Back to Journals » OncoTargets and Therapy » Volume 13
Is there a CDKN2A-centric network in pancreatic ductal adenocarcinoma?
Authors Wu C, Yang P, Liu B, Tang Y
Received 25 September 2019
Accepted for publication 19 February 2020
Published 27 March 2020 Volume 2020:13 Pages 2551—2562
DOI https://doi.org/10.2147/OTT.S232464
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr XuYu Yang
Chu Wu, Ping Yang, Bingxue Liu, Yunlian Tang
Cancer Research Institute, Key Laboratory of Tumor Cellular & Molecular Pathology, Medical College of Hengyang, University of South China, Hengyang, Hunan 421001, People’s Republic of China
Correspondence: Yunlian Tang
Cancer Research Institute, Key Laboratory of Tumor Cellular & Molecular Pathology, Medical College of Hengyang, University of South China, Hengyang, Hunan 421001, People’s Republic of China
Tel +86 734-8281075
Fax +86 734-8282901
Email [email protected]
Abstract: Pancreatic cancer has a high mortality rate and its incidence has risen rapidly in recent years. Meanwhile, the diagnosis and treatment of this cancer remain challenging. Pancreatic ductal adenocarcinoma (PDAC) is the most common type of pancreatic cancer, but, currently, no sufficiently effective modalities for its treatment exist. The early diagnosis rate of pancreatic cancer is low and most patients have reached an advanced stage at the time of diagnosis. PDAC evolves from precancerous lesions and is highly aggressive and metastatic. It is essential to understand how the disease progresses and metastasizes. CDKN2A mutations are very common in PDAC. Therefore, here, we have performed a literature review and discuss the role of CDKN2A and some related genes in the development of PDAC, as well as the basis of gene targeting with a correlation coefficient of CDKN2A above 0.9 on the STRING website. It is noteworthy that the interaction of CDKN2A with each gene has been reported in the literature. The role of these genes and CDKN2A in PDAC may provide new directions that will advance the current knowledge base and treatment options since cancer progression is realized through interactions among cells. Our findings provide new insights into the treatment of PADC that can, to some extent, improve the diagnosis rate and quality of life of patients.
Keywords: PDAC, CDKN2A, cell cycle, genes, biomarkers
Pancreatic Ductal Adenocarcinoma (PDAC)
In the industrial age, pancreatic cancer is the fourth most common cause of cancer deaths in the world that is expected to become the second leading cause in the next few years.1,2 The survival prognosis of patients with pancreatic cancer is worse thanother cancer types due to the low rate of early diagnosis, the high invasiveness and metastatic potential, and the resistance to chemotherapy, as well as the absence of effective treatment for refractory pancreatic cancer.3,4
Pancreatic ductal adenocarcinoma (PDAC) is the most common type of pancreatic cancer. Approximately 90% of pancreatic solid tumors are PDAC, which are usually diagnosed in the late stage.5,6 PDAC has one of the worst prognoses among all solid tumors. The median survival time of postoperative patients is 8–12 months and the 5-year survival rate is less than 10%.7,8 The probability of PDAC metastasis to distant organs is high, mainly in the liver, peritoneum, and the lung. PDAC is usually asymptomatic at an early stage and current screening methods fail to achieve the effectiveness and ubiquity of early diagnosis without invasive surgery, and thus early-stage diagnosis and the standard resection of this cancer are critical to the survival and prognosis of patients.9,10 Studies have shown that most PDACs are characterized by continuous genetic changes as a result of long-lasting accumulation including of genes, such as KRAS and TP53, followed by CDKN2A and SMAD4.11 More attention is to be focused on understanding the molecular mechanisms of pancreatic cancer development and progression.12 Therefore, further discovery and identification of predictive biomarkers for therapy are highly necessary for the provision of a rational basis for priority in the treatment of pancreatic cancer patients.13–15
CDKN2A
CDKN2A was discovered in 1993 and was a cyclin-dependent kinase inhibitor (CDKI) consisting of three exons. CDKN2A has an alternative splicing exon (El-β) and is located on some 9p21 regions in the chromosome.
CDKN2A encodes four products: p16INK4a, p14 alternate reading frame (p14ARF, mouse p19ARF), cyclin-dependent kinase4 p15 (p15INK4A) and long-chain non-coding RNA (lncRNA) ANRIL (also known as CDKN2B-AS), products involved in cell cycle regulation, differentiation, senescence and apoptosis.16,17 p16INK4a is one of the important coding products of CDKN2A and an inhibitor of the cyclin-dependent kinase family (CDK), whose amount increases during the aging process of many tissues including islets.18 The lack of CDKN2A isolates CDK4/6 and prevents its binding to D-cyclin, so that the tumor suppressor protein retinoblastoma (RB) binds to the transcription factor E2F and the loss of protein activity results in cell cycle arrest and cell senescence.19 The lack of expression of p16INK4a leads to overexpression of CDK4 and proliferation of B cells, rising insulin secretion and causing pancreatic hyperplasia.20 p16INK4a regulates cyclin D1 expression, and D1/CDK4 is critically involved in cellular metabolism and cell cycle progression, which provides therapeutic potential for inhibiting the progression of pancreatic cancer by cell cycle suppression.21
CDKN2A Mutations
In cell immortalization and subsequent transformation in many types of cancer, CDKN2A has high-frequency loss of heterozygosity (LOH) or mutation.22 CDKN2A involves the function regulation of islets, fat, muscle, liver and immune cells, and even the whole process of uterine development.23 CDKN2A affects the risk of human vascular disease, including coronary artery disease, aneurysm, ischemic stroke, glaucoma, Alzheimer’s disease, endometriosis and periodontitis.16,24 Additionally, CDKN2A mutations are involved in a variety of cancers.25 Evidence exists that CDKN2A mutations are strongly associated with the recurrence of melanoma.26 Significant deletion of CDKN2A can be used as a biological target in a cell line for early identification of human head and neck squamous cell carcinoma (HNSCC).27 In early metaplasia, the methylation of the CDKN2A promoter is a very common event in esophageal adenocarcinoma.28 Dysregulation of CDKN2A in bladder cancer is also frequent.29 Furthermore, the homozygous deletion of CDKN2A is associated with a more aggressive prognosis of mesothelioma.30 The mutations of CDKN2A are also involved in the development of primary breast cancer.31
CDKN2A in PDAC
Hypermethylation of the CDKN2A promoter was confirmed as a marker of CDKN2A inactivation. Moreover, the hypermethylation of CpG islands in the CDKN2A promoter was found to be mediated by DNA methyltransferase, which is a transcriptional mechanism of silencing tumor suppressor genes, thereby promoting cancer metastasis.32 CpG methylation-mediated gene inactivation in the CDKN2A promoter region is one of the determinants of malignant tumor development.33 Transcriptional silencing plays a key role in the development of PDAC, thus CDKN2A is considered a target for this cancer.34 Promoter methylation provides a new insight into better understanding the role of CDKN2A and opens up new directions for improving the diagnosis and treatment of patients with PDAC.
Various defects in DNA repair genes, such as genes involved in DNA repair pathways or cell cycle regulation (such as CDKN2A), may be involved in the development of PDAC.35 CDKN2A mutation occurs in early PDAC, and the regions of significant loss or expansion of CDKN2A can be established using specific calculation methods.35,36 Patients with a CDKN2A mutation in familial atypical multiple melanoma syndrome (also known as pancreatic cancer-melanoma syndrome) have a 20-fold higher risk of pancreatic cancer than individuals without mutations.37 It is noteworthy that patients with a CDKN2A gene deletion had shorter overall survival (P = 0.002), this indicates that CDKN2A mutations can be used as a prognostic marker for PDAC.34
CDKN2A negatively regulates the cell cycle regulator CDK4/6 in the development of PDAC. PDAC has four major driving genes: KRAS, TP53, CDKN2A and SMAD4.38 Chromosome changes in PDACs lead to the loss of tumor suppressor factors, such as CDKN2A, TP53 and SMAD4.38 Multiple gene mutations occur throughout the development of PDAC, which are critical for more effective screening and treating of pancreatic cancer and other invasive tumors. Investigations on the mutations of these genes and how they affect the cell cycle to promote disease progression and metastatic phenotype are thus of substantial significance.
