Back to Journals » OncoTargets and Therapy » Volume 11
Is APC hypermethylation a diagnostic biomarker for bladder cancer? A meta-analysis
Authors Han W, Wang Y, Fan J, Wang C
Received 19 June 2018
Accepted for publication 20 August 2018
Published 27 November 2018 Volume 2018:11 Pages 8359—8369
DOI https://doi.org/10.2147/OTT.S177601
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Faris Farassati
Wei Han,1 Yutao Wang,2 Jingli Fan,2 Chunlei Wang2
1Department of Pharmacy, Central Hospital of Zibo Mining Group Limited Liability Company, Zibo, China; 2Shandong Institute of Prevention and Control for Endemic Disease, Thyroid Disease Prevention and Control Center, Jinan, China
Objective: Numerous studies have been performed to investigate the association between APC promoter hypermethylation and bladder cancer risk. Nevertheless, the conclusion was uncertain due to small sample size, different ethnicities, and tumor subtype. Hence, to accurately assess the effect of APC promoter hypermethylation on the risk of bladder cancer, we performed the meta-analysis.
Materials and methods: We retrieved the relevant literatures from electronic databases such as PubMed, Web of Science, Wanfang, Vapp, and CNKI (Chinese National Knowledge Infrastructure). 95% CI and OR were calculated to evaluate the associations of APC promoter hypermethylation with risk and clinical features of bladder cancer. Heterogeneity among studies was assessed with Q test and I2 statistic. In addition, the diagnostic sensitivity, specificity, and area under the curve (AUC) value of APC hypermethylation for bladder cancer were calculated.
Results: In total, 14 articles with 531 controls and 1,293 cases were included to assess the associations of APC promoter hypermethylation with the risk and clinical characteristics of bladder cancer. The significant association between APC promoter hypermethylation and bladder cancer risk was detected (OR =17.01, CI =7.40–39.07). Furthermore, the results revealed that APC promoter hypermethylation was significantly correlated with the grade of bladder tumor (pTNM stage: OR =1.84, CI =0.87–3.93; grade: OR =4.11, CI =1.62–10.43). According to the results of diagnostic evaluation, the diagnostic sensitivity, specificity, and AUC value of APC hypermethylation for bladder cancer risk were 0.52 (95% CI =0.41–0.63), 0.98 (95% CI =0.90–1.00), and 0.80 (95% CI =0.76–0.83), respectively.
Conclusion: This meta-analysis revealed that APC promoter hypermethylation was a risk factor for bladder cancer risk. In addition, APC promoter hypermethylation was significantly associated with the grade of bladder cancer. APC hypermethylation might be a useful biomarker for the clinical diagnosis of bladder cancer.
Keywords: APC, promoter hypermethylation, bladder carcinoma, meta-analysis
Introduction
Bladder cancer is one of the most common cancers worldwide, with 429,000 newly diagnosed bladder cancer patients in 2012.1 It is reported that North America, Europe, and North Africa had the higher incidence of bladder cancer than other regions.2 In addition, some studies have found that some environmental factors such as smoking, exposure to toxic industrial chemicals and gases, several medications of bladder cancer treatment, and genetic variations were all the risk factors for bladder cancer.3,4 Painful urination, blood in urine, and frequent urination were the early symptoms of bladder cancer. Thirty percent of new bladder cancer patients were initially diagnosed with a lethal phenotype of muscle invasive, while approximately 70% of bladder tumors were non-muscle invasive. In addition, approximately 25% non-muscle-invasive bladder cancer would progress to muscle-invasive disease. Because metastatic and muscle-invasive bladder cancer patients had higher mortality, surgery, radiation therapy, and chemotherapy were usually used in the multidisciplinary care.5 Cisplatin-based chemotherapy, methotrexate, vinblastine, adriamycin, and cisplatin (M-VAC) regimen, and the doublet of gemcitabine and cisplatin (GC) represented the common therapeutic regimen for the bladder cancer patients in advanced stage.6 In recent years, targeted therapeutics have been studied and applied in several malignancies such as gastric cancer, colorectal cancer, and lung cancer. For example, VEGF inhibitors and EGFR inhibitors were often used to treat several cancers. Although environmental factors played an