Back to Journals » Clinical Ophthalmology » Volume 11
Investigation of associations of ARMS2, CD14, and TLR4 gene polymorphisms with wet age-related macular degeneration in a Greek population
Authors Sarli A, Skalidakis I, Velissari A, Koutsandrea C, Stefaniotou M, Petersen MB, Kroupis C , Kitsos G, Moschos MM
Received 12 February 2017
Accepted for publication 13 June 2017
Published 26 July 2017 Volume 2017:11 Pages 1347—1358
DOI https://doi.org/10.2147/OPTH.S134538
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Scott Fraser
Antonia Sarli,1 Iosif Skalidakis,2 Aliki Velissari,1 Chryssanthi Koutsandrea,2 Maria Stefaniotou,3 Michael B Petersen,4,5 Christos Kroupis,1,* George Kitsos,3,* Marilita M Moschos2,*
1Department of Clinical Biochemistry, Attikon University General Hospital, 21st Department of Ophthalmology, “G. Gennimatas” General Hospital, Medical School, National and Kapodistrian University of Athens, Athens, 3Department of Ophthalmology, Ioannina University General Hospital, University of Ioannina, Ioannina, Greece; 4Department of Clinical Genetics, Aalborg University Hospital, Aalborg, Denmark; 5Department of Clinical Medicine, Aalborg University Hospital, Aalborg, Denmark
*These authors contributed equally to this work
Background: Age-related macular degeneration (AMD) is a multifactorial degenerative ocular disease that leads to loss of central vision. Functional gene polymorphisms have already been associated with the disease (for example, ARMS2 A69S, rs10490924).
Aim: The goal of our study was to verify the correlation of the aforementioned ARMS2 variation with the disease, to examine, for the first time, the role of the CD14 C260T variation (rs2569190), and to investigate the association of two TLR4 polymorphisms (Asp299Gly or rs4986790 and Thr399Ile or rs4986791) in a Greek population with the wet form of AMD.
Patients and methods: Genomic DNAs were isolated from blood samples of 103 healthy controls and 120 Greek patients with wet AMD who were age- and sex-matched, and all of whom were clinically evaluated. For the genotyping of all selected polymorphisms, polymerase chain reaction–restriction fragment length polymorphism analysis was performed.
Results and conclusions: This study confirmed the association between the ARMS2 variation and AMD, detecting the T risk allele in a significantly higher frequency in the patient group, compared with the control subjects (45% vs 29.13%, P<0.001, odds ratio [OR] 1.99, confidence interval 1.34–2.95). For the CD14 polymorphism, no statistically significant correlation was observed. As for the TLR4 polymorphisms, the percentage of heterozygotes increased from 2.9% to 11.7% in the patient population for Asp299Gly and from 1.9% to 10% for the Thr399Ile polymorphism (ORs 4.40 [P=0.01] and 5.61 [P=0.0088], respectively). Although our ARMS2 and CD14 results provided definite conclusions, the role of innate immunity TLR4 gene awaits further investigation in larger AMD populations with more clinical data collected on past microbial infections.
Keywords: AMD, ARMS2, CD14, TLR4, SNPs, PCR-RFLP
Introduction
Age-related macular degeneration (AMD) is a multifactorial, heterogeneous, degenerative disorder of the human eye that affects patients over 50 years of age and can lead to severe loss of central vision.1 It is considered to be the leading cause of irreversible blindness in Western societies and, given the fact that life expectancy is constantly rising, the prevalence of the disease is expected to increase significantly.2 Clinically, two forms of AMD are recognized: the dry form, which affects 80% of the patients and the more severe and wet form, which affects the remaining 20%. Choroidal neovascularization (CNV) is a typical sign of wet AMD.3