CDK4, CDK6, MYC, TP53 and MDM2 in PDAC
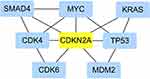
We searched STRING for genes highly correlated with CDKN2A (correlation coefficient more than 0.9 and top 5), including CDK4, CDK6, MYC, MDM2 and TP53 as can be seen in Figure 1.
CDK4
CDK4 is a major regulator of insulin signaling.39 The activity of CDK4 in mouse islets is critical for the proliferation of differentiated B cells, which is the primary mechanism for regulating islet mass to accommodate the steady-state requirements of insulin secretion, and similar situations may apply to the regulation of adult pituitary cell proliferation.40 The absence of CDK4 completely inhibited the development of pituitary and islets, and pituitary defects caused male and female infertility.41 Deletion of CDK4 in islet cell line inhibits Rb hyperphosphorylation, and the CDK4-Rb pathway is critically involved in controlling neuroendocrine cell proliferation.41 CDK4 activation is essential for the early stage of the carcinogenic process of neuroendocrine cell types and a close relationship exists between cancer development and CDK4 activation.
Cell proliferation is dependent on D-cyclin and CDKs, which are key nodes controlling the progression from the G phase to the S phase. Both cyclin D2 and CDK4 proteins are essential for pancreatic cell growth and replication. CDKN2A inhibits CDK4, which may be a key function of PDAC cell cycle regulation.42 Furthermore, CDK4 is involved in hepatic glucose metabolism, and the induction of gluconeogenesis in the liver predetermines the potential for the application of CDK4 inhibitors in the treatment of hyperglycemia in hepatic cancer patients. CDK4 also affects muscle mitochondrial oxidative metabolism, which is essential for maintaining normal physiological functions of the human body.43 CDK4 affects fat cell function and cell proliferation and metabolism. Its expression level can indirectly influence cell aging and the occurrence and development of various diseases. Future research will address the metabolic function of CDK4 in PDAC and its therapeutic targets in metabolic diseases.43 Affecting metabolic pathways, CDK4-targeted therapy can regulate the key mechanisms of various cellular processes and environmental cues during the primary, secondary and circulating cancer cell (CTC) transport, thereby controlling PDAC progression.44 Therefore, CDK4 regulation will have important therapeutic significance for the personalized treatment of pancreatic cancer in the future.
CDK6
CDK6, first reported in 1994 by Meyerson and Harlow, is similar in structure and function to CDK4. It belongs to the same CDKs, which are regulators of D-cyclin kinase and the G1 phase. CDK6 and CDK4 are involved in the regulation of G1 to S cell cycle progression, transcription, differentiation and other biological processes.45 Activation of CDK4/6 requires binding to cyclin D1, D2, or D3, and the phosphorylation of CDK6 regulates Rb activity, exerting control of the cell cycle, and also blocks the transcriptional regulation of differentiation.46 The availability of nutrients initiates downstream signals, after which CDK6 is involved in signaling pathways that stimulate metabolism and cell proliferation, promoting cell cycle progression.47 The transcriptional role of CDK6 is involved in the production of blood vessels in cancers. CDK6 downregulation is associated with the release of Runt-associated transcription factor 1 (Runx1, an important blood-regulating gene, transcription factor required for chromatin opening) in hematopoietic cells.48 Most of the CDK6 is localized in the cytoplasm, which facilitates the obtaining of more comprehensive knowledge on the role of CDK6 in cell cycle control, differentiation and cancer development.
CDK6 is involved in the negative regulation of cell differentiation. Some cancer cells require CDK6 for their proliferation. CDK6 also promotes the growth of cancer cells and their upregulated expression in various cancer types. The overexpression of CDK6 in lymphoma can be used as a specific therapeutic target for malignant lymphoma.49 Additionally, the overexpression of CDK6 in pancreatic cancer promotes the growth and proliferation of pancreatic cancer cells, affects tumor transformation and progression of cell cycle events by participating in the progression of the cell cycle, a process that may be involved in other endocrine cancers.50 The knockout of CDK6 expression in colorectal cancer COLO320 cells significantly inhibited tumor cell growth, indicates that CDK6 inhibitors present potential therapeutic benefits for colorectal cancer patients, this indicates that CDK6 is a key target for anti-tumor therapy.51 Inhibition of CDK6 blocks tumor cell proliferation and suppresses the supply of tumors to blood demand, which provides new insights into CDK6 transcription, this also inhibits cell cycle and terminates the progression of pancreatic cancer.52
The combination of CDK6 and cyclin D3 (D3-CDK6) plays a unique role in glucose metabolism and promotes cancer cell survival, which may become a strong target for cancer treatment.53 Clinically, PDACs are particularly sensitive to drugs that reproduce p16IN4a activity. CDKN2A-encoded p16INK4a protein inhibits the kinase activity of CDK4 and CDK6. Therefore, studying the effects of the CDK4/6 combination will increase the positive influence of drugs on patients with PDAC.47,54,55 Given the wide application and effect of CDK4 and CDK6 inhibitors in clinical trials, the use of D3-CDK6 inhibitors for anticancer treatment is expected to improve the clinical efficacy of anti-CDK4/6 treatment, thus positively affecting the treatment and prognosis of PDAC patients.53
MYC
MYC is a promoter and oncogenes which is involved in the regulation of various cellular functions, including cell growth, differentiation, adhesion, migration, invasion and apoptosis.12 MYC controls metabolic pathways, promotes glucose metabolism, glutamine metabolism, fatty acid synthesis, oxidative phosphorylation, nucleotide synthesis and ribosomal biogenesis.56 The loss of MYC leads to the misuse of fat as a cell energy source and eventually results in mitochondrial dysfunction.57 Many genes involved in the glucose metabolism are target genes of MYC, which promotes glucose metabolism reprogramming of cancer cells through different targets, providing bioenergy to ensure cell growth and metabolism the unlimited proliferation of cells.58,59 MYC-induced cell growth and bioenergy accumulation depend on the production of appropriate ATP in mitochondria.60 MYC controls glycolysis and thus regulates the overall mitochondrial function, increasing the resistance of cells to individual metabolic disorders.61
MYC selectively activates many genes involved in cellular processes, including DNA replication and transcription, translation, chromatin modification and protein synthesis and degradation. Moreover, MYC coordinates the metabolism and promotes cell growth, enhancing RNA extension throughout the genome and interacting with other transcription factors.62–65 MYC exists in open chromatin regions in almost all genomes. The level of amplification entirely depends on the pattern of open chromatin in the gene.66,67 Acute MYC activation causes chromatin changes and promotes gene transcription by inhibiting RNA polymerase promoter pauses.68 Cancer cells have a stronger ability to replicate and proliferate than normal cells.69 In rapidly proliferating cells, especially cancer cells, MYC directly regulates protein synthesis and responds efficiently to growth signals.70 The activation of mutations in MYC induces a normal short-circuit of the cell’s mitotic signal, forming a wound that cannot heal and promoting the development of cancer.36,71 MYC has an indispensable role in cancer and the deletion of MYC inhibited cell proliferation in a human Burkitt’s lymphoma cell line.72 Moreover, MYC is one of the most frequently amplified oncogenes.73 The abnormal expression of MYC in many cancers, including PDAC, is usually not due to dysfunction of the MYC gene itself, but is caused, for example, by amplification, chromosomal translocation, or upstream carcinogenic signaling disorders, as well as by a loss of tumor suppressor factors.74,75 MYC is required for the maturation and maintenance of embryonic and adult acinar differentiation, and the activation of MYC signaling induces cancerous changes in pancreatic tissue.76 MYC overexpression occurs in up to 42% of late PDAC, which is associated with poor clinical outcomes, increased probability of recurrence, worsening disease, and decreased survival in patients with PDAC.36 In addition, MYC was found to protect pancreatic cancer cells from failure and inhibits their differentiation, indicates that MYC is a critical therapeutic target in PDAC.77
