrared spectroscopy
The FT-IR spectrum for the two compounds exhibited absorption bands, which indicated the presence of NH, aromatic C–H stretching, carbonyl group, and C=O groups in the structure, respectively, at Vmax 3,345 cm−1 and 3,257 cm−1 (NH2 group and O–H stretching), 2,901 cm−1 (aromatic C–H stretching), 1,684 cm−1 (carbonyl group), 1,343 cm−1 (C–H bending), 652 cm−1 (carbon-chlorine bonds), 1,092 and 1,121 (C–N stretch), and 972 cm−1 (H–C–H – asymmetric stretch) as shown in Table S2 and Figures S12 and S13.
Discussion
The aim of this study was to screen the antagonistic activity of Streptomyces SUK 25 isolated from roots of Z. spectabile against drug-resistant pathogen, MRSA. The primary screening results indicated that SUK 25 has the ability to produce secondary metabolites in nutrient agar, which diffused through the agar that prevented and inhibited the growth of three MRSA strains, namely, MRSA ATCC 43300, MRSA ATCC 33591, and MRSA ATCC 49476. The IZ was ~12 mm from the edge of growth of SUK 25. A secondary screening using disk diffusion method had 20 mm IZ against MRSA ATCC 43300, which is similar with previous study conducted by Morakchi et al.37 Cho et al38 also reported the antibacterial activity of Streptomyces sp. CS392 against MRSA and vancomycin-resistant enterococci. Marine Streptomyces sp. VITBRK2 also has the ability to produce active secondary metabolites against MRSA and vancomycin-resistant enterococci strains.39 Furthermore, strain Streptomyces MUSC 135T, which was isolated from a soil sample collected from a mangrove forest located on the east coast of Peninsular Malaysia, exhibited a broad-spectrum bacitracin against MRSA ATCC BAA-44.40 In addition, Streptomyces sp. HUST012 was isolated from the stems of the medicinal plant Dracaena cochinchinensis Lour. This strain produced secondary metabolites, namely, SPE-B11.8 and SPE-B5.4, which showed antimicrobial activity against MRSA ATCC 25923 with MIC 62.5 and 0.04, respectively.41
The results of the analysis translated through phylogenetic tree NJ method of 16S rRNA sequences of SUK 25. (1,450 nucleotides) was closely related to several strains of Streptomyces sp. from culture collections in the database NCBI GenBank. Analysis of 16S rRNA showed a similarity level of 99% within S. omiyaensis NBRC 13449T, which was isolated from Omiya city in Japan and showed a potential activity to produce CAP.42 Chemotaxonomic analyses showed that the cell wall of strain SUK 25 contained DAP, indicating that it was of cell-wall type I.43 Production of secondary metabolites by SUK 25 for large-scale fermentation was optimized through Thronton’s media at 12 days incubation with pH 7 and aerated at 140 rpm as reported by Junaidah et al44 with some modification on incubation time from 7 days to 12 days according to the optimization of growth curve of SUK 25. From the results of bioautography, there were five active fractions against MRSA ATCC 43300. The most active fraction was the fraction number VII, which had a significant IZ of 22 mm by disk diffusion assay compared to only 18 mm of vancomycin at the same concentration of 30 μg/disk. Furthermore, two active compounds F2 and F3 were isolated from fraction number VII. Separation, purification, and isolation of fraction number VII and its subfractions in pure form were carried out using TLC, open CC, analytical and RP-HPLC, HR-MS, and FT-IR. Chemical structure elucidation was carried out by 1D and 2D NMR to confirm the structure of compounds. The first compound (F2) was isolated as cyclo-(L-tryptophanyl-L-prolyl), also known as brevianamide F, with m/z 283.1321 [M+] and molecular formula C16H17N3O2. It is a form of DKPs tryptophan–proline derivative. It has been described, isolated, and structurally characterized for the first time from Penicillium brevicompactum.34 This compound has been reported by Mahmoud45 and Amar et al.36 It has insecticidal properties46 and has been reported to possess nematocidal activity.47 Furthermore, it has anti-inflammatory properties,48 and the antibacterial activity of this compound has been recorded against Escherichia coli, Pseudomonas aeruginosa, Klebsiella pneumoniae, S. aureus, Bacillus subtilis, and Streptococcus pneumonia.49,50 It also has antimicrobial effect against Micrococcus luteus LB 14110 and S. aureus ATCC 6538 with IZ 14 mm.51 In this study, the IZ was 15 mm against MRSA ATCC 43300. It also found that this compound has antifouling potential activity and showed potent activity against larval settlement of Bugula neritina.52