AMD is a highly complex disease resulting from interactions between genetic susceptibility, aging, oxidative damage due to a surplus of oxygen and light in the eye, environmental influences, and underlying diseases.4,5 Initially through family-based and then, with genome-wide association studies the strong genetic component of the disease has been verified (eg, in the most recent 2016 study with the largest number of patients and healthy controls so far), 52 single-nucleotide polymorphisms (SNPs) in 34 genes have been associated with the disease with a P-value <10−8.6 However, since 2005, two loci stand out in most studies, and their polymorphisms have been verified in most populations and are associated with the highest odds ratios (OR) depending on the population:5,7 the CFH gene is involved in the alternative pathway regulation with the Y402H polymorphism (rs1061170)8,9 and a genomic locus at chromosome 10q26 (LOC387715) where the ARMS2 gene is located. The ARMS2 gene encodes for a small 107-amino acid 12 kDa protein and A69S SNP (rs10490924) in exon 1 of the ARMS2 gene, resulting from a G to T change (NM_001099667.1:c205G>T), is a mutation associated with subsequent mitochondrial dysfunction, generation of reactive oxygen species (ROS) and accumulation of somatic mutations in mitochondrial DNA.10 According to a meta-analysis study, TT mutant homozygotes carry a 7.5-fold increased AMD risk, and the TG heterozygotes carry a 2.4-fold increased AMD risk when compared to the wild-type GG homozygotes.11 However, there is an ongoing debate whether it is this gene or the adjacent HTRA1 gene that shows true association.12 Evidence for the ARMS2 gene include reverse-transcriptase polymerase chain reaction (PCR) experiments – which have demonstrated that ARMS2 variant transcripts are expressed in the ellipsoid region of the photoreceptors in the human retina, where the majority of mitochondria are located – and immunohistochemistry experiments, showing co-localization with a mitochondrial marker, anti-MTCO2.13,14
Other genes that are involved in inflammation, immune response, cholesterol transport, ubiquitin proteolytic system, and oxidative stress seem to play a key role in the development of AMD.5,12 Therefore, SNPs in genes that are involved in the aforementioned conditions are worthy of investigation.
Toll-like receptors (TLRs) are members of a family that recognizes conserved microbial components in pathogens and contributes to the activation of immune response.15 TLR4 is located on chromosome 9q33.1, contains 4 exons that encode an 839 amino acid protein of 96 kDa molecular mass. It is highly expressed on lymphocytes, monocytes, and neutrophils and mediates for lipopolysaccharide (LPS) recognition. This interaction requires the presence of CD14-MD2 co-receptors and produces proinflammatory cytokines through adaptor molecules (eg, MAL, MyD88).16 Allelic variations such as Asp299Gly, resulting from an A to G change (rs4986790, NM_138554.4:c.896A>G), and Thr399Ile, resulting from a C to T change (rs4986791, NM_138554.4:c.1196C>T), have been associated with atherosclerosis, cardiovascular disease, and other inflammatory diseases;17–20 the role of these TLR4 gene polymorphisms in AMD susceptibility was first mentioned in Zareparsi et al.21 The frequencies of Gly and Ile amino acids in the aforementioned SNPs were found to be increased in patients, compared to controls, indicating that carriers of the changed amino acids had a 2.0- and 2.4-fold increased risk of developing AMD, respectively.
The co-receptor of TLR4, CD14, is a glycosylphosphatidylinositol (GPI) receptor that mediates the inflammatory response to bacterial products by binding low doses of LPS22 present and transfer it to the TLR4–MD2 complex in order to initiate the transduction of the NF-κB pathway.23,24 The CD14 gene is located on chromosome 5q31.1, contains three exons, and encodes for a 356 amino-acid glycoprotein that is expressed mainly in the liver and is anchored to the cellular membrane through the GPI linkage. CD14 exists in two forms – the membrane-expressed (mCD14) present on the surface of mature myeloid cells and macrophages, as well as the soluble molecule (sCD14) that mediates LPS activation of epithelial cells.25 The allelic variant C260T (rs2569190, NM_000591.3:c-260 C to T change at position −260 to the start codon) in the region of the gene promoter is associated with enhanced transcriptional activity and results in a higher density of the receptor.18,22,26
Thus far, no other study has examined the role of CD14 gene in AMD, to the best of our knowledge. Considering its role in innate immunity and its role in cooperation with TLR4, we examined in this study, for the first time, whether C260T variation in CD14 gene is correlated with AMD, and we investigated TLR4 Asp299Gly and Thr399Ile variations in a Greek population with the wet AMD form. In the same population, we additionally tried to verify the association of A69S variation in the ARMS2 gene with AMD.