MYC is downstream of many growth-promoting signaling pathways, including signaling pathways initiated by growth factor-stimulated receptor tyrosine kinases, T-cell receptors and WNT signaling.12 MYC is an important downstream effector of KRAS in PDAC. KRAS expression disorder is common in PDAC, and MYC may be involved in the early development and progression of this cancer.78,79 Aerobic glycolysis is a metabolic feature of invasive cancer, in which MYC promotes the progression of PDAC.80 The knockout of the MYC gene reduces LDL expression, lactate production, glucose consumption and alternative approaches that focus on interfering with MYC-mediated downstream effectors may provide new therapeutic avenues for PDAC.81 Besides, MYC regulates the ability of related stem cells, blocks cellular senescence and differentiation, and coordinates the changes in the cancer microenvironment, including activation of angiogenesis and inhibition of host immune responses. Given that MYC regulates multiple components of the cellular process, further research is required on the MYC-regulated pathways to ensure normal cell function while preventing the occurrence of ineffective cycles.82
TP53
More than 50% of the malignant cancers have mutations in TP53, and the frequency of these mutations varies considerably, depending on the specific cancer type. TP53 mutation leads to the loss of its tumor suppressor function, contributing to the development of solid and hematopoietic cancer.83 PDAC is often associated with dense interstitial fibrosis, which leads to drug resistance, cancer growth and metastasis. TP53 induces lipid accumulation and plays a key role in fibrosis, exerting anticancer effects through a paracrine mechanism in the cancer microenvironment.84 Evidence exists that TP53 and MDM2 are involved in the AURORA signaling pathway. Hence, affecting the AURORA signaling pathway by influencing TP53 and MDM2 can indirectly lead to changes in the occurrence and progression of PDAC.85 In the past, many signaling pathways, nutrients and natural products interacted with pathways involved in TP53 and drug resistance.86
TP53-positive chronic pancreatitis is considered inefficient in inducing carcinogenesis. On the contrary, in the absence of TP53, the genetic instability affects the latency required for tumor formation, DNA damage, etc., resulting in increased pancreatitis-induced carcinogenesis.87 In an examination, the expression of TP53 in PDAC tissues was significantly lower than that in normal tissues and benign tissues, the overall survival rate of patients lacking TP53 was poor. The low expression of TP53 in people was associated with an increased risk of PDAC.88 Mainly due to DNA amplification of genes in PDAC tissues, a high proportion of oncogenes MDM2, MDM4 and WIP1 are overexpressed in cancer cells. These gene changes attenuate the function of the cancer suppressor TP53, which results in TP53 mutation. These changes also contribute to the development of cancer, indicates that the dysfunction of the TP53 pathway may be an important mechanism for the proliferation and progression of PDAC.89 The deletion of TP53 in mice induces cancer formation and the recovery of its expression level will rapidly resolve the formed carcinoma in situ. This indicates that TP53 has the therapeutic potential to repair cancer tissues and new therapeutic strategies can be developed to combat cancers lacking TP53, which will have a therapeutic impact.90–92
Small-molecule activation of TP53 is an attractive therapeutic strategy for the restoration of the wild-type TP53 function in the treatment of PDAC. Based on these strategies, MDM2 inhibitors had a good beneficial effect on TP53 wild-type cancer in vivo and in vitro.93 Nonetheless, some evidence suggested that TP53 inactivation or dysfunction can directly or indirectly lead to the promotion of tumorigenesis. However, most mutations of TP53 lead to the loss of function, resulting in apoptosis and senescence. Moreover, further validation and functional assessment of these genetic variants, targeting TP53 can also improve the prognosis of patients with PDAC to some extent.88,94
MDM2
MDM2 (mouse double microsome 2) is an oncogene found in a locus amplified on the double microsome of a tumor-bearing mouse cell line (3T3-DM) in 1992. The function of TP53 in cells is mainly regulated by its inhibitor, MDM2. The latter directly binds to the N-terminus of TP53 by regulating the position, stability and activity of TP53, and derives TP53 from the nucleus, continuously promoting the degradation of TP53 transforming it into a short-lived protein.95,96 Cellular stress, such as oncogene activation or DNA damage, disrupts the mutual regulation relationship between MDM2 and TP53, and MDM2 itself is polyubiquitinated and degraded. Meanwhile, the level and activity of TP53 increase, inducing a cellular response to cancer cells, including cell cycle arrest, apoptosis and aging.97,98 The MDM2 gene contains the TP53 promoter, which is regulated by TP53 transcription, TP53 acts as a transcription factor of MDM2, which induces the synthesis of MDM2.98 This regulatory mechanism is subject to a feedback loop in which MDM2 inhibits TP53 activity, which in turn regulates MDM2 transcription levels, resulting in a delicate balance between the amount of TP53 and that of MDM2. The two form a negative feedback loop, that is, the automatic adjustment loop.99
MDM2 is overexpressed in many tumors, including PDAC, usually due to the expansion of chromosome segments, and its amplification is closely related to tumor metastasis. MDM2 ubiquitinates and degrades E-cadherin, which is associated with EMT and tumor grade. This is an important step in the development of metastatic tumors.100 Approximately 50–75% of PDACs carry TP53 mutations, and the MDM2–TP53 pathway in PDAC is frequently affected.101 A mouse model with a cancer-associated single nucleotide polymorphism had high levels of MDM2 expression, decreased TP53 function and increased tumor incidence.102 The overexpression of MDM2 inhibited the transcriptional activity of TP53, elevated the proliferation and invasion of cancer cells, suppressed apoptosis and induced resistance to chemotherapy.103,104 MDM2 was involved in DNA double-strand break (DSB) repair. Delayed the expression of MDM2 in a mouse PDAC model, the cells repair DNA was damaged, caused an arrest of the cell cycle and slowed down the growth of PDAC cells.105 Furthermore, the overexpression of MDM2 in PDAC was associated with diverse clinicopathological features, such as invasiveness, high differentiation, advanced stage, translocation, recurrence and poor prognosis.106 Patients with MDM2 expression had a shorter median (OS) and progression-free survival (PFS) than others. Thus, MDM2 expression is an unfavorable prognostic factor for PDAC.104,107
The TP53 mutation in PDAC occurs in its late stage, which results in a loss of the DNA binding ability and induces transcriptional activation of the gene. There is a TP53 mutation in PDAC, which may invalidate MDM2 inhibitors. TP53 mutations facilitate the escape of damaged cells from cell cycle checkpoints and their transformation into carcinogens with an increased cancer potential for metastasis and invasion to other organs.108,109 To avoid infections and the formation of TP53 wild-type cancers, the treatment design is crucial to be based on a comprehensive understanding of the effects of MDM2 on tumors. Hence, the identification of a new TP53-independent therapeutic agent that inhibits MDM2 is highly necessary.110,111 Different TP53 mutations may have diverse activities and appropriate customized treatments need to be considered along with a timely genetic diagnosis. A better understanding of oncogenes and tumor suppressor genes will lay the foundation for prompt diagnosis and effective treatment of PDAC.