important role in the occurrence of bladder cancer, many genetic alterations were also found in many studies of bladder cancer. Genomic alterations in fibroblast growth factor receptor 3 (FGFR3) and other receptor tyrosine kinase (RTK) activation were found in the non-muscle-invasive bladder cancers.6 In addition, there was evidence that the signal of PI3K/AKT/mTOR often produced many genetic alternations in bladder cancer cells.7 Previous studies have identified a lot of somatic copy-number alterations such as E2F3, PPARG, EGFR, MDM2, CCND1, CDKN2A, and RB1.8,9 In addition, studies of sequencing in bladder cancer have identified many gene mutations in TP53, PIK3CA, TSC1, HRAS, and RB1.8,9 In these candidate genes, KDM6A, EP300, CREBBP, and ARID1A, chromatin remodeling genes, processed a frequency of >10% in bladder cancer.10 These advances in genetics of cancers improved the clinical management and prognosis of bladder cancer. In addition, the molecular biomarkers have been used for the early diagnosis and treatment of solid tumor. However, more diagnostic molecular biomarkers are still needed and combined to find the bladder cancer in the early stage.
APC gene consists of 15 exons, locating on 5q21-q22 and encoding a large multidomain protein which contains 2,843 amino acids, which are involved in the Wnt signaling pathway.11 This gene was found in 1991 which was responsible for familial adenomatous polyposis (FAP).12 The multifunctional protein was expressed widely in various tissues, especially in intestine and central nervous tissues. APC protein can combine with glycogen synthesis kinase 3β (GSK-3β) to form a complex named “destruction complex”, which resulted in the degradation of β-catenin in the cytoplasm.13 Immunoprecipitation was first used to explore the function of APC protein, and the precipitation of β-catenin was found, which suggested an important role of APC protein in Wnt signaling pathway.13 Furthermore, APC protein could act as a regulatory factor to regulate cell migration, cell cycle, DNA replication and repair, and cell apoptosis.14 In addition, APC protein can act as a checkpoint from the G0/G1 phase to the S phase in the process of cell cycle and repress the infinite proliferation of cancer cells.14 Therefore, APC protein possessed the function of tumor inhibition, and its inactivation was often found in many cancers such as thyroid cancer, breast cancer, and colorectal cancer.15
Although many studies that evaluated the association of APC promoter methylation with bladder cancer risk had been published, it was still needed to summarize these published data to get a more accurate result. Furthermore, evidence was still lacking from the present studies that assessed the role of APC promoter hypermethylation in bladder cancer risk. As a consequence, to assess the potential role of APC promoter hypermethylation in the risk and pathogenesis of bladder cancer, we performed this meta-analysis.
Materials and methods
Search strategy
According to the PRISMA statement, literatures that assessed the associations of APC promoter hypermethylation with bladder cancer risk and clinical characteristics were searched from the relevant databases including PubMed, Web of Science, Wanfang, Vapp, and CNKI (Chinese National Knowledge Infrastructure).16 The following strategies were used: (bladder cancer OR bladder carcinoma OR bladder neoplasms OR carcinoma of urinary bladder) AND (APC OR Adenomatous polyposis coli OR Adenomatous polyposis coli gene). Furthermore, the retrieval words were applied with “methylation”, “hypermethylation”, and “epigenetic”. Meanwhile, the relevant references of included original articles and reviews were scanned to identify additional eligible studies. The latest update of literature searching was conducted in March 2017.
Inclusion and exclusion criteria
The inclusion criteria for the eligible studies were as follows: 1) original case–control studies that assessed the association between APC promoter hypermethylation and bladder cancer risk and contained sufficient data of APC promoter hypermethylation frequency in controls and cases; 2) studies that were published in English or Chinese language; 3) eligible studies that contained diagnostic criteria of bladder cancer patients; and 4) studies that declared source of controls. In addition, duplicate or studies that did not obey the inclusion criteria were excluded.