Patients and methods
Patients
The study was conducted in a cohort of 103 healthy controls and 120 Greek patients, with the wet form of AMD. Blood samples were collected in EDTA tubes in Athens “G. Gennimatas” General Hospital and Ioannina University General Hospital (40-patient enrollment) during a 4-year period (2010–2013) and stored in −20°C until DNA extraction. All participants were selected after ophthalmologic evaluation and all clinical data were collected on cataract surgery and smoking habits, and conditions such as glaucoma, diabetes, arterial hypertension, and heart disease that, wherever observed, were being treated with an appropriate medication regimen (Table 1). Ophthalmologic evaluation included visual acuity, ocular pressure measurement, slit-lamp anterior and posterior segment examination, optical coherence tomography (OCT), and fundus angiography (fluorescein or indocyanine). Inclusion criteria were: CNV in the fovea/perifoveal area due to AMD, active leakage of the new choroidal blood vessels, and decreased visual acuity. Exclusion criteria were: age <50 years, Snellen visual acuity better than 7/10 or worse than 1/10, and any other pathology leading to CNV (eg, angioid streaks, high myopia, presumed ocular histoplasmosis syndrome, choroidal rupture). The samples were obtained after approvals from the “G. Gennimatas” General Hospital and Ioannina University General Hospital ethics committees and after obtaining a signed informed consent from each participant.
  | Table 1 Clinical characteristics of all 223 participants in the study |
Genomic DNA isolation
Genomic DNA was isolated from 200 μL blood by the NucleoSpin Blood kit (Macherey-Nagel, Duren, Germany) according to the manufacturer’s instructions. DNA purity and quantity were determined by fluorescence readings with the use of Quant-iT dsDNA-BR kit in the Qubit 1.0 fluorometer (ThermoFisher Invitrogen, Waltham, MA, USA).
Analysis of ARMS2-A69S, CD14-C260T, TLR4-Asp299Gly, and Thr399Ile gene polymorphisms
For the genotyping of all selected polymorphisms, PCR – restriction fragment length polymorphism analysis was performed on a fragment amplified using primers from the literature27,28 (Table S1). Primers were synthesized by IDT (Integrated DNA Technologies, Inc. Coralville, IA, USA).
ARMS2 (A69S) genotyping
The PCR was performed in 0.2 mL tubes in a Primus 25 Advanced PCR engine (PEQLAB Biotechnologie GmbH, Erlangen, Germany). The amplification mixture of a total 20 μL volume included 1 μL of 20 pmol/μL LOC1-forward primer and 1 μL of 20 pmol/μL LOC2-reverse primer, 1× Go Taq Green Master Mix (Promega, USA) and ~2 μL of genomic DNA (≥50 ng). Sterile water was used to supplement up to 20 μL. The cycling protocol for ARMS2 comprised a pre-incubation step at 94°C for 10 min hot-start polymerase activation, followed by 40 cycles of denaturation at 94°C for 30 s, annealing at 56°C for 30 s, extension at 72°C for 45 s, and a final extension step at 72°C for 7 min.
CD14 (C260T) genotyping
The PCR protocol for the C260T polymorphism was the same as described earlier (with CD14_F and CD14_R primers instead); the cycling conditions were: initial denaturation at 95°C for 5 min, followed by 35 cycles at 92°C for 40 s, 62°C for 35 s, and 72°C for 50 s. The final extension step was prolonged to 5 min.
TLR4 (Asp299Gly and Thr399Ile) genotyping
The PCR protocol was analogous as mentioned previously (with TLR4 primers 1–2 and 3–4, respectively) and cycling conditions: 95°C for 5 min followed by 35 cycles at 95°C for 30 s, 55°C for 30 s, 72°C for 30 s, and a final incubation at 72°C for 5 min.
Restriction enzyme incubations
After confirming proper amplifications by running 5 μL PCR products in a 1.5% agarose gel, the rest of the PCR products (15 μL) were digested with the appropriate restriction enzymes for 4 h at 37°C.
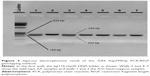
After PvuII digestion (ThermoFisher Fermentas, Lithuania), the 449-bp PCR amplicon for A69S in the ARMS2 gene was cut into fragments of 259 and 190 bp in the case of the wild-type G allele whereas it was left uncut in the case of the altered (mutant) T allele (449 bp). Therefore, a G/T heterozygote sample was cut into three fragments of 449, 259, and 190 bp (Figure 1).
The 561-bp amplified region for CD14 was digested with HaeIII restriction enzyme (NEB, USA). The wild-type C allele separated into fragments of 204, 201 (run as one band), and 156 bp. The T minor allele showed a loss of one HaeIII cleavage site, resulting in the presence of fragments of 360 and 201 bp. Therefore the C/T heterozygote was cut into three fragments of 360, 204/201, and 156 bp (Figure 2).