KRAS and SAMD4 in PDAC
KRAS
In many cancers, KRAS is an oncogene.112 The development of pancreatic ductal adenocarcinoma PDAC is a slow process and most often begins with mutations of the oncogene KRAS in pancreatic cells.113,114 About 90% of pancreatic tumors express mutations in KRAS, the earliest genetic changes in most pancreatic cancers.115,116 KRAS overexpression induces up-regulation of EMT signals such as vimentin and inhibition of E-cadherin, driving the occurrence of early PDAC tumors.117,118 Some cases with KRAS mutation and TP53 deletion, early PDAC converted to invasive PDAC.119 KRAS mutations allow cell cycle arrest to enter the aging process and are usually established and maintained by TP53 and its upstream regulators CDKN1A and CDKN2A in combination with activation of retinoblastoma (RB) tumor suppressor pathways.73,120 The oncogene KRAS plays a central role in PDAC cell proliferation, and its mutation requires CDKN2A, TP53 and MYC are also regulated by KRAS.117 KRAS mutations depend on CDKN2A inactivation and KRAS mutations are the preferred evolutionary pathway for CDKN2A inactivation.121 After KRAS mutation, KRAS downstream signal MYC is amplified and CDKN2A inactivation may occur at the same stage.121 MYC is an important mediator of metabolic changes in pancreatic cancer cells induced by KRAS.73 Inactivation of TP53 and SMAD4 occurs mainly in true invasive cancers.122 MYC amplification can induce TP53 and SMAD4 silencing, mutation or loss and promote further development of PDAC.123,124
SMAD4
SMAD4 is a core component of the TGF-β signaling pathway.125 Changes in genes related to the TGF-β signaling pathway are present in almost all PDAC cases. In the advanced stages of cancer, genetic changes may accelerate disease progression, for example by inducing epithelial-mesenchymal transition (EMT).126 The loss of SMAD4 may prevent the occurrence of EMT.127 Many studies have shown that SMAD4 deletion or mutation is a negative prognostic factor for PDAC.128 In cases of SMDA4-deficient PDACs, TP53 usually has a missense mutation.129 Both TP53 and SMAD4 inactivation occur in the late stages of PDAC, but TP53 mutations occur earlier than SMAD4 loss. TP53 mutations and SMAD4 loss occur mainly in aggressive PDACs and they may be markers of high metastatic potential for tumors.122,123,130 Both SMAD4 deletions and mutations contribute to the inactivation of SMAD4 and the role of SMAD4 in cancer progression has only been partially reported.131,132 Some researchers have reported that SMAD4 deletion is more common in early or resectable tumors.133 Reports show that SMAD4 is lost more frequently in patients with PDAC (72%) with extensive metastatic disease than in patients with locally destructive disease (35%, p = 0.007).130 This findings may have potential significance for patients with PDAC and provide new ideas for treatment options to reduce local or distant tumor growth.129
Interactions in PDAC
Genomic analysis reveals repeated mutations in many genes in PDAC and the oncogenic point mutations of individual genes aggregate into core molecular pathways.108 Using targeted sequencing to detect the number of mutation-driven genes and subsequent sequence analysis may be a promising genomic biomarker approach for predicting postoperative and prognosis in patients with PDAC.134 In previous examinations, gene sequencing confirmed that chromatin remodeling, DNA repair, cell cycle, WNT-β-catenin and NOTCH signaling pathways may be disrupted in PDAC cells. The interaction between these pathways is complex and most genes of these pathways would alter actively or be altered passively.135,136 CDKN2A and the aforementioned genes participate in the metabolic pathways of cells and can develop effective antimetabolites to target each cancer according to the differences between specific metabolic pathways.58 Therapy for a specific gene or pathway will have a positive impact on improving the patient’s survival prognosis.136,137
D-cyclin (D1, D2 and D3) and the associated cyclin-dependent kinases (CDK4 and CDK6) are part of the core cell cycle machinery that drives cell proliferation. CDK4 is involved in the G1-S phase, CDK6 is a regulator of the G1 phase and the product of CDKN2A, p16INK4a, participates in tumor angiogenesis by regulating CDK4/6, promoting cell cycle and participating in metabolism. MYC only amplifies existing transcriptional programs in cells and it is unclear how MYC regulates the ratio between growth-promoting and growth-inhibiting genes to stimulate growth and proliferation.65 MDM2 and TP53 interact by automatic negative feedback adjustment. Mutations of CDKN2A have been detected in many human cancer types. In this respect, CDKN2A is similar to the typical tumor suppressor gene TP53. TP53 prevents cell cycle progression by increasing the expression of the paralogous gene of the cyclin-dependent kinase inhibitor CDKN2A, CDKN1A.138 TP53 plays an important role in maintaining genetic stability including DNA repair, cell cycle and apoptosis.139 MDM2 phosphorylation can affect CDKN2A expression changes.140 CDKN1A, MDM2, TP53 are included in the TP53 signal pathway in PDAC.129 In the process of cell division, the major transcription factor of the TP53 pathway and its cofactor CDKN2A prevent the proliferating cells from repairing the damage by strongly fighting the MYC pathway, or initiate an orderly suicide if the damage cannot be repaired, in order to protect the integrity of the entire tissue.141,142 TP53 and CDKN2A are often biallelically inactivated in human malignancies, which has a significant impact on treatment.143,144 MYC induces the inactivation or loss of TP53 by activating the tumor suppressor p19ARF, at this time p19ARF and MYC have a significant carcinogenic synergy.145 Disruption of the TP53 pathway in PDAC may lose the intrinsic proliferation pathway triggered by MYC activation through mechanisms other than apoptosis inhibition and promote tumorigenesis.145 MYC can fight growth inhibition mediated by the CDK inhibitor CDKN2A.146 CDK4 can inhibit the activity of CDKN2A, which is a transcription target of MYC.146 After CDKN2A deletion, MYC amplification and non-silent mutations and/or deletions are significantly enhanced, which contributes to the metastatic function of PDAC cells.135,147 Subsequently, MYC regulates the rapid rise of CDK4 mRNA levels through four highly conserved MYC binding sites in the CDK4 promoter, promoting tumorigenicity and cell cycle regulation.146 Cells are deficient in MYC, the amount of CDK4 protein is reduced and cell cycle progression is delayed.146 After MYC deletion, the level of apoptotic regulator TP53 was significantly up-regulated and the proliferation rate of cancer cells was slowed down, possibly due to the presence of TP53 mutation.135 The downregulation of MYC may trigger other cell death mechanisms besides apoptosis, such as autophagic cell death, which is of great significance for the treatment of PDAC.148
Conclusion
Of the relationship between CDKN2A and five genes for which we searched on the STRING webpage and KRAS and TP53, a CDKN2A-centric network regulation may exist in PDAC, although the relationships between CDK6 and MYC in PDAC require further research for their better understanding. Early identification of this cancer is of critical importance to better prognosis and survival. Since no specific biomarkers have been identified to date, novel and more sensitive biomarkers need to be discovered and developed to diagnose early this deadly disease.149,150 Both CDKN2A and these genes involved in the cell cycle or oncogenic transcriptional machinery can be used for tumor therapy by regulating the oncogenic transcriptional regulation mechanism and cell cycle progression.151 Understanding the precise cell cycle and metabolic characteristics, as well as the convergent effects of oncogenic pathways in different types of cancer and exploring the relationship between CDKN2A and these genes is critical for determining the optimal combination strategy and using the advances in tumor biology for achieving more effective treatment of PDAC.58,152
Data Sharing Statement
- The way we get the network regulation network in this paper includes the following steps.
- Open the STRING website.
- Enter the Protein Name “CDKN2A”, select the Organism “Homo sapiens”, press the blue button “Select” and press “SEARCH”.
- Select the first default option on the website and press the blue button “Continue” of the upper right corner on this website.
- Select the minimum required interaction score “highest confidence (0.900)” and 1st shell of the max number of interactors to show “no more than five interactors”, press “UPDATE”.