The second compound (F3) was identified as CAP with m/z 322 and molecular formula C11H12Cl2N2O5, which was reported by some of the previous studies by Pfenning et al53 and Gantverg et al.54 Previously, CAP was obtained from Streptomyces venezuelae by Ehrlich et al,55 which was the first research that reported the isolation of the new CAP from a soil sample from Caracas, Venezuela. It was then isolated and identified by other researchers such as Smith et al56 and Umezawa et al.57 At the same time, in 1948, Okami reported ~14 strains of Streptomyces pheochromogenus var chloromyceticus that have the ability to produce this antibiotic. In addition, S. Omiyaensis, which was isolated from Omiya city, showed a potential activity to produce this antibiotic.42 Furthermore, Streptosporangium viridogriseum var kofuense isolated from the soil has the ability to produce CAP.58 The findings from this study were also similar to other previous studies by Mosher et al59 and Aouiche et al,60 where they isolated and identified CAP from Streptomyces lividans M252 and Saccharothrix sp. PAL54, respectively. This study compared the data from F3 with standard commercial CAP from Sigma-Aldrich Co. using FT-IR, HR-LC–MS, and NMR data and showed that all data were compatible for both of them. These findings confirmed that this fraction was similar to commercial CAP as shown in Figures S3–S6. This antibiotic has been designated as a broad-spectrum antibiotic effective against Gram-positive and Gram-negative bacteria.61 Its mode of action was reported as a potent protein inhibitor, which is inhibiting bacterial growth by binding to their 50S ribosomal subunit.62 However, several previous researchers found that this antibiotic has some side effects, such as aplastic anemia or reproductive/hepatotoxic effects, in human and animals.63,64 Therefore, the European Union has not authorized the use of this antibiotic in food-producing animals to protect public and animal health.65 Despite its side effects, it is still been used in the treatment of serious infections, such as meningitis, brain abscesses, severe Haemophilus influenzae infections, and typhoid fever.66 This is because it has excellent cellular penetration, and it is extensively distributed in the body.67 In 1948, Woodward et al evaluated the effectiveness of CAP as a treatment for typhoid and paratyphoid fevers. All the patients in their trials responded well to this antibiotic, where the mortality rate of typhoid fever has been reduced from 20% to <1%, and the duration of fever decreased from 2–4 weeks to 4–5 days. It has provided the gold-standard against which other antibiotics have been compared at that time. The MICs of this antibiotic against Salmonella typhi were between 0.75 μg/mL and 5 μg/mL.68
The MIC and MBC values of F2 were equal to 16 μg/mL and 32 μg/mL, which were compatible with the previous study by Kumar et al.50 In addition, the MIC and MBC values of F3 were equal to 8 μg/mL and 64 μg/mL, respectively, which were compatible with the previous study by Fayyaz et al.69 In relation to the ratio of MBCs to MICs of the compounds, it was <4 when F2 was used, which indicates that this compound has bactericidal effects that kill most of the bacteria,70 whereas this ratio was >4 when F3 was used, which indicates bacteriostatic effects of this compound that only inhibit the growth of bacterial cells but do not kill completely.71–73
Conclusion
This study identified two natural bioactive compounds, cyclo-(L-tryptophanyl-L-prolyl) and CAP, from SUK 25 that were potent against MRSA. This indicates that this bacterium is a potential source for bioactive natural products in the pharmacological industry against MRSA.