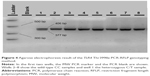
With regard to the TLR4 amplicons after digestion with the NcoI (Takara, Japan) restriction enzyme for the 299 residue and with HinfI (NEB, USA) for the 399 residue, fragment sizes for carriers of the polymorphic allele decreased from 249 (wild-type) to 223 bp for the 299 residue and from 406 (wild-type) to 377 bp for the 399 residue (Figures 3 and 4). All restricted amplicons were analyzed by 3% agarose gel electrophoresis (2:1 Nusieve/Seakem, Lonza, Basel, Switzerland), visualized by ethidium bromide and sized by a MW marker (PCR Marker or φχ174 HaeIII, both New England Biolabs, Ipswich, MA, USA).
DNA sequencing
For the verification of the PCR-restriction fragment length polymorphism results, the gold standard method of DNA sequencing was used for method comparison (Figures S1–S3). After purification of the amplicons of conventional PCR (High Pure PCR Cleanup Micro kit, Roche Applied Science, Penzberg, Germany), cycle sequencing reaction was performed with the Big Dye 1.1 reagent in both directions with the use of either the forward or the reverse primer (ThermoFisher Scientific, Waltham, MA, USA); 10 μL of the purified cycle sequencing reactions (by NucleoSeq columns, Macherey-Nagel, Germany) were heated at 95°C for 2 min and cooled immediately at 4°C for 2 min with 10 μL formamide and then run in capillaries of the ABI Prism 310 Genetic Analyzer. For the analysis of DNA sequencing electropherograms, the Chromas 2.01 software was used (Technelysium Pty Ltd, Brisbane, QLD, Australia) and results were compared with the expected gene sequences with the NCBI BLAST (https://blast.ncbi.nlm.nih.gov).
Statistical analysis
Adequacy of the number of total samples and statistical power for all χ2 tests were estimated with the G*Power 3.1.9.2 software.29
Genotyping statistical analysis was performed through the SNPStats Internet platform (http://bioinfo.iconcologia.net/snpstats/start.htm).30 The variants were tested for Hardy–Weinberg equilibrium (HWE) in either patients with AMD or controls for each studied SNP, and OR and 95% confidence intervals (CIs) for all genotypes were calculated for all inheritance models (dominant, co-dominant, recessive, or log-additive). The one with the lower Akaike information criterion was preferred.30 Furthermore, haplotype analysis and linkage disequilibrium estimation was performed for the two variants of TLR4 gene.
Analysis of the association between AMD and risk factors (and genotypes in the multivariate model) was performed with the SPSS statistical software (version 21.0, IBM, Armonk, NY, USA). Age distribution was tested initially for normality with the Kolmogorov–Smirnov test and, subsequently, the Mann–Whitney U-test for median comparison was used. For gender and for clinical and environmental categorical variables, comparisons between percentages of groups were performed with the χ2 test. To evaluate the risk of developing AMD, binary logistic regression analysis was adjusted for the presence of specific polymorphisms, age, sex, visual acuity, smoking habits, diabetes, glaucoma, surgical cataract, hypertension, and heart disease status. All tests of significance were two-sided and P-values <0.05 were considered statistically significant.
Results
The total number of samples provided sufficient statistical power (92%) to even detect medium-sized effects (0.3) in all χ2-tests performed as estimated by the G*Power software. In the group of patients with wet AMD, 20 subjects (16.7%) also had geographic atrophy. Age distribution was not normal in patients and controls; therefore, medians were compared between the two groups and no significance difference was detected; the median age of the patients was 77 years and, for the controls, it was 78 (interquartile range 73–81 and 75–82), respectively. Sex distribution was the same: males comprised 55% of the patients and 57% of the controls (P>0.05). When all covariates were studied, in two parameters, glaucoma and diabetes, a reduction was observed in patients with AMD (Table 1).
All genotypes in all genes were in HWE equilibrium for both controls and patients (P>0.05). The ARMS2 genetic variation and its distribution in the studied cohort are shown in Table 2. The T risk allele was detected in 108 of 240 patient total alleles; in a much higher frequency (45%) than in control subjects (29.13%, 60 out of 206 control alleles, P<0.001, odds ratio [OR] 1.99, 95% CI 1.34−2.95) and TT homozygotes doubled from controls to patients with AMD (9.70%−20.8%). ORs and levels of significance for each one of all possible inheritance models were calculated and a robust statistically significant correlation was found a between AMD and the A69S SNP. In the model with the lower Akaike information criterion, the log-additive model, the calculated OR is 1.94 (95% CI 1.31−2.88).