- In the end you will see a CDKN2A-related interaction network picture on this website.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (No.81272182), the Key Project of Hunan Provincial Education Department (16K077), Hunan Province Key Laboratory of Tumor Cellular & Molecular Pathology (2016TP1015), Hunan Province Cooperative Innovation Center for Molecular Target New Drug Study (2014-405).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Balachandran VP, Beatty GL, Dougan SK. Broadening the impact of immunotherapy to pancreatic cancer: challenges and opportunities. Gastroenterology. 2019;156(7):2056–2072. doi:10.1053/j.gastro.2018.12.038
2. Collisson EA, Bailey P, Chang DK, et al. Molecular subtypes of pancreatic cancer. Nat Rev Gastroenterol Hepatol. 2019;16(4):207–220. doi:10.1038/s41575-019-0109-y
3. Vincent A, Herman J, Schulick R, et al. Pancreatic cancer. Lancet. 2011;378(9791):607–620. doi:10.1016/S0140-6736(10)62307-0
4. The Lancet Oncology. Pancreatic cancer in the spotlight. Lancet Oncol. 2014;15(3):241. doi:10.1016/S1470-2045(14)70097-X
5. Nagrath S, Jack RM, Sahai V, et al. Opportunities and challenges for pancreatic circulating tumor cells. Gastroenterology. 2016;151(3):412–426. doi:10.1053/j.gastro.2016.05.052
6. Tiriac H, Belleau P, Engle DD, et al. Organoid profiling identifies common responders to chemotherapy in pancreatic cancer. Cancer Discov. 2018;8(9):1112–1129. doi:10.1158/2159-8290.CD-18-0349
7. Kleeff J, Korc M, Apte M, et al. Pancreatic cancer. Nat Rev Dis Primers. 2016;2(1):16022. doi:10.1038/nrdp.2016.22
8. Neesse A, Algül H, Tuveson DA, et al. Stromal biology and therapy in pancreatic cancer: a changing paradigm. Gut. 2015;64(9):1476–1484. doi:10.1136/gutjnl-2015-309304
9. Evans DB, Varadhachary GR, Crane CH, et al. Preoperative gemcitabine-based chemoradiation for patients with resectable adenocarcinoma of the pancreatic head. J Clin Oncol. 2008;26(21):3496–3502. doi:10.1200/JCO.2007.15.8634
10. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30. doi:10.3322/caac.21387
11. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7–30. doi:10.3322/caac.21442
12. Dang CV. MYC on the path to cancer. Cell. 2012;149(1):22–35. doi:10.1016/j.cell.2012.03.003
13. Neoptolemos JP, Palmer DH, Ghaneh P, et al. Comparison of adjuvant gemcitabine and capecitabine with gemcitabine monotherapy in patients with resected pancreatic cancer (ESPAC-4): a multicentre, open-label, randomised, Phase 3 trial. Lancet. 2017;389(10073):1011–1024. doi:10.1016/S0140-6736(16)32409-6
14. Sinn M, Bahra M, Liersch T, et al. CONKO-005: adjuvant chemotherapy with gemcitabine plus erlotinib versus gemcitabine alone in patients after R0 resection of pancreatic cancer: a multicenter randomized Phase III trial. J Clin Oncol. 2017;35(29):3330–3337. doi:10.1200/JCO.2017.72.6463
15. Dreyer SB, Chang DK, Bailey P, et al. Pancreatic cancer genomes: implications for clinical management and therapeutic development. Clin Cancer Res. 2017;23(7):1638–1646. doi:10.1158/1078-0432.CCR-16-2411
16. Hannou SA, Wouters K, Paumelle R, et al. Functional genomics of the CDKN2A/B locus in cardiovascular and metabolic disease: what have we learned from GWASs? Trends Endocrinol Metab. 2015;26(4):176–184. doi:10.1016/j.tem.2015.01.008
17. Sharpless NE, Sherr CJ. Forging a signature of in vivo senescence. Nat Rev Cancer. 2015;15(7):397–408. doi:10.1038/nrc3960
18. Krishnamurthy J, Ramsey MR, Ligon KL, et al. p16INK4a induces an age-dependent decline in islet regenerative potential. Nature. 2006;443(7110):453–457. doi:10.1038/nature05092
19. Ohtani N, Zebedee Z, Huot TJG, et al. Opposing effects of Ets and Id proteins on p16INK4a expression during cellular senescence. Nature. 2001;409(6823):1067–1070. doi:10.1038/35059131
20. Pal A, Potjer TP, Thomsen SK, et al. Loss-of-function mutations in the cell-cycle control gene CDKN2A impact on glucose homeostasis in humans. Diabetes. 2016;65(2):527–533. doi:10.2337/db15-0602
21. Al-khalaf HH, Colak D, Al-saif M, et al. p16(INK4a) positively regulates cyclin D1 and E2F1 through negative control of AUF1. PLoS One. 2011;6(7):e21111. doi:10.1371/journal.pone.0021111
22. Liggett WH
23. Kong Y, Sharma RB, Nwosu BU, et al. Islet biology, the CDKN2A/B locus and type 2 diabetes risk. Diabetologia. 2016;59(8):1579–1593. doi:10.1007/s00125-016-3967-7
24. Burdon KP, Macgregor S, Hewitt AW, et al. Genome-wide association study identifies susceptibility loci for open angle glaucoma at TMCO1 and CDKN2B-AS1. Nat Genet. 2011;43(6):574–578. doi:10.1038/ng.824
25. Vengoechea J, Tallo C. A germline deletion of 9p21.3 presenting as familial melanoma, astrocytoma and breast cancer: clinical and genetic counselling challenges. J Med Genet. 2017;54(10):682–684. doi:10.1136/jmedgenet-2017-104690
26. Helgadottir H, Höiom V, Tuominen R, et al. Germline CDKN2A mutation status and survival in familial melanoma cases. J Natl Cancer Inst. 2016;108(11):djw135. doi:10.1093/jnci/djw135
27. Gadhikar MA, Zhang J, Shen L, et al. CDKN2A/p16 deletion in head and neck cancer cells is associated with CDK2 activation, replication stress, and vulnerability to CHK1 inhibition. Cancer Res. 2018;78(3):781–797. doi:10.1158/0008-5472.CAN-17-2802
28. Bian YS, Osterheld M, Fontolliet C, et al. p16 inactivation by methylation of the CDKN2A promoter occurs early during neoplastic progression in Barrett’s esophagus. Gastroenterology. 2002;122(4):1113–1121. doi:10.1053/gast.2002.32370
29. Choi W, Ochoa A, McConkey DJ, et al. Genetic alterations in the molecular subtypes of bladder cancer: illustration in the cancer genome atlas dataset. Eur Urol. 2017;72(3):354–365. doi:10.1016/j.eururo.2017.03.010
30. Illei PB, Rusch VW, Zakowski MF, et al. Homozygous deletion of CDKN2A and codeletion of the methylthioadenosine phosphorylase gene in the majority of pleural mesotheliomas. Clin Cancer Res. 2003;9(6):2108–2113.
31. Borg A, Sandberg T, Nilsson K, et al. High frequency of multiple melanomas and breast and pancreas carcinomas in CDKN2A mutation-positive melanoma families. J Natl Cancer Inst. 2000;92(15):1260–1266. doi:10.1093/jnci/92.15.1260
32. Cui C, Gan Y, Gu L, et al. P16-specific DNA methylation by engineered zinc finger methyltransferase inactivates gene transcription and promotes cancer metastasis. Genome Biol. 2015;16(1):252. doi:10.1186/s13059-015-0819-6
33. Huang X, Wu C, Fu Y, et al. Methylation analysis for multiple gene promoters in non-small cell lung cancers in high indoor air pollution region in China. Bull Cancer. 2018;105(9):746–754. doi:10.1016/j.bulcan.2018.05.004
34. Tang B, Li Y, Qi G, et al. Clinicopathological significance of CDKN2A promoter hypermethylation frequency with pancreatic cancer. Sci Rep. 2015;5(1):13563. doi:10.1038/srep13563
35. Borazanci E, Dang CV, Robey RW, et al. Pancreatic cancer: “a riddle wrapped in a mystery inside an enigma”. Clin Cancer Res. 2017;23(7):1629–1637. doi:10.1158/1078-0432.CCR-16-2070
36. Witkiewicz AK, McMillan EA, Balaji U, et al. Whole-exome sequencing of pancreatic cancer defines genetic diversity and therapeutic targets. Nat Commun. 2015;6(1):6744. doi:10.1038/ncomms7744
37. Goldstein AM, Fraser MC, Struewing JP, et al. Increased risk of pancreatic cancer in melanoma-prone kindreds with p16INK4 mutations. N Engl J Med. 1995;333(15):970–975. doi:10.1056/NEJM199510123331504
38. Notta F, Chan-seng-yue M, Lemire M, et al. A renewed model of pancreatic cancer evolution based on genomic rearrangement patterns. Nature. 2016;538(7625):378–382. doi:10.1038/nature19823
39. Lagarrigue S, Lopez-mejia IC, Denechaud P-D, et al. CDK4 is an essential insulin effector in adipocytes. J Clin Invest. 2016;126(1):335–348. doi:10.1172/JCI81480
40. Dor Y, Brown J, Martinez OI, et al. Adult pancreatic β-cells are formed by self-duplication rather than stem-cell differentiation. Nature. 2004;429(6987):41–46. doi:10.1038/nature02520
41. Gillam MP, Nimbalkar D, Sun L, et al. MEN1 tumorigenesis in the pituitary and pancreatic islet requires Cdk4 but not Cdk2. Oncogene. 2015;34(7):932–938. doi:10.1038/onc.2014.3
42. Zeggini E, Weedon MN, Lindgren CM, et al. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science. 2007;316(5829):1336–1341. doi:10.1126/science.1142364
43. Lee Y, Dominy JE, Choi YJ, et al. Cyclin D1-Cdk4 controls glucose metabolism independently of cell cycle progression. Nature. 2014;510(7506):547–551. doi:10.1038/nature13267
44. Chou A, Froio D, Nagrial AM, et al. Tailored first-line and second-line CDK4-targeting treatment combinations in mouse models of pancreatic cancer. Gut. 2018;67(12):2142–2155. doi:10.1136/gutjnl-2017-315144
45. Eser S, Reiff N, Messer M, et al. Selective requirement of PI3K/PDK1 signaling for Kras oncogene-driven pancreatic cell plasticity and cancer. Cancer Cell. 2013;23(3):406–420. doi:10.1016/j.ccr.2013.01.023
46. Sherr CJ. Cancer cell cycles. Science. 1996;274(5293):1672–1677. doi:10.1126/science.274.5293.1672
47. Asghar U, Witkiewicz AK, Turner NC, et al. The history and future of targeting cyclin-dependent kinases in cancer therapy. Nat Rev Drug Discov. 2015;14(2):130–146. doi:10.1038/nrd4504
48. Fujimoto T, Anderson K, Jacobsen SE, et al. Cdk6 blocks myeloid differentiation by interfering with Runx1 DNA binding and Runx1-C/EBPalpha interaction. EMBO J. 2007;26(9):2361–2370. doi:10.1038/sj.emboj.7601675
49. Brito-babapulle V, Gruszka-westwood AM, Platt G, et al. Translocation t (2;7)(p12;q21-22) with dysregulation of the CDK6 gene mapping to 7q21-22 in a non-Hodgkin’s lymphoma with leukemia. Haematologica. 2002;87(4):357–362.