Data extraction
According to the inclusion criteria, two researchers independently extracted the data from the eligible studies. The frequency of APC promoter hypermethylation in cases and controls was extracted to calculate OR and 95% CI. The additional information was extracted, such as first author, publication year, pTNM stage and grade of bladder cancer patients, detection method of APC promoter hypermethylation, study design type, sample type, and ethnicity. The Newcastle–Ottawa Scale (NOS; http://www.ohri.ca/programs/clinical_epidemiology/oxford.asp) was used to evaluate the qualities of included studies.17 Studies with scores 7–9 were considered as high-quality studies, and those with scores 4–6 were defined as medium-quality studies.
Statistical analyses
The frequency of APC promoter hypermethylation in healthy controls and cases was accumulated to calculate OR and 95% CI. Subgroup analysis based on ethnicity and sample type was performed to investigate the strength of correlation between APC promoter hypermethylation and bladder cancer risk. Q statistic and I2 value were applied to evaluate the statistical heterogeneity among studies.17,18 If P-value of Q test was <0.05 which indicated a significant heterogeneity, the random-effects model was used. If P-value of Q test was ≥0.05, the fixed-effects model was selected.19,20 The possibility of publication bias was assessed with Begg’s test and Egger’s test (P<0.05 indicated a significant publication bias).21,22 To validate the stability of results and eliminate the individual heterogeneous study, sensitivity analysis was carried out by sequentially removing each study. If the overall OR had a big change after one study was omitted, this study would be eliminated and the overall OR would be recalculated. In addition, the summarized sensitivity, specificity, and the summary receiver operator characteristic (SROC) curve (area under the curve, AUC) values were calculated to evaluate the value of the diagnostic value of APC promoter hypermethylation in bladder cancer.23 All statistical analyses were carried out using STATA 12.0 (StataCorp LP, College Station, TX, USA) software, and P-values were two sided in this meta-analysis.
Results
Study characteristics
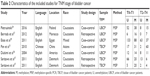
In total, 229 literatures were identified from the initial searching, in which 100 articles were duplicate and were excluded. In the remaining 129 articles, 11 articles were reviewed and removed. Then, 118 studies were remained after titles and abstracts were scanned. Of the 118 articles with full-text format, 94 literatures were excluded since they did not involve the association between APC promoter hypermethylation and bladder cancer risk. In the process of data extracting, 10 articles were eliminated due to insufficient data of APC promoter hypermethylation in controls and cases. Thus, 14 studies were enrolled including 531 controls and 1,293 cases in the final meta-analysis. The main characteristics of eligible studies are presented in Table 1. From the 14 studies, 13 articles were in English language and one article was in Chinese language.24–37 Eleven studies were conducted in Caucasians, two studies were in Asians, and one study was in mixed population. Thirteen studies were included for the diagnostic meta-analysis.24–36 The sample type of case group included serum, urine, and tissue of bladder cancer patients (TBCPs); the sample in the control group consisted of serum, urine, and tissue of healthy people. Methylation-specific PCR (MSP) was adopted in most included studies, and only one study applied quantitative MSP (qMSP; Figure 1).
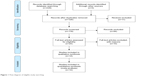  | Figure 1 Flow diagram of eligible study searching. |
Quality score assessment
Table 1 summarizes the methodological quality of the included studies. Thirteen studies that obtained scores of 6–9 were regarded as medium or high quality. However, one study did not contain healthy control; therefore, the evaluation of methodological quality was not carried out in that study.