The association for C260T variation in the CD14 gene did not reach any statistical significance (P=0.14) and is shown in Table 3.
The Asp299Gly and Thr399Ile polymorphisms in the TLR4 gene are rare in the total population, but certainly, the heterozygotes are quite enriched in the patient population (Table 4): from 2.9% in controls to 11.7% in patients in the first SNP and, in the latter, from 1.9% in controls to 10% in patients. No homozygote was detected in both polymorphisms (GG in 299 and TT in the 399 TLR4 polymorphisms respectively). The calculated ORs were 4.40 (P=0.01) and 5.61 (P=0.0088), respectively (but with broad CIs). Haplotype analysis was performed only for the TLR4 gene (Table 5) because both genetic variations are located on the same chromosome and showed that the risk GT haplotype is again correlated with the disease (OR =8.66, P=0.045), but the two polymorphisms are not linked (r2=0.398, not shown). No difference in the percentage of genotype distribution in all studied genes exists within the subgroups of the clinical variables (all χ2 tests with P>0.05).
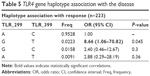  | Table 5 TLR4 gene haplotype association with the disease |
When examining the effect of all significant SNPs determined in this study (ARMS2 and both SNPs of the TLR4 genes) and all clinical variables with binary logistic regression for the prediction of AMD, it was found that, among the three genes, only ARMS2 shows a strong effect on the development of AMD due to the rarity of TLR4 risk alleles (Table 6). If we also include the results for CFH and FCGR2A SNPs in the same population that were obtained from our previous study,31 these two SNPs and ARMS2 remain strong in the model of AMD prediction, albeit with reduced ORs (Table S2).
Concerning the clinical variables, the majority have no effect on the development and progression of the disease, except for glaucoma and diabetes that seem protective not only as shown in the logistic regression of Table 6 but also as expected from the χ2 tests of Table 1.
Discussion
AMD is a complex, degenerative disease of the human eye that apparently fits the multi-hit “threshold model”.5 Numerous defects in genes affecting chronic inflammation, lipid transfer and recycling, response to oxidative stress, proteasome system functionality and extracellular matrix remodeling will eventually lead to drusen creation and Bruch’s membrane breakdown and take their toll on photoreceptors and retinal pigment epithelial cells. The progress toward geographic atrophy and/or CNV will depend on the content of these hits.4,5,12 However, even in the latest and largest of the genetic association studies, there is still an estimate of about 42% “missing heritability” (in the highest disease prevalence group of people above 85 years of age).6 Therefore, it is worthwhile investigating every plausible lead and testing it in various populations because minor allele frequencies and corresponding OR can differ significantly as seen, for example, in the CFH Y402H polymorphism between Caucasian and Asian populations.5,7
So far, in the Greek population genetic studies exist on ARMS2, CFH, FCGR2A, C3, C2, and BF genes,31–33 but there were no data on TLR4 (Asp299Gly and Thr399Ile) and CD14 (C260T) allele frequencies in Greek patients with AMD. Both genes operate in the same pathway of inflammatory response, and the TLR4 variations Asp299Gly and Thr399Ile affect the extracellular domain of the corresponding protein and result in a differential response to LPS.34 In this study, findings for the aforementioned SNPs and investigation of their association with AMD susceptibility are reported. No other study has examined the role of the CD14 SNP in AMD so far. The study was implemented on 103 control subjects and 120 Greek patients with wet AMD who were all evaluated with a full ocular examination. Our results showed no association with the CD14 SNP but demonstrated that both TLR4 polymorphisms are linked with an increased risk of wet AMD with ORs (95% CI), 4.40 (1.23–15.78) and 5.61 (1.23–25.68), respectively. In this way, our study corroborates the results from Zareparsi et al’s investigation.21 However, there exist other and larger studies that have presented contradictory conclusions concerning the association between TLR4 variants and AMD.6,35,36 Our TLR4 findings in our control subjects are within the variation seen in various populations for these rare SNPs (data from dbSNP and ExAC browsers); however, our AMD well-ascertained population showed differences that cannot be explained unless there is a particular patient subpopulation. As TLR4 has a role in triggering innate immune defense against microbial agents, a limitation of our study might be the lack of clinical data concerning past infections, especially those that affected the eyes of some of our patients. This argument awaits further investigation in larger sample sizes in the future.