Acknowledgments
The researchers are grateful to the Ministry of Higher Education (MOHE) for financial support through the research grant code FRGS/1/2011/ST/UKM/02/1. The authors would like to thank Professor Dr Jean Frederic Faizal Weber Abdullah, Atta-ur-Rahman Institute (UiTM), Centre for Research & Instrumentation (CRIM), Faculty of Dentistry (Universiti Kebangsaan Malaysia), and Siti Junaidah Binti Ahmad for analytical assistance.
Author contributions
Muhanna M Alshaibani, Juriyati Jalil, and Noraziah M Zin designed and performed the whole experiment, analyzed the data, and contributed to manuscript preparation. Nik M Sidik and Ruangelie Edrada-Ebel assisted the experimental work and performed data acquisition. All authors contributed toward designed the experiment, data analysis, drafting and critically revising the paper and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
Ji HF, Li XJ, Zhang HY. Natural products and drug discovery. EMBO Rep. 2009;10(3):194–200. | ||
Pan L, Chai H-B, Kinghorn AD. Discovery of new anticancer agents from higher plants. Front Biosci. 2012;4:142. | ||
Tayung K, Sarkar M, Baruah P. Endophytic fungi occurring in Ipomoea carnea tissues and their antimicrobial potentials. Braz Arch Biol Technol. 2012;55(5):653–660. | ||
Strobel G, Daisy B. Bioprospecting for microbial endophytes and their natural products. Microbiol Mol Biol Rev. 2003;67(4):491–502. | ||
Shukla Y, Singh M. Cancer preventive properties of ginger: a brief review. Food Chem Toxicol. 2007;45(5):683–690. | ||
El-Ghorab AH, Nauman M, Anjum FM, Hussain S, Nadeem M. A comparative study on chemical composition and antioxidant activity of ginger (Zingiber officinale) and cumin (Cuminum cyminum). J Agric Food Chem. 2010;58(14):8231–8237. | ||
Kundu JK, Na H-K, Surh Y-J. Ginger-derived phenolic substances with cancer preventive and therapeutic potential. Forum Nutr. 2009;61:182–192. | ||
Khalid MH, Akhtar MN, Mohamad AS, et al. Antinociceptive effect of the essential oil of Zingiber zerumbet in mice: possible mechanisms. J Ethnopharmacol. 2011;137(1):345–351. | ||
Lechevalier H, Lechevalier MP. Introduction to the order Actinomycetales. In: Starr MP, Stolp H, Trüper HG, Balows A, Schlegel HG, editors. The Prokaryotes. Vol. 2. Germany: Springer-Verlag Berlin; 1981:1915–1922. | ||
McCormick JR, Flärdh K. Signals and regulators that govern Streptomyces development. FEMS Microbiol Rev. 2012;36(1):206–231. | ||
Williams S, Goodfellow M, Alderson G, Wellington E, Sneath P, Sackin M. Numerical classification of Streptomyces and related genera. J Gen Microbiol. 1983;129(6):1743–1813. | ||
Gao B, Gupta RS. Phylogenetic framework and molecular signatures for the main clades of the phylum Actinobacteria. Microbiol Mol Biol Rev. 2012;76(1):66–112. | ||
Tanaka Y, Omura S. Agroactive compounds of microbial origin. Annu Rev Microbiol. 1993;47:57–87. | ||
Berdy J. Bioactive microbial metabolites. J Antibiot. 2005;58(1):1–26. | ||
Weber T, Welzel K, Pelzer S, Vente A, Wohlleben W. Exploiting the genetic potential of polyketide producing streptomycetes. J Biotechnol. 2003;106(2):221–232. | ||
Zin N, Loi C, Sarmin N, Rosli A. Cultivation-dependent characterization of endophytic actinomycetes. Res J Microbiol. 2010;5(8):717–724. | ||