Meta-analysis results
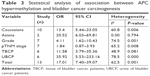
The cumulative results of 13 studies, involving in 1,189 bladder cancer patients, indicated a significant relationship between APC promoter hypermethylation and bladder cancer risk (OR =17.01, 95% CI =7.40–39.07). This result revealed that people with APC promoter hypermethylation had an elevated bladder cancer risk than those with APC promoter hypomethylation. Statistical heterogeneity was observed in the meta-analysis (I2 =62.5%, P=0.001); thus, random-effects model was selected. Stratified analysis based on ethnicity and sample type was also performed to decrease the effect of heterogeneity on the overall OR. Significant differences were found in the pooled OR of two subgroups based on sample type (urine of bladder cancer patients [UBCPs]: OR =35.92, 95% CI =5.53–233.16; TBCPs: OR =11.58, 95% CI =3.79–35.36). In addition, the pooled meta-analysis showed that individuals with APC promoter hypermethylation had a higher grade than individuals with APC promoter hypomethylation (OR =4.11, 95% CI =1.62–10.43). No association between APC promoter hypermethylation and pTNM stage of bladder cancer was observed (OR =1.84, 95% CI =0.87–3.93). In the analysis about clinical information, random-effects model and subgroup analysis based on sample type were used because of the significant statistical heterogeneity. The heterogeneity was reduced in the stratified analysis, and the results are shown in Figures 2–4 (Tables 2 and 3). In addition, we also discussed the relationship between the grade of bladder cancer and APC gene promoter hypermethylation in non-muscle-invasive bladder cancer and muscle-invasive bladder cancer. But no significant associations were found (non-muscle-invasive bladder cancer: OR =1.14, 95% CI =0.61–2.14; muscle-invasive bladder cancer: OR =1.54, 95% CI =0.81–2.94).
Publication bias and sensitivity analyses
The funnel plot of the relationship between APC promoter hypermethylation and bladder cancer risk is shown in Figure 4. P-value of Begg’s test and Egger’s test was 0.088 and 0.001, respectively. Meanwhile, P-value of Begg’s test and Egger’s test, assessing the association between APC promoter hypermethylation and grade of bladder cancer, was 0.099 and 0.007, respectively. In addition, publication bias was found based on the result of Egger’s test, while the result of Begg’s test indicated that no publication bias was found. Thus, we carried out sensitivity analyses by omitting individual study at each time. Interestingly, the overall OR was between 13.91 (95% CI =6.42–30.15) and 20.87 (95% CI =9.28–46.95), which indicated that the positive result was stable. According to the results of subgroup analysis, sample type and ethnicity might partly lead to the publication bias. In the subgroup analysis of TBCPs for bladder cancer risk, P-value of Begg’s test and Egger’s test was 0.497 and 0.270, respectively. In addition, in the subgroup analysis of Caucasians for the grade of bladder cancer, P-value of Begg’s test and Egger’s test was 0.188 and 0.036, respectively. No significant publication bias was found in the analysis of association between APC promoter hypermethylation and the pTNM stage of bladder cancer (Begg’s test, P=0.652; Egger’s test, P=0.208). Finally, to eliminate the publication bias and achieve an accurate and stable pooled OR, we conducted the trim-and-fill analysis using random-effects model. The adjusted OR was 7.70 with 95% CI of 3.60–16.47, which indicated a significant association between APC promoter hypermethylation and bladder cancer risk (Figure 5).
  | Figure 5 Trim-and-fill funnel plot of the association between APC promoter hypermethylation and bladder cancer risk. |
Diagnostic capacity of APC hypermethylation in bladder cancer vs normal controls
The diagnostic value of APC hypermethylation as a potential biomarker in the bladder cancer was assessed. In addition, the pooled sensitivity, specificity, and AUC values of APC hypermethylation were 0.52 (95% CI =0.41–0.63), 0.98 (95% CI =0.90–1.00), and 0.80 (95% CI =0.76–0.83), respectively. The SROC curve with an AUC value and the forest plot of sensitivity and specificity are shown in Figures 6 and 7, respectively. Furthermore, the likelihood ratio scattergram was drawn to evaluate the diagnostic value of APC hypermethylation in the bladder cancer patients and controls (Figure 8).