Concerning the ARMS2 A69S variation, our results verify its association with AMD in any of the inheritance models as seen in many other studies worldwide.10,11 In our study, the association reaches a P-value of 7×10−4 in the log-additive model with an OR of 1.94 (95% CI 1.31−2.88).
The binary logistic regression analysis including all the studied polymorphisms and all clinical parameters implicate that the ARMS2 A69S polymorphism remains a strong, independent, and provisional factor for the development of the disease, whereas the two TLR4 polymorphisms show weak predictability due to the rareness of these variations.
Furthermore, taking into consideration the results from our previous study on AMD that includes the CFH and FCGR2A genes31 and the clinical and environmental factors, now we have a much broader view of the effect that all genes have on the development of wet AMD (Table S2). The strong genetic component in AMD is clearly demonstrated when only genes remain as strong predictors: in the logistic regression model, the ARMS2 gene shows the strongest association with the disease followed by FCGR2A – an innate immunity gene studied for the first time in AMD from our research team31 and, finally, the CFH gene, another established factor. Similar study for the aforementioned genes must be replicated for the dry form of AMD as well. Our results from both studies highlight the role of innate immunity and it will be of interest to investigate the relationship between AMD and other genes that are involved in this inflammatory response in order to achieve a comprehensive picture of the genetic background.
So far, results from genetic analysis of patients with AMD have had limited clinical utility: ARMS2 risk alleles show higher OR toward CNV progression compared to CFH risk alleles37 whereas both SNPs are used in prediction models of progression to advanced AMD forms with or without body mass index and smoking.38 Moreover, there have been numerous conflicting reports with regard to their use in selecting nutritional supplementation therapy based on the number of ARMS2 and/or CFH risk alleles.39,40
Toward this direction of incorporating genetic findings in AMD clinical practice, the novel next-generation sequencing (NGS) approach will be useful, because this technique allows the targeted massive parallel sequencing of several genes in a large number of samples altogether in a cost-effective way, because each specimen can be identified by a unique “barcode”. Genetically, AMD is highly heterogeneous and offers an ideal model in which to investigate its complexities with NGS. There are already several NGS studies in AMD that show promising results in this field, and it seems that this technique will increase the use of molecular diagnostics in AMD and the identification of rare causative variants.41,42 Besides the analysis of genomic DNA, another approach that shows promise in AMD is the miRNA analysis in peripheral blood. miRNAs are small single-stranded non-coding RNA molecules containing 19–25 nucleotides that can recognize sequences in the 3′untranslated region of selected mRNAs and result in their translational repression or even cleavage. They can target multiple genes, and some of them are already implicated in ocular AMD.43 Several groups have already identified profiles of miRNAs in plasma of peripheral blood of patients with AMD and can even differentiate dry from wet cases.44,45 miRNAs in peripheral blood demonstrate stability and are not susceptible to RNAase degradation. Therefore, both DNA gene SNPs and miRNAs in peripheral blood could become very useful as biomarkers in the prediction or progression of AMD disease in the near future.
Disclosure
The authors report no conflicts of interest in this work.
References
Katta S, Kaur I, Chakrabarti S. The molecular genetic basis of age-related macular degeneration: an overview. J Genet. 2009;88(4):425–449. | ||
Gehrs KM, Anderson DH, Johnson LV, Hageman GS. Age-related macular degeneration – emerging pathogenetic and therapeutic concepts. Ann Med. 2006;38(7):450–471. | ||