50. Tomita T. Cyclin-dependent kinase (cdk6) and p16 in pancreatic endocrine neoplasms. Pathology. 2004;36(6):566–570. doi:10.1080/00313020400011342
51. Li C, Qi L, Bellail AC, et al. PD-0332991 induces G1 arrest of colorectal carcinoma cells through inhibition of the cyclin-dependent kinase-6 and retinoblastoma protein axis. Oncol Lett. 2014;7(5):1673–1678. doi:10.3892/ol.2014.1957
52. Kollmann K, Sexl V. CDK6 and p16INK4A in lymphoid malignancies. Oncotarget. 2013;4(11):1858–1859. doi:10.18632/oncotarget.1541
53. Wang H, Nicolay BN, Chick JM, et al. The metabolic function of cyclin D3-CDK6 kinase in cancer cell survival. Nature. 2017;546(7658):426–430. doi:10.1038/nature22797
54. Serrano M, Gomez-lahoz E, DePinho R, et al. Inhibition of ras-induced proliferation and cellular transformation by p16INK4. Science. 1995;267(5195):249–252. doi:10.1126/science.7809631
55. Serrano M, Lin AW, McCurrach ME, et al. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88(5):593–602. doi:10.1016/S0092-8674(00)81902-9
56. Cole MD, Cowling VH. Specific regulation of mRNA cap methylation by the c-Myc and E2F1 transcription factors. Oncogene. 2009;28(9):1169–1175. doi:10.1038/onc.2008.463
57. Edmunds LR, Sharma L, Kang A, et al. c-Myc programs fatty acid metabolism and dictates acetyl-CoA abundance and fate. J Biol Chem. 2014;289(36):25382–25392. doi:10.1074/jbc.M114.580662
58. Hsieh AL, Walton ZE, Altman BJ, et al. MYC and metabolism on the path to cancer. Semin Cell Dev Biol. 2015;43:11–21. doi:10.1016/j.semcdb.2015.08.003
59. Duvel K, Yecies JL, Menon S, et al. Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Mol Cell. 2010;39(2):171–183. doi:10.1016/j.molcel.2010.06.022
60. Wirth M, Schneider G. MYC: a stratification marker for pancreatic cancer therapy. Trends Cancer. 2016;2(1):1–3. doi:10.1016/j.trecan.2015.12.002
61. Le A, Lane A, Hamaker M, et al. Glucose-independent glutamine metabolism via TCA cycling for proliferation and survival in B cells. Cell Metab. 2012;15(1):110–121. doi:10.1016/j.cmet.2011.12.009
62. Dang CV. Gene regulation: fine-tuned amplification in cells. Nature. 2014;511(7510):417–418. doi:10.1038/nature13518
63. Walz S, Lorenzin F, Morton J, et al. Activation and repression by oncogenic MYC shape tumour-specific gene expression profiles. Nature. 2014;511(7510):483–487. doi:10.1038/nature13473
64. Wolf E, Lin CY, Eilers M, et al. Taming of the beast: shaping Myc-dependent amplification. Trends Cell Biol. 2015;25(4):241–248. doi:10.1016/j.tcb.2014.10.006
65. Sabo A, Kress TR, Pelizzola M, et al. Selective transcriptional regulation by Myc in cellular growth control and lymphomagenesis. Nature. 2014;511(7510):488–492. doi:10.1038/nature13537
66. Nie Z, Hu G, Wei G, et al. c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell. 2012;151(1):68–79. doi:10.1016/j.cell.2012.08.033
67. Lin CY, Lovén J, Rahl P, et al. Transcriptional amplification in tumor cells with elevated c-Myc. Cell. 2012;151(1):56–67. doi:10.1016/j.cell.2012.08.026
68. Rahl PB, Lin CY, Seila AC, et al. c-Myc regulates transcriptional pause release. Cell. 2010;141(3):432–445. doi:10.1016/j.cell.2010.03.030
69. Carugo A, Genovese G, Seth S, et al. In vivo functional platform targeting patient-derived xenografts identifies WDR5-Myc association as a critical determinant of pancreatic cancer. Cell Rep. 2016;16(1):133–147. doi:10.1016/j.celrep.2016.05.063
70. van Riggelen J, Yetil A, Felsher DW. MYC as a regulator of ribosome biogenesis and protein synthesis. Nat Rev Cancer. 2010;10(4):301–309. doi:10.1038/nrc2819
71. Biankin AV, Waddell N, Kassahn KS, et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature. 2012;491(7424):399–405. doi:10.1038/nature11547
72. Yustein JT, Liu Y-C, Gao P, et al. Induction of ectopic Myc target gene JAG2 augments hypoxic growth and tumorigenesis in a human B-cell model. Proc Natl Acad Sci U S A. 2010;107(8):3534–3539. doi:10.1073/pnas.0901230107
73. Ying H, Kimmelman A, Lyssiotis C, et al. Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell. 2012;149(3):656–670. doi:10.1016/j.cell.2012.01.058
74. Beroukhim R, Mermel CH, Porter D, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463(7283):899–905. doi:10.1038/nature08822
75. Evan GI, Hah N, Littlewood TD, et al. Re-engineering the pancreas tumor microenvironment: a “regenerative program” hacked. Clin Cancer Res. 2017;23(7):1647–1655. doi:10.1158/1078-0432.CCR-16-3275
76. Hessmann E, Schneider G, Ellenrieder V, et al. MYC in pancreatic cancer: novel mechanistic insights and their translation into therapeutic strategies. Oncogene. 2016;35(13):1609–1618. doi:10.1038/onc.2015.216
77. Korc M. Beyond Kras: MYC rules in pancreatic cancer. Cell Mol Gastroenterol Hepatol. 2018;6(2):223–224. doi:10.1016/j.jcmgh.2018.04.009
78. Diersch S, Wirth M, Schneeweis C, et al. Kras (G12D) induces EGFR-MYC cross signaling in murine primary pancreatic ductal epithelial cells. Oncogene. 2016;35(29):3880–3886. doi:10.1038/onc.2015.437
79. Mazur PK, Herner A, Mello SS, et al. Combined inhibition of BET family proteins and histone deacetylases as a potential epigenetics-based therapy for pancreatic ductal adenocarcinoma. Nat Med. 2015;21(10):1163–1171. doi:10.1038/nm.3952
80. Carroll PA, Diolaiti D, McFerrin L, et al. Deregulated Myc requires MondoA/Mlx for metabolic reprogramming and tumorigenesis. Cancer Cell. 2015;27(2):271–285. doi:10.1016/j.ccell.2014.11.024
81. Ji S, Qin Y, Liang C, et al. FBW7 (F-box and WD repeat domain-containing 7) negatively regulates glucose metabolism by targeting the c-Myc/TXNIP (Thioredoxin-binding protein) axis in pancreatic cancer. Clin Cancer Res. 2016;22(15):3950–3960. doi:10.1158/1078-0432.CCR-15-2380
82. Cunningham JT, Moreno M, Lodi A, et al. Protein and nucleotide biosynthesis are coupled by a single rate-limiting enzyme, PRPS2, to drive cancer. Cell. 2014;157(5):1088–1103. doi:10.1016/j.cell.2014.03.052
83. Hollstein M, Sidransky D, Vogelstein B, et al. p53 mutations in human cancers. Science. 1991;253(5015):49–53. doi:10.1126/science.1905840
84. Du Y, Liu Z, You L, et al. Pancreatic cancer progression relies upon mutant p53-induced oncogenic signaling mediated by NOP14. Cancer Res. 2017;77(10):2661–2673. doi:10.1158/0008-5472.CAN-16-2339