  | Figure 6 The SROC curve based on APC hypermethylation. |
  | Figure 7 Forest plot of sensitivity and specificity of APC hypermethylation for the diagnosis of bladder cancer. |
Discussion
Promoter hypermethylation of APC gene might result in the downregulation of APC protein expression and increase the cancer risk. It has been reported that the inactivity of APC protein was associated with the accumulation of β-catenin, which affected the growth of cancer cells.38 Previous studies have explored the association between APC promoter hypermethylation and cancer risk, including colorectal cancer, breast cancer, prostate cancer, and non-small-cell lung cancer.39–42 According to the study of breast cancer, APC promoter hypermethylation had a significant association with the grade of breast cancer.43 Meanwhile, the downregulation of APC expression or APC promoter hypermethylation was related to the formation of colorectal cancer.44 Therefore, APC gene, as a well-characterized tumor suppressor gene, was an effective diagnostic factor for various tumors. Although several studies were performed to explore the correlation between APC promoter hypermethylation and bladder cancer risk, no cumulative meta-analysis was performed. Therefore, we conducted this meta-analysis to investigate the potential role of APC promoter hypermethylation in bladder cancer risk.
The results based on the overall pooled OR suggested that the frequency of APC promoter hypermethylation in cases was higher than that in healthy controls. However, evident heterogeneity of included studies was presented in this meta-analysis. As a consequence, subgroup analysis based on sample type and ethnicity was performed to explore the source of heterogeneity. The results showed that ethnicity and sample type contributed a lot to the heterogeneity among studies. In subgroup analysis based on sample type and significant association between APC promoter hypermethylation and bladder cancer pathogenesis was found in tissue sample and urine sample. In previous reports, DNA was extracted from tissue sample and urine sample to detect the level of APC promoter methylation; however, these results often produced false-negative and false-positive results in urine sample compared with tissue sample.27,36 Therefore, the frequency of APC promoter hypermethylation in tissue and urine had a tiny difference, in which the pooled OR was 35.92 in tissue sample and 11.58 in urine sample. The subgroup analysis based on methylation detection methods was not performed since most studies used MSP and only one study applied the detection method of qMSP. Furthermore, subgroup analysis based on ethnicity was conducted, which indicated a significant association between APC promoter hypermethylation and bladder cancer risk in both Caucasians and Asians. The results of sensitivity analysis showed that although many environmental factors and other genetic factors existed, the positive correlation was still robust and stable. The trim-and-fill analysis was performed due to significant publication bias, which showed a significant adjusted OR. In addition, we found that the status of APC promoter hypermethylation in bladder cancer patients with advanced stage was significantly increased compared with early-stage patients. This result demonstrated that APC promoter hypermethylation might be associated with the malignant evolution of bladder cancer. The similar results were found in breast cancer patients.38 As mentioned earlier, the results of four studies indicated a significant association of APC promoter hypermethylation with grade of bladder cancer.25,27,31,37 In addition, three studies detected negative results.24,29,36 From the meta-analysis, we found no significant association between APC promoter hypermethylation and pTNM stage of bladder cancer in Caucasians and Asians. Surprisingly, only P-value of Egger’s linear regression was <0.05, which showed an evident publication bias. Egger et al45 reported that Egger’s test was more sensitive than Begg’s test. The publication bias might result from many factors such as sample type, smoking, source of funding, other genetic alternations, used medicine in the treatment of bladder cancer, and other complications. It is worth mentioning that non-English articles were not included in this meta-analysis, which might partly lead to the publication bias in this meta-analysis. However, the large sample size of included studies ensured the reliability of results and reduced the publication bias. In addition, according to the published articles, we summed up an interaction profiling of proteins with APC protein. These genes might be changed in different degrees in the development of bladder cancer. For instance, the hypermethylation of ARHGEF4 gene promoter was found in the sample of bladder cancer.46 On the basis of the published literatures, KCTD1 decreased the expression of β-catenin in Hela cell, and APC protein affected the process.45 Moreover, some studies also found that genetic variants in the wnt/β-catenin (CTNNB1) signaling pathway have been implicated in the development of bladder cancer.48 So, according to the present result of the meta-analysis and the published articles, these proteins including ARHGEF4, CTNNB1, KCTD1, CTNNA1, KHDRBS1, IQGAP1, and FRAT1 might have close relationships with APC protein and might be involved in the development of bladder cancer.46–51
To explore whether APC hypermethylation is an effective diagnostic marker, diagnostic analysis was also conducted. From the results, significant statistical differences were found in the analysis of sensitivity and specificity. According to the summarized results of likelihood ratio scattergram, the APC hypermethylation was really useful for the diagnosis of bladder cancer. However, APC gene was commonly studied as a tumor suppressor gene in several cancers. Therefore, APC hypermethylation may be a universal biomarker for several cancers. In the future studies, it is very necessary to perform more studies evaluating the associations between APC hypermethylation and the development of some common tumors. In addition, the most important is that we should systematically analyze the role of APC hypermethylation in various cancers.