Junaidah AS, Suhaini S, Sidek HM, Basri DF, Zin NM. Anti-methicillin resistant Staphylococcus aureus activity and optimal culture condition of Streptomyces sp. SUK 25. Jundishapur J Microbiol. 2015;8(5):e16784. | ||
David MZ, Daum RS. Community-associated methicillin-resistant Staphylococcus aureus: epidemiology and clinical consequences of an emerging epidemic. Clin Microbiol Rev. 2010;23(3):616–687. | ||
Stapleton PD, Taylor PW. Methicillin resistance in Staphylococcus aureus: mechanisms and modulation. Sci Prog. 2002;85(pt 1):57. | ||
Prasad C. Bioactive cyclic dipeptides. Peptides. 1995;16(1):151–164. | ||
Park YC, Gunasekera SP, Lopez JV, McCarthy PJ, Wright AE. Metabolites from the marine-derived fungus Chromocleista sp. isolated from a deep-water sediment sample collected in the Gulf of Mexico. J Nat Prod. 2006;69(4):580–584. | ||
Martins MB, Carvalho I. Diketopiperazines: biological activity and synthesis. Tetrahedron. 2007;63(40):9923–9932. | ||
Ahmed Z, Vining L. Evidence for a chromosomal location of the genes coding for chloramphenicol production in Streptomyces venezuelae. J Bacteriol. 1983;154(1):239–244. | ||
Thakur D, Bora T, Bordoloi G, Mazumdar S. Influence of nutrition and culturing conditions for optimum growth and antimicrobial metabolite production by Streptomyces sp. 201. J Med Mycol. 2009;19(3):161–167. | ||
Williston EH, Zia-Walrath P, Youmans GP. Plate methods for testing antibiotic activity of actinomycetes against virulent human type tubercle bacilli. J Bacteriol. 1947;54(5):563–568. | ||
Alexander M. Introduction to Soil Microbiology. Hoboken, NJ: John Wiley & Sons; 1977. | ||
Zin NM, Sarmin NI, Ghadin N, et al. Bioactive endophytic streptomycetes from the Malay Peninsula. FEMS Microbiol Lett. 2007;274(1):83–88. | ||
Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. Practical Streptomyces Genetics. Norwich: John Innes Foundation; 2000. | ||
Coombs JT, Franco CMM. Isolation and identification of actinobacteria from surface-sterilized wheat roots. Appl Environ Microbiol. 2003;69(9):5603–5608. | ||
Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4(4):406–425. | ||
Staneck JL, Roberts GD. Simplified approach to identification of aerobic actinomycetes by thin-layer chromatography. Appl Microbiol. 1974;28(2):226–231. | ||
CLSI (Clinical and Laboratory Standards Institute). Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically. Approved Standard. M07-A8. 8th ed. Wayne, PA: CLSI; 2011. | ||
Magiorakos AP, Srinivasan A, Carey R, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18(3):268–281. | ||
Birch A, Wright J. The brevianamides: a new class of fungal alkaloid. J Chem Soc D. 1969;12:644b–645b. | ||
Kobayashi D, Palumbo J. Bacterial endophytes and their effects on plants and uses in agriculture. In: Bacon CW, White JF, editors. Microbial Endophytes. New York, NY: Marcel Dekker, Inc.; 2000:199–233. | ||
Amar MB, Elleuch L, Abd-Alla HI, et al. The new Streptomyces sp. TN605 strain secretes simultaneously three active compounds and a high level of the interesting pharmaceutical industry intermediate: 2-hydroxyphenylacetic acid. Bull Environ Pharmacol Life Sci. 2012;1:48–56. | ||
Morakchi H, Ayari A, Taok M, Kirane D, Cochet N. Characterization of Streptomyces strain SLO-105 isolated from Lake Oubeira sediments in North-East of Algeria. Afr J Biotechnol. 2009;8(22):6332–6336. | ||