Chakravarthy U, Wong TY, Fletcher A, et al. Clinical risk factors for age-related macular degeneration: a systematic review and meta-analysis. BMC Ophthalmol. 2010;10:31. | ||
Chiras D, Kitsos G, Petersen MB, Skalidakis I, Kroupis C. Oxidative stress in dry age-related macular degeneration and exfoliation syndrome. Crit Rev Clin Lab Sci. 2015;52(1):12–27. | ||
Fritsche LG, Fariss RN, Stambolian D, Abecasis GR, Curcio CA, Swaroop A. Age-related macular degeneration: genetics and biology coming together. Annu Rev Genomics Hum Genet. 2014;15:151–171. | ||
Fritsche LG, Igl W, Bailey JN, et al. A large genome-wide association study of age-related macular degeneration highlights contributions of rare and common variants. Nat Genet. 2016;48(2):134–143. | ||
Tan PL, Bowes RC, Katsanis N. AMD and the alternative complement pathway: genetics and functional implications. Hum Genomics. 2016;10(1):23. | ||
Edwards AO, Ritter R 3rd, Abel KJ, Manning A, Panhuysen C, Farrer LA. Complement factor H polymorphism and age-related macular degeneration. Science. 2005;308(5720):421–424. | ||
Klein RJ, Zeiss C, Chew EY, et al. Complement factor H polymorphism in age-related macular degeneration. Science. 2005;308(5720):385–389. | ||
Rivera A, Fisher SA, Fritsche LG, et al. Hypothetical LOC387715 is a second major susceptibility gene for age-related macular degeneration, contributing independently of complement factor H to disease risk. Hum Mol Genet. 2005;14(21):3227–3236. | ||
Tong Y, Liao J, Zhang Y, Zhou J, Zhang H, Mao M. LOC387715/HTRA1 gene polymorphisms and susceptibility to age-related macular degeneration: A HuGE review and meta-analysis. Mol Vis. 2010;16:1958–1981. | ||
Gorin MB. Genetic insights into age-related macular degeneration: controversies addressing risk, causality, and therapeutics. Mol Aspects Med. 2012;33(4):467–486. | ||
Fritsche LG, Loenhardt T, Janssen A, et al. Age-related macular degeneration is associated with an unstable ARMS2 (LOC387715) mRNA. Nat Genet. 2008;40(7):892–896. | ||
Kanda A, Chen W, Othman M, et al. A variant of mitochondrial protein LOC387715/ARMS2, not HTRA1, is strongly associated with age-related macular degeneration. Proc Natl Acad Sci U S A. 2007;104(41):16227–16232. | ||
Cook DN, Pisetsky DS, Schwartz DA. Toll-like receptors in the pathogenesis of human disease. Nat Immunol. 2004;5(10):975–979. | ||
Akira S, Sato S. Toll-like receptors and their signaling mechanisms. Scand J Infect Dis. 2003;35(9):555–562. | ||
Kiechl S, Lorenz E, Reindl M, et al. Toll-like receptor 4 polymorphisms and atherogenesis. N Engl J Med. 2002;347(3):185–192. | ||
Vainas T, Stassen FR, Bruggeman CA, et al. Synergistic effect of Toll-like receptor 4 and CD14 polymorphisms on the total atherosclerosis burden in patients with peripheral arterial disease. J Vasc Surg. 2006;44(2):326–332. | ||
Konstantinidou MK, Goutas N, Vlachodimitropoulos D, et al. TLR-4 and CD14 genotypes and soluble CD14: could they predispose to coronary atherosclerosis? J Cardiovasc Dev Dis. 2016;3(1):9. | ||
Savva A, Kanni T, Damoraki G, et al. Impact of Toll-like receptor-4 and tumour necrosis factor gene polymorphisms in patients with hidradenitis suppurativa. Br J Dermatol. 2013;168(2):311–317. | ||
Zareparsi S, Buraczynska M, Branham KE, et al. Toll-like receptor 4 variant D299G is associated with susceptibility to age-related macular degeneration. Hum Mol Genet. 2005;14(11):1449–1455. | ||
Zanoni I, Granucci F. Role of CD14 in host protection against infections and in metabolism regulation. Front Cell Infect Microbiol. 2013;3:32. | ||
Gioannini TL, Teghanemt A, Zhang D, et al. Isolation of an endotoxin-MD-2 complex that produces Toll-like receptor 4-dependent cell activation at picomolar concentrations. Proc Natl Acad Sci U S A. 2004;101(12):4186–4191. | ||
Zanoni I, Ostuni R, Marek LR, et al. CD14 controls the LPS-induced endocytosis of Toll-like receptor 4. Cell. 2011;147(4):868–880. | ||
Antal-Szalmás P. Evaluation of CD14 in host defence. Eur J Clin Invest. 2000;30(2):167–179. | ||
LeVan TD, Bloom JW, Bailey TJ, et al. A common single nucleotide polymorphism in the CD14 promoter decreases the affinity of Sp protein binding and enhances transcriptional activity. J Immunol. 2001;167(10):5838–5844. | ||