Although significant association between APC hypermethylation and bladder cancer was found, the present study still had some limitations. First, the sample size in Asian populations was too small to evaluate the association between APC promoter hypermethylation and bladder cancer risk. Second, no environmental factors were included in this study because of few environmental information in original articles. Third, although we collected all data from eligible studies, the combined sample size was still small, which might increase the risk of publication bias and lower the stability of results. Fourth, the methodological quality of included studies was inconsistent, which might result in an obvious heterogeneity among eligible studies. Thus, future large-scale and higher-quality studies, including more environmental information, should be performed.
Conclusion
This was the first meta-analysis to evaluate the association between APC promoter hypermethylation and bladder cancer risk. The current evidence revealed that APC promoter hypermethylation was significantly associated with the risk of bladder cancer. In addition, there was a significant correlation between APC promoter hypermethylation and the grade of bladder cancer. More importantly, we discovered that APC hypermethylation has the potential to be a biomarker to diagnose bladder cancer according to the present meta-analysis. Therefore, more studies with larger size and clinical information are still needed to be performed to confirm these results in the future.
Disclosure
The authors report no conflicts of interest in this work.
References
Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136(5):E359–E386. | ||
Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127(12):2893–2917. | ||
Burger M, Catto JW, Dalbagni G, et al. Epidemiology and risk factors of urothelial bladder cancer. Eur Urol. 2013;63(2):234–241. | ||
Milowsky MI, Kim WY. The geriatrics and genetics behind bladder cancer. Am Soc Clin Oncol Educ Book. 2014:e192–5. | ||
Svatek RS, Hollenbeck BK, Holmäng S, et al. The economics of bladder cancer: costs and considerations of caring for this disease. Eur Urol. 2014;66(2):253–262. | ||
Sternberg CN, Yagoda A, Scher HI, et al. M-VAC (methotrexate, vinblastine, doxorubicin and cisplatin) for advanced transitional cell carcinoma of the urothelium. J Urol. 1988;139(3):461–469. | ||
Cancer Genome Atlas Research Network. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature. 2014;507(7492):315–322. | ||
Williams SV, Hurst CD, Knowles MA. Oncogenic FGFR3 gene fusions in bladder cancer. Hum Mol Genet. 2013;22(4):795–803. | ||
Wu YM, Su F, Kalyana-Sundaram S, et al. Identification of targetable FGFR gene fusions in diverse cancers. Cancer Discov. 2013;3(6):636–647. | ||
Gui Y, Guo G, Huang Y, et al. Frequent mutations of chromatin remodeling genes in transitional cell carcinoma of the bladder. Nat Genet. 2011;43(9):875–878. | ||
Schirosi L, Pellegrino M, Tarantino P, Mauro S, Tinelli A, Greco M. A new germline stop codon mutation in exon 15 of the APC gene predisposing to familial adenomatous polyposis. Int J Biol Markers. 2013;28(4):405–408. | ||
Groden J, Thliveris A, Samowitz W, et al. Identification and characterization of the familial adenomatous polyposis coli gene. Cell. 1991;66(3):589–600. | ||