Cho SS, Choi YH, Simkhada JR, Mander P, Park da J, Yoo JC. A newly isolated Streptomyces sp. CS392 producing three antimicrobial compounds. Bioprocess Biosyst Eng. 2012;35(1–2):247–254. | ||
Rajan BM, Kannabiran K. Extraction and identification of antibacterial secondary metabolites from marine Streptomyces sp. VITBRK2. Int J Mol Cell Med. 2014;3(3):130. | ||
Lee L-H, Zainal N, Azman A-S, et al. Streptomyces pluripotens sp. nov., a bacteriocin-producing streptomycete that inhibits meticillin-resistant Staphylococcus aureus. Int J Syst Evol Microbiol. 2014;64(pt 9):3297–3306. | ||
Khieu T-N, Liu M-J, Nimaichand S, et al. Characterization and evaluation of antimicrobial and cytotoxic effects of Streptomyces sp. HUST012 isolated from medicinal plant Dracaena cochinchinensis Lour. Front Microbiol. 2015;6:574. | ||
Okami Y. Studies on the characters of antibiotic Streptomyces I. on the characters of a chloromycetin-producing strain (O-163). Jpn Med J. 1948;1(6):499–503. | ||
Lechevalier MP, Lechevalier H. Chemical composition as a criterion in the classification of aerobic actinomycetes. Int J Syst Bacteriol. 1970;20(4):435–443. | ||
Junaidah AS, Lian HH, Basri DF, Zin NM. Mode of action of endophytic Streptomyces sp., SUK 25 extracts against MRSA; microscopic, biochemical and time-kill analysis. Int J Pharm Sci Rev Res. 2014;30(1):11–17. | ||
Mahmoud MAS. Bioactive secondary metabolites from marine and terrestrial bacteria: isoquinolinequinones, bacterial compounds with a novel pharmacophor, PhD Thesis, Göttingen University, 2005. | ||
Gloer JB. The chemistry of fungal antagonism and defense. Can J Botany. 1995;73(S1):1265–1274. | ||
Shiomi K, Omura S. Antiparasitic agents produced by microorganisms. Proc Jpn Acad Ser B. 2004;80(6):245–258. | ||
Rand TG, Giles S, Flemming J, Miller JD, Puniani E. Inflammatory and cytotoxic responses in mouse lungs exposed to purified toxins from building isolated Penicillium brevicompactum Dierckx and P. chrysogenum Thom. Toxicol Sci. 2005;87(1):213–222. | ||
Graz M, Hunt A, Jamie H, Grant G, Milne P. Antimicrobial activity of selected cyclic dipeptides. Pharmazie. 1999;54(10):772–775. | ||
Kumar SN, Mohandas C, Nambisan B. Purification, structural elucidation and bioactivity of tryptophan containing diketopiperazines, from Comamonas testosteroni associated with a rhabditid entomopathogenic nematode against major human-pathogenic bacteria. Peptides. 2014;53:48–58. | ||
Ben Ameur Mehdi R, Shaaban KA, Rebai IK, Smaoui S, Bejar S, Mellouli L. Five naturally bioactive molecules including two rhamnopyranoside derivatives isolated from the Streptomyces sp. strain TN58. Nat Prod Res. 2009;23(12):1095–1107. | ||
Zhang X-Y, Xu X-Y, Peng J, et al. Antifouling potentials of eight deep-sea-derived fungi from the South China Sea. J Ind Microbiol Biotechnol. 2014;41(4):741–748. | ||
Pfenning A, Turnipseed S, Roybal J, et al. Confirmation of chloramphenicol residues in crawfish by electrospray LC/MS. US FDA – Laboratory Information Bulletin No. 2002;4289(18):8. | ||
Gantverg A, Shishani I, Hoffman M. Determination of chloramphenicol in animal tissues and urine: liquid chromatography-tandem mass spectrometry versus gas chromatography-mass spectrometry. Anal Chim Acta. 2003;483(1):125–135. | ||