85. Hori Y, Miyabe K, Yoshida M, et al. Impact of TP53 codon 72 and MDM2 SNP 309 polymorphisms in pancreatic ductal adenocarcinoma. PLoS One. 2015;10(3):e0118829. doi:10.1371/journal.pone.0118829
86. McCubrey JA, Lertpiriyapong K, Steelman LS, et al. Effects of resveratrol, curcumin, berberine and other nutraceuticals on aging, cancer development, cancer stem cells and microRNAs. Aging (Albany NY). 2017;9(6):1477–1536. doi:10.18632/aging.v9i6
87. Swidnicka-siergiejko AK, Gomez-chou SB, Cruz-monserrate Z, et al. Chronic inflammation initiates multiple forms of K-Ras-independent mouse pancreatic cancer in the absence of TP53. Oncogene. 2017;36(22):3149–3158. doi:10.1038/onc.2016.461
88. Feng Y, Liu H, Duan B, et al. Potential functional variants in SMC2 and TP53 in the AURORA pathway genes and risk of pancreatic cancer. Carcinogenesis. 2019;40(4):521–528. doi:10.1093/carcin/bgz029
89. Grochola LF, Taubert H, Greither T, et al. Elevated transcript levels from the MDM2 P1 promoter and low p53 transcript levels are associated with poor prognosis in human pancreatic ductal adenocarcinoma. Pancreas. 2011;40(2):265–270. doi:10.1097/MPA.0b013e3181f95104
90. Martins CP, Brown-swigart L, Evan GI. Modeling the therapeutic efficacy of p53 restoration in tumors. Cell. 2006;127(7):1323–1334. doi:10.1016/j.cell.2006.12.007
91. Ventura A, Kirsch DG, McLaughlin ME, et al. Restoration of p53 function leads to tumour regression in vivo. Nature. 2007;445(7128):661–665. doi:10.1038/nature05541
92. Xue W, Zender L, Miething C, et al. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature. 2007;445(7128):656–660. doi:10.1038/nature05529
93. Cheok CF, Verma CS, Baselga J, et al. Translating p53 into the clinic. Nat Rev Clin Oncol. 2011;8(1):25–37. doi:10.1038/nrclinonc.2010.174
94. Cui Y, Guo G. Immunomodulatory function of the tumor suppressor p53 in host immune response and the tumor microenvironment. Int J Mol Sci. 2016;17(11):1942. doi:10.3390/ijms17111942
95. Kubbutat MH, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nature. 1997;387(6630):299–303. doi:10.1038/387299a0
96. Haupt Y, Maya R, Kazaz A, et al. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387(6630):296–299. doi:10.1038/387296a0
97. Vousden KH, Prives C. Blinded by the light: the growing complexity of p53. Cell. 2009;137(3):413–431. doi:10.1016/j.cell.2009.04.037
98. Brooks CL, Gu W. p53 ubiquitination: mdm2 and beyond. Mol Cell. 2006;21(3):307–315. doi:10.1016/j.molcel.2006.01.020
99. Kastan MB. Wild-type p53: tumors can’t stand it. Cell. 2007;128(5):837–840. doi:10.1016/j.cell.2007.02.022
100. Yang J-Y, Zong CS, Xia W, et al. MDM2 promotes cell motility and invasiveness by regulating E-cadherin degradation. Mol Cell Biol. 2006;26(19):7269–7282. doi:10.1128/MCB.00172-06
101. Hidalgo M. Pancreatic cancer. N Engl J Med. 2010;362(17):1605–1617. doi:10.1056/NEJMra0901557
102. Post SM, Quintás-cardama A, Pant V, et al. A high-frequency regulatory polymorphism in the p53 pathway accelerates tumor development. Cancer Cell. 2010;18(3):220–230. doi:10.1016/j.ccr.2010.07.010
103. Wade M, Li YC, Wahl GM. MDM2, MDMX and p53 in oncogenesis and cancer therapy. Nat Rev Cancer. 2013;13(2):83–96. doi:10.1038/nrc3430
104. Karni-schmidt O, Lokshin M, Prives C. The roles of MDM2 and MDMX in cancer. Annu Rev Pathol. 2016;11(1):617–644. doi:10.1146/annurev-pathol-012414-040349
105. Zhao Y, Aguilar A, Bernard D, et al. Small-molecule inhibitors of the MDM2-p53 protein-protein interaction (MDM2 Inhibitors) in clinical trials for cancer treatment. J Med Chem. 2015;58(3):1038–1052. doi:10.1021/jm501092z
106. Zhao Y, Yu H, Hu W. The regulation of MDM2 oncogene and its impact on human cancers. Acta Biochim Biophys Sin (Shanghai). 2014;46(3):180–189. doi:10.1093/abbs/gmt147
107. Gu L, Zhang H, Liu T, et al. Discovery of dual inhibitors of MDM2 and XIAP for cancer treatment. Cancer Cell. 2016;30(4):623–636. doi:10.1016/j.ccell.2016.08.015
108. Waddell N, Pajic M, Patch A-M, et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature. 2015;518(7540):495–501. doi:10.1038/nature14169
109. Makohon-moore A, Iacobuzio-donahue CA. Pancreatic cancer biology and genetics from an evolutionary perspective. Nat Rev Cancer. 2016;16(9):553–565. doi:10.1038/nrc.2016.66
110. Wang W, Qin -J-J, Voruganti S, et al. Discovery and characterization of dual inhibitors of MDM2 and NFAT1 for pancreatic cancer therapy. Cancer Res. 2018;78(19):5656–5667. doi:10.1158/0008-5472.CAN-17-3939
111. Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408(6810):307–310. doi:10.1038/35042675
112. Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat Med. 2004;10(8):789–799. doi:10.1038/nm1087
113. Kopp JL, Von figura G, Mayes E, et al. Identification of Sox9-dependent acinar-to-ductal reprogramming as the principal mechanism for initiation of pancreatic ductal adenocarcinoma. Cancer Cell. 2012;22(6):737–750. doi:10.1016/j.ccr.2012.10.025
114. Bryant KL, Mancias JD, Kimmelman AC, et al. KRAS: feeding pancreatic cancer proliferation. Trends Biochem Sci. 2014;39(2):91–100. doi:10.1016/j.tibs.2013.12.004
115. Liu J, Ji S, Liang C, et al. Critical role of oncogenic KRAS in pancreatic cancer (Review). Mol Med Rep. 2016;13(6):4943–4949. doi:10.3892/mmr.2016.5196
116. Bauer TM, Dhir T, Strickland A, et al. Genetic drivers of pancreatic cancer are identical between the primary tumor and a secondary lesion in a long-term (>5 years) survivor after a whipple procedure. J Pancreat Cancer. 2018;4(1):81–87. doi:10.1089/pancan.2018.0015
117. Scully KM, Lahmy R, Signaevskaia L, et al. E47 governs the MYC-CDKN1B/p27 KIP1 -RB network to growth arrest PDA cells independent of CDKN2A/p16 INK4A and wild-type p53. Cell Mol Gastroenterol Hepatol. 2018;6(2):181–198. doi:10.1016/j.jcmgh.2018.05.002
118. Mueller S, Engleitner T, Maresch R, et al. Evolutionary routes and KRAS dosage define pancreatic cancer phenotypes. Nature. 2018;554(7690):62–68. doi:10.1038/nature25459
119. Rosenfeldt MT, O’prey J, Morton JP, et al. p53 status determines the role of autophagy in pancreatic tumour development. Nature. 2013;504(7479):296–300. doi:10.1038/nature12865
120. Campisi J. F. d’Adda di Fagagna cellular senescence: when bad things happen to good cells. Nat Rev Mol Cell Biol. 2007;8(9):729–740.
121. Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi:10.1038/nprot.2008.211
122. Hosoda W, Chianchiano P, Griffin JF, et al. Genetic analyses of isolated high-grade pancreatic intraepithelial neoplasia (HG-PanIN) reveal paucity of alterations in TP53 and SMAD4. J Pathol. 2017;242(1):16–23. doi:10.1002/path.2017.242.issue-1
123. Yachida S, White CM, Naito Y, et al. Clinical significance of the genetic landscape of pancreatic cancer and implications for identification of potential long-term survivors. Clin Cancer Res. 2012;18(22):6339–6347. doi:10.1158/1078-0432.CCR-12-1215
124. Singh P, Srinivasan R, Wig JD. Major molecular markers in pancreatic ductal adenocarcinoma and their roles in screening, diagnosis, prognosis, and treatment. Pancreas. 2011;40(5):644–652. doi:10.1097/MPA.0b013e31821ff741
125. Heldin CH, Moustakas A. Role of Smads in TGFbeta signaling. Cell Tissue Res. 2012;347(1):21–36. doi:10.1007/s00441-011-1190-x
126. Trager MM, Dhayat SA. Epigenetics of epithelial-to-mesenchymal transition in pancreatic carcinoma. Int J Cancer. 2017;141(1):24–32. doi:10.1002/ijc.v141.1
127. Levy L, Hill CS. Smad4 dependency defines two classes of transforming growth factor {beta} (TGF-{beta}) target genes and distinguishes TGF-{beta}-induced epithelial-mesenchymal transition from its antiproliferative and migratory responses. Mol Cell Biol. 2005;25(18):8108–8125. doi:10.1128/MCB.25.18.8108-8125.2005
128. Blackford A, Serrano OK, Wolfgang CL, et al. SMAD4 gene mutations are associated with poor prognosis in pancreatic cancer. Clin Cancer Res. 2009;15(14):4674–4679. doi:10.1158/1078-0432.CCR-09-0227
129. Liszka L. Ductal adenocarcinoma of the pancreas usually retained SMAD4 and p53 protein status as well as expression of epithelial-to-mesenchymal transition markers and cell cycle regulators at the stage of liver metastasis. Pol J Pathol. 2014;65(2):100–112. doi:10.5114/pjp.2014.43959
130. Schutte M, Hruban RH, Hedrick L, et al. DPC4 gene in various tumor types. Cancer Res. 1996;56(11):2527–2530.
131. Wilentz RE, Su GH, Dai JL, et al. Immunohistochemical labeling for dpc4 mirrors genetic status in pancreatic adenocarcinomas: a new marker of DPC4 inactivation. Am J Pathol. 2000;156(1):37–43. doi:10.1016/S0002-9440(10)64703-7
132. Singh P, Srinivasan R, Wig JD. SMAD4 genetic alterations predict a worse prognosis in patients with pancreatic ductal adenocarcinoma. Pancreas. 2012;41(4):541–546. doi:10.1097/MPA.0b013e318247d6af
133. Biankin AV, Morey AL, Lee C-S, et al. DPC4/Smad4 expression and outcome in pancreatic ductal adenocarcinoma. J Clin Oncol. 2002;20(23):4531–4542. doi:10.1200/JCO.2002.12.063
134. Hayashi H, Kohno T, Ueno H, et al. Utility of assessing the number of mutated KRAS, CDKN2A, TP53, and SMAD4 genes using a targeted deep sequencing assay as a prognostic biomarker for pancreatic cancer. Pancreas. 2017;46(3):335–340. doi:10.1097/MPA.0000000000000760
135. Lin W-C, Rajbhandari N, Liu C, et al. Dormant cancer cells contribute to residual disease in a model of reversible pancreatic cancer. Cancer Res. 2013;73(6):1821–1830. doi:10.1158/0008-5472.CAN-12-2067
136. Knudsen ES, O’reilly EM, Brody JR, et al. Genetic diversity of pancreatic ductal adenocarcinoma and opportunities for precision medicine. Gastroenterology. 2016;150(1):48–63. doi:10.1053/j.gastro.2015.08.056
137. Kamisawa T, Wood LD, Itoi T, et al. Pancreatic cancer. Lancet. 2016;388(10039):73–85. doi:10.1016/S0140-6736(16)00141-0
138. Bates S, Ryan KM, Phillips AC, et al. Cell cycle arrest and DNA endoreduplication following p21Waf1/Cip1 expression. Oncogene. 1998;17(13):1691–1703. doi:10.1038/sj.onc.1202104
139. Vogelstein B, Kinzler KW. p53 function and dysfunction. Cell. 1992;70(4):523–526. doi:10.1016/0092-8674(92)90421-8
140. Wood NT, Meek DW, Mackintosh C. 14-3-3 binding to pim-phosphorylated Ser166 and Ser186 of human Mdm2–Potential interplay with the PKB/Akt pathway and p14(ARF). FEBS Lett. 2009;583(4):615–620. doi:10.1016/j.febslet.2009.01.003
141. Molchadsky A, Rivlin N, Brosh R, et al. p53 is balancing development, differentiation and de-differentiation to assure cancer prevention. Carcinogenesis. 2010;31(9):1501–1508. doi:10.1093/carcin/bgq101
142. Zheng H, Ying H, Yan H, et al. p53 and Pten control neural and glioma stem/progenitor cell renewal and differentiation. Nature. 2008;455(7216):1129–1133. doi:10.1038/nature07443
143. Enane FO, Saunthararajah Y, Korc M. Differentiation therapy and the mechanisms that terminate cancer cell proliferation without harming normal cells. Cell Death Dis. 2018;9(9):912. doi:10.1038/s41419-018-0919-9
144. Hata T, Suenaga M, Marchionni L, et al. Genome-wide somatic copy number alterations and mutations in high-grade pancreatic intraepithelial neoplasia. Am J Pathol. 2018;188(7):1723–1733. doi:10.1016/j.ajpath.2018.03.012
145. Finch A, Prescott J, Shchors K, et al. Bcl-xL gain of function and p19 ARF loss of function cooperate oncogenically with Myc in vivo by distinct mechanisms. Cancer Cell. 2006;10(2):113–120. doi:10.1016/j.ccr.2006.06.017
146. Hermeking H, Rago C, Schuhmacher M, et al. Identification of CDK4 as a target of c-MYC. Proc Natl Acad Sci U S A. 2000;97(5):2229–2234. doi:10.1073/pnas.050586197
147. Ouyang H, Mou L-J, Luk C, et al. Immortal human pancreatic duct epithelial cell lines with near normal genotype and phenotype. Am J Pathol. 2000;157(5):1623–1631. doi:10.1016/S0002-9440(10)64800-6
148. Levine B, Yuan J. Autophagy in cell death: an innocent convict? J Clin Invest. 2005;115(10):2679–2688. doi:10.1172/JCI26390
149. Zhang L, Sanagapalli S, Stoita A. Challenges in diagnosis of pancreatic cancer. World J Gastroenterol. 2018;24(19):2047–2060. doi:10.3748/wjg.v24.i19.2047
150. Zhu H, Li T, Du Y, et al. Pancreatic cancer: challenges and opportunities. BMC Med. 2018;16(1):214. doi:10.1186/s12916-018-1215-3
151. Mishra VK, Wegwitz F, Kosinsky RL, et al. Histone deacetylase class-I inhibition promotes epithelial gene expression in pancreatic cancer cells in a BRD4-and MYC-dependent manner. Nucleic Acids Res. 2017;45(11):6334–6349. doi:10.1093/nar/gkx212
152. Franco J, Balaji U, Freinkman E, et al. Metabolic reprogramming of pancreatic cancer mediated by CDK4/6 inhibition elicits unique vulnerabilities. Cell Rep. 2016;14(5):979–990. doi:10.1016/j.celrep.2015.12.094
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.