Baldwin AT, Phillips BT. The tumor suppressor APC differentially regulates multiple β-catenins through the function of axin and CKIα during C. elegans asymmetric stem cell divisions. J Cell Sci. 2014;127(Pt 12):2771–2781. | ||
Lesko AC, Goss KH, Prosperi JR. Exploiting APC function as a novel cancer therapy. Curr Drug Targets. 2014;15(1):90–102. | ||
Liu Q, Li X, Li S, et al. Three novel mutations of APC gene in Chinese patients with familial adenomatous polyposis. Tumour Biol. 2016;37(8):11421–11427. | ||
Liberati A, Altman DG, Tetzlaff J, et al. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate health care interventions: explanation and elaboration. PLoS Med. 2009;6(7):e1000100. | ||
Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol. 2010;25(9):603–605. | ||
Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327(7414):557–560. | ||
Dersimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7(3):177–188. | ||
Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22(4):719–748. | ||
Song F, Gilbody S. Bias in meta-analysis detected by a simple, graphical test. Increase in studies of publication bias coincided with increasing use of meta-analysis. BMJ. 1998;316(7129):471. | ||
Peters JL, Sutton AJ, Jones DR, Abrams KR, Rushton L. Comparison of two methods to detect publication bias in meta-analysis. JAMA. 2006;295(6):676–680. | ||
Reitsma JB, Glas AS, Rutjes AW, Scholten RJ, Bossuyt PM, Zwinderman AH. Bivariate analysis of sensitivity and specificity produces informative summary measures in diagnostic reviews. J Clin Epidemiol. 2005;58(10):982–990. | ||
Pietrusiński M, Kȩpczyński Ƚ, Jȩdrzejczyk A, et al. Detection of bladder cancer in urine sediments by a hypermethylation panel of selected tumor suppressor genes. Cancer Biomark. 2017;18(1):47–59. | ||
Bilgrami SM, Qureshi SA, Pervez S, Abbas F. Promoter hypermethylation of tumor suppressor genes correlates with tumor grade and invasiveness in patients with urothelial bladder cancer. Springerplus. 2014;3:178. | ||
Hauser S, Kogej M, Fechner G, et al. Serum DNA hypermethylation in patients with bladder cancer: results of a prospective multicenter study. Anticancer Res. 2013;33(3):779–784. | ||
Berrada N, Amzazi S, Ameziane El Hassani R, et al. Epigenetic alterations of adenomatous polyposis coli (APC), retinoic acid receptor beta (RARβ) and survivin genes in tumor tissues and voided urine of bladder cancer patients. Cell Mol Biol. 2012;Suppl 58:OL1744–OL1751. | ||
Zuiverloon TC, Beukers W, van der Keur KA, et al. A methylation assay for the detection of non-muscle-invasive bladder cancer (NMIBC) recurrences in voided urine. BJU Int. 2012;109(6):941–948. | ||
Eissa S, Swellam M, El-Khouly IM, et al. Aberrant methylation of RARbeta2 and APC genes in voided urine as molecular markers for early detection of bilharzial and nonbilharzial bladder cancer. Cancer Epidemiol Biomarkers Prev. 2011;20(8):1657–1664. | ||
Chen PC, Tsai MH, Yip SK, et al. Distinct DNA methylation epigenotypes in bladder cancer from different Chinese sub-populations and its implication in cancer detection using voided urine. BMC Med Genomics. 2011;4:45. | ||
Pan J, Chen LW, Wang WW, Qu H, Wang SZ. Clinical Implications of Methylation Status of Apaf-1 and APC Gene in Bladder Carcinoma. J Sun Yat-Sen Univ. 2010;31(3):397–405. | ||
Yates DR, Rehman I, Abbod MF, et al. Promoter hypermethylation identifies progression risk in bladder cancer. Clin Cancer Res. 2007;13(7):2046–2053. | ||