Lorenz E, Frees KL, Schwartz DA. Determination of the TLR4 genotype using allele-specific PCR. Biotechniques. 2001;31(1):22–24. | ||
Hubacek JA, Rothe G, Pit’ha J, et al. C(-260)→T polymorphism in the promoter of the CD14 monocyte receptor gene as a risk factor for myocardial infarction. Circulation. 1999;99(25):3218–3220. | ||
Faul F, Erdfelder E, Lang AG, Buchner A. G*Power 3: a flexible statistical power analysis program for the social, behavioral, and biomedical sciences. Behav Res Methods. 2007;39(2):175–191. | ||
Solé X, Guinó E, Valls J, Iniesta R, Moreno V. SNPStats: a web tool for the analysis of association studies. Bioinformatics. 2006;22(15):1928–1929. | ||
Velissari A, Skalidakis I, Oliveira SC, et al. Novel association of FCGR2A polymorphism with age-related macular degeneration (AMD) and development of a novel CFH real-time genotyping method. Clin Chem Lab Med. 2015;53(10):1521–1529. | ||
Marioli DI, Pharmakakis N, Deli A, Havvas I, Zarkadis IK. Complement factor H and LOC387715 gene polymorphisms in a Greek population with age-related macular degeneration. Graefes Arch Clin Exp Ophthalmol. 2009;247(11):1547–1553. | ||
Havvas I, Marioli DI, Deli A, Zarkadis IK, Pharmakakis N. Complement C3, C2, and factor B gene polymorphisms and age-related macular degeneration in a Greek cohort study. Eur J Ophthalmol. 2014;24(5):751–760. | ||
Arbour NC, Lorenz E, Schutte BC, et al. TLR4 mutations are associated with endotoxin hyporesponsiveness in humans. Nat Genet. 2000;25(2):187–191. | ||
Cho Y, Wang JJ, Chew EY, et al. Toll-like receptor polymorphisms and age-related macular degeneration: replication in three case-control samples. Invest Ophthalmol Vis Sci. 2009;50(12):5614–5618. | ||
Edwards AO, Chen D, Fridley BL, et al. Toll-like receptor polymorphisms and age-related macular degeneration. Invest Ophthalmol Vis Sci. 2008;49(4):1652–1659. | ||
Fritsche LG, Chen W, Schu M, et al; AMD Gene Consortium. Seven new loci associated with age-related macular degeneration. Nat Genet. 2013;45(4):433–439. | ||
Seddon JM, Francis PJ, George S, Schultz DW, Rosner B, Klein ML. Association of CFH Y402H and LOC387715 A69S with progression of age-related macular degeneration. JAMA. 2007;297(16):1793–1800. | ||
Awh CC, Hawken S, Zanke BW. Treatment response to antioxidants and zinc based on CFH and ARMS2 genetic risk allele number in the Age-Related Eye Disease Study. Ophthalmology. 2015;122(1):162–169. | ||
Chew EY, Klein ML, Clemons TE, Agrón E, Abecasis GR. Genetic testing in persons with age-related macular degeneration and the use of the AREDS supplements: to test or not to test? Ophthalmology. 2015;122(1):212–215. | ||
Ratnapriya R, Swaroop A. Genetic architecture of retinal and macular degenerative diseases: the promise and challenges of next-generation sequencing. Genome Med. 2013;5(10):84. | ||
Yu Y, Triebwasser MP, Wong EK, et al. Whole-exome sequencing identifies rare, functional CFH variants in families with macular degeneration. Hum Mol Genet. 2014;23(19):5283–5293. | ||
Hill JM, Pogue AI, Lukiw WJ. Pathogenic microRNAs common to brain and retinal degeneration; recent observations in Alzheimer’s disease and age-related macular degeneration. Front Neurol. 2015;6:232. | ||
Szemraj M, Bielecka-Kowalska A, Oszajca K, et al. Serum microRNAs as potential biomarkers of AMD. Med Sci Monit. 2015;21:2734–2742. | ||
Ertekin S, Yildirim O, Dinç E, Ayaz L, Fidanci SB, Tamer L. Evaluation of circulating miRNAs in wet age-related macular degeneration. Mol Vis. 2014;20:1057–1066. |
Supplementary materials
  | Figure S1 DNA sequencing analysis of a wild-type GG, a heterozygote G/T, and a mutant TT around the ARMS2 A69S SNP area (forward primer). |
  | Figure S2 DNA sequencing analysis of a wild-type GG, a heterozygote G/T, and a mutant TT around the CD14 C260T SNP area (reverse primer). |
  | Figure S3 DNA sequencing analysis of a wild-type AA and a heterozygote A/G around the TLR4 Asp299Gly SNP area (forward primer). |
 © 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.