Ehrlich J, Bartz QR, Smith RM, Joslyn DA, Burkholder PR. Chloromycetin, a new antibiotic from a soil actinomycete. Science. 1947;106(2757):417–417. | ||
Smith RM, Joslyn DA, Gruhzit OM, McLean IW Jr, Penner MA, Ehrlich J. Chloromycetin: biological studies. J Bacteriol. 1948;55(3):425. | ||
Umezawa H, Tazaki T, Okami Y, Fukuyama S. On the new source of chloromycetin, Streptomyces omiyaensis. Jpn Med J. 1949;2(4):207–211. | ||
Tamura A, Takeda I, Naruto S, Yoshimura Y. Chloramphenicol from Streptosporangium viridogriseum var. kofuense. J Antibiot. 1971;24(4):270–270. | ||
Mosher RH, Camp DJ, Yang K, Brown MP, Shaw WV, Vining LC. Inactivation of chloramphenicol by o-phosphorylation a novel resistance mechanism in Streptomyces venezuelae isp5230, a chloramphenicol producer. J Biol Chem. 1995;270(45):27000–27006. | ||
Aouiche A, Sabaou N, Meklat A, et al. Saccharothrix sp. PAL54, a new chloramphenicol-producing strain isolated from a Saharan soil. World J Microbiol Biotechnol. 2012;28(3):943–951. | ||
Seth S. Textbook of pharmacology. Indian J Pharmacol. 1998;30(1):58. | ||
McKee E, Ferguson M, Bentley A, Marks T. Inhibition of mammalian mitochondrial protein synthesis by oxazolidinones. Antimicrob Agents Chemother. 2006;50(6):2042–2049. | ||
Kirk RW, Osborne C, Murtaugh R. Kirk’s Current Veterinary Therapy XII. Small Animal Practice. Philadelphia, PA: WB Saunders Company; 1995. | ||
Plumb D. Veterinary Drug Handbook. 4th ed. White Bear Lake, MN: Pharma Vet Publishing; 1991. | ||
EFSA. Scientific opinion on chloramphenicol in food and feed. EFSA J. 2014;12(11):3907,3145. | ||
Yaffe SJ, Aranda JV. Neonatal and Pediatric Pharmacology: Therapeutic Principles in Practice. Philadelphia, PA: Lippincott Williams & Wilkins; 2010. | ||
White NJ. Salmonella typhi (typhoid fever) and S. paratyphi (paratyphoid fever). Antimicrob Ther Vaccine. 2002. Available from http://www.antimicrobe.org/b106.asp. Accessed December 23, 2015. | ||
Woodward TE, Smadel JE. Management of typhoid fever and its complications. Ann Intern Med. 1964;60(1):144–157. | ||
Fayyaz M, Mirza IA, Ahmed Z, Abbasi SA, Hussain A, Ali S. In vitro susceptibility of chloramphenicol against methicillin-resistant Staphylococcus aureus. J Coll Physicians Surg Pak. 2013;23(9):637–640. | ||
French G. Bactericidal agents in the treatment of MRSA infections – the potential role of daptomycin. J Antimicrob Chemother. 2006;58(6):1107–1117. | ||
Berche P, Gaillard JL, Simonet M. Bactériologie: Bactéries des Infections Humaines. [Nosocomial Infections Caused by Bacteria and Their Prevention in Bacteriology]. Paris: Flammarion Médecine-Sciences; 1988. | ||
Konaté K, Mavoungou JF, Lepengué AN, et al. Antibacterial activity against β-lactamase producing methicillin and ampicillin-resistants Staphylococcus aureus: fractional inhibitory concentration index (FICI) determination. Ann Clin Microbiol Antimicrob. 2012;11(1):18. | ||
Cha JO, Park YK, Lee YS, Chung GT. In vitro biofilm formation and bactericidal activities of methicillin-resistant Staphylococcus aureus clones prevalent in Korea. Diagn Microbiol Infect Dis. 2011;70(1):112–118. |
 © 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.