Hoque MO, Begum S, Topaloglu O, et al. Quantitation of promoter methylation of multiple genes in urine DNA and bladder cancer detection. J Natl Cancer Inst. 2006;98(14):996–1004. | ||
Neuhausen A, Florl AR, Grimm MO, Schulz WA. DNA methylation alterations in urothelial carcinoma. Cancer Biol Ther. 2006;5(8):993–1001. | ||
Yates DR, Rehman I, Meuth M, Cross SS, Hamdy FC, Catto JW. Methylational urinalysis: a prospective study of bladder cancer patients and age stratified benign controls. Oncogene. 2006;25(13):1984–1988. | ||
Dulaimi E, Uzzo RG, Greenberg RE, Al-Saleem T, Cairns P. Detection of bladder cancer in urine by a tumor suppressor gene hypermethylation panel. Clin Cancer Res. 2004;10(6):1887–1893. | ||
Serizawa RR, Ralfkiaer U, Steven K, et al. Integrated genetic and epigenetic analysis of bladder cancer reveals an additive diagnostic value of FGFR3 mutations and hypermethylation events. Int J Cancer. 2011;129(1):78–87. | ||
Zhou D, Tang W, Wang W, Pan X, An HX, Zhang Y. Association between aberrant APC promoter methylation and breast cancer pathogenesis: a meta-analysis of 35 observational studies. PeerJ. 2016;4:e2203. | ||
Chen Y, Li J, Yu X, et al. APC gene hypermethylation and prostate cancer: a systematic review and meta-analysis. Eur J Hum Genet. 2013;21(9):929–935. | ||
Sparks AB, Morin PJ, Vogelstein B, Kinzler KW. Mutational analysis of the APC/beta-catenin/Tcf pathway in colorectal cancer. Cancer Res. 1998;58(6):1130–1134. | ||
Ding Z, Jiang T, Piao Y, Han T, Han Y, Xie X. Meta-analysis of the association between APC promoter methylation and colorectal cancer. Onco Targets Ther. 2015;8:211–222. | ||
Nemecek R, Berkovcova J, Radova L, et al. Mutational analysis of primary and metastatic colorectal cancer samples underlying the resistance to cetuximab-based therapy. Onco Targets Ther. 2016;9:4695–4703. | ||
Matsuda Y, Schlange T, Oakeley EJ, Boulay A, Hynes NE. WNT signaling enhances breast cancer cell motility and blockade of the WNT pathway by sFRP1 suppresses MDA-MB-231 xenograft growth. Breast Cancer Res. 2009;11(3):R32. | ||
Ashktorab H, Rahi H, Wansley D, et al. Toward a comprehensive and systematic methylome signature in colorectal cancers. Epigenetics. 2013;8(8):807–815. | ||
Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315(7109):629–634. | ||
Kitchen MO, Bryan RT, Emes RD, et al. Quantitative genome-wide methylation analysis of high-grade non-muscle invasive bladder cancer. Epigenetics. 2016;11(3):237–246. | ||
Li X, Chen C, Wang F, et al. KCTD1 suppresses canonical Wnt signaling pathway by enhancing β-catenin degradation. PLoS One. 2014;9(4):e94343. | ||
Pierzynski JA, Hildebrandt MA, Kamat AM, et al. Genetic Variants in the Wnt/β-Catenin Signaling Pathway as Indicators of Bladder Cancer Risk. J Urol. 2015;194(6):1771–1776. | ||
Zhang Z, Yu C, Li Y, Jiang L, Zhou F. Utility of SAM68 in the progression and prognosis for bladder cancer. BMC Cancer. 2015;15(15):364. | ||
Watanabe T, Wang S, Noritake J, et al. Interaction with IQGAP1 links APC to Rac1, Cdc42, and actin filaments during cell polarization and migration. Dev Cell. 2004;7(6):871–883. | ||
Jin R, Liu W, Menezes S, et al. The metastasis suppressor NDRG1 modulates the phosphorylation and nuclear translocation of β-catenin through mechanisms involving FRAT1 and PAK4. J Cell Sci. 2014;127(Pt 14):3116–3130. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.