Back to Journals » Infection and Drug Resistance » Volume 14
Intrapatient Development of Multi-Class Drug Resistance in an Individual Infected with HIV-1 CRF01_AE
Authors Peng X, Xu Y, Huang Y, Zhu B
Received 21 June 2021
Accepted for publication 6 August 2021
Published 25 August 2021 Volume 2021:14 Pages 3441—3448
DOI https://doi.org/10.2147/IDR.S323762
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Suresh Antony
Xiaorong Peng,* Yufan Xu,* Ying Huang, Biao Zhu
State Key Laboratory for Diagnosis and Treatment of Infectious Diseases, The First Affiliated Hospital, School of Medicine, Zhejiang University, Hangzhou, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Biao Zhu
State Key Laboratory for Diagnosis and Treatment of Infectious Diseases, The First Affiliated Hospital, School of Medicine, Zhejiang University, Hangzhou, People’s Republic of China
Email [email protected]
Abstract: The rapid expansion of access to antiretroviral therapy (ART) has led to the emergence of multi-class drug resistance (MDR) in people living with HIV (PLWH). However, the viral evolutionary dynamics of the development of MDR has not been well documented. For this study, plasma and peripheral blood mononuclear cells (PBMC) were longitudinally collected at different time points from a PLWH who suffered several periods of ART failure. Next generation sequencing (NGS) was used to analyze the distribution and percent of drug resistance mutations in PBMC and plasma. The results showed the gradual replacement of the wild type protease and integrase genotype by protease inhibitors (PI) and integrase strand transfer inhibitor (INSTI) drug resistant mutations when patient’s ART regimen was changed – driving the increase of genetic variability in HIV DNA. Sampling for this study was initiated after the patient was first diagnosed with ART failure, five years after ART treatment was first initiated. By that time, mutants resistant to the reverse transcriptase inhibitor nevirapine (NVP) had already replaced almost 100% of wild type. After the introduction of the protease inhibitor lopinavir/ritonavir (LPV/r) to the patient’s ART, resistant protease inhibitor (PI) mutants developed slowly. After one month, none were found in PMBC DNA; after sixteen months, less than 20% were mutants; and after three years (two months prior to the patient’s death) PI mutants were still under 50%. However, integrase strand transfer inhibitor (INSTI) mutations evolved much more quickly, replacing approximately 75% of the wild genotype in HIV DNA one year after addition of the integrase inhibitor raltegravir to the patient’s ART, and almost 100% after two years. In summary, our dataset provides the first analysis of the distribution and percent of drug resistance mutations in PBMC and plasma during the development of a four-class drug resistant HIV-1 CRF01_AE virion. The study also showed that months before drug resistant mutants could be found in plasma, NGS identified them in HIV DNA, demonstrating that this can be a very effective tool for early detection of the development of drug resistance.
Keywords: multi-class drug resistance, HIV, CRF01_AE, ART, NGS, HIV DNA
Introduction
The lifelong administration of combination antiretroviral therapy (ART) can effectively suppress viral replication and reduce morbidity and mortality of people living with HIV (PLWH).1 There are multiple classes of ART drugs, including nucleoside reverse transcriptase inhibitors (NRTI) including lamivudine (3TC) and azidothymidine (AZT), non-nucleoside reverse transcriptase inhibitors (NNRTI) including nevirapine (NVP), protease inhibitors (PI) including Lopinavir/Ritonavir (LPV/r) and the integrase strand transfer inhibitor (INSTI) raltegravir (RAL).2 In recent years, the rapid expansion of access to ART has led to the emergence of multi-class drug resistance (MDR), defined as a virus mutant with resistance to at least three different drug classes.3
A previous study of a large cohort of cART-experienced patients in Italy showed a dramatic drop in drug resistance from 80–85% in 1999 to around 36% in 2018. In recent years (2011–18), the percentage of isolates with at least three classes of drug resistance has remained stable at around 5% (range 3–6%).4 The majority of these patients have been found to have a long history of HIV infection, with previous exposure to suboptimal therapies, and to have, over time, accumulated many mutations resistant to several drug classes.4 The viral evolutionary dynamics within these patients that leads to the development of MDR has not been well documented.
Most drug resistance data have been collected from patients infected with HIV-1 subtype B in the United States, Oceania, and Europe. When ART has become increasingly available in new geographic areas, drug resistance in a diverse group of M subtypes and distinct circulating recombinant forms (CRFs) has evolved. CRF01_AE emerged in Southeast Asia in the 1990s, expanded rapidly in China, and is now the most prevalent HIV-1 form in Southeast Asia.5,6 Previous studies have identified a 9–20% higher resistance mutation frequency at reverse transcriptase positions in CRF01_AE than in subtype B, and a 12–18% higher predicted cross-resistance to future therapy options.7 The influence of genetic variation across subtypes has therefore become an active area of research into resistance evolution and disease progression.
In our previous study of the evolutionary patterns during ART failure, plasma and peripheral blood mononuclear cells (PBMC) were longitudinally sampled at different time points from a single patient who suffered several periods of ART failure before successful reduction of viral load. The different intrapatient evolutionary dynamics patterns of env and pol viral segments witness not only the emergence of drug resistant mutants, but also the switch of tropism.8
In the current study, the same longitudinal approach was applied to learn more about the viral evolutionary dynamics during the development of four-class MDR in a single patient infected with the CRF01_AE experiencing ART failure and subsequent mortality. The distribution and percent of drug resistance mutants in the reverse transcriptase (RT), protease (PR) and integrase (IN) genes were determined by next generation sequencing, and the demographic history of the HIV DNA reservoir in PBMC was reconstructed by applying phylodynamics methods.
Case Presentation
A 27-year-old patient was diagnosed as HIV-positive in August 2008. PBMC and plasma samples were collected at different time points from September, 2013 to June, 2017 (Figure 1). The study was approved by the institutional review boards of the First Affiliated Hospital, School of Medicine, Zhejiang University (Reference Number: 2020265). Written informed consent was provided by the patient to allow the case details and any accompanying images to be published.
Plasma samples were tested for viral load during treatment. The patient had been diagnosed as HIV-positive in August 2008 and initiated ART with 3TC+AZT+NVP. The ART regimen was switched to 3TC+AZT+LPV/r in August 2013 because of unsuppressed viral load (1*105 copies/mL) and detection of reverse transcriptase resistant mutations, both to NRTI and NNRTI. One month later, in September, 2013, viral load decreased to about 1.4*103 copies/mL, and was under the detection limit (50 copies/mL) from March, 2014 to December 2014. In March, 2015, the ART regimen was changed, to 3TC+AZT+LPV/r+RAL, again due to unsuppressed viral load (5.4*104 copies/mL). By June, 2017, two years later, the viral load had increased, to 1.8*105 copies/mL (Table 1), and was followed by the patient’s death in August, 2017.
 |
Table 1 Characteristics of Drug-Resistant Mutant Sequences Isolated from Plasma |
Sanger Sequencing and Next Generation Sequencing
All collected samples during treatment were sequenced by Sanger sequencing and Next Generation Sequencing (NGS) techniques. The purified PR/RT amplicon and IN amplicon were randomly interrupted by Covaris ultrasonic breaker and then used for library preparation (NEBNext® Ultra™ II DNA Library Prep Kit for Illumina) according to manufacturer’s instructions. Sequencing was carried out by the Illumina high-throughput sequencing platform (Nova-Seq). After data processing and quality filtering performed to obtain clean data, fastq files were aligned and generated the codon frequency tables using fastq2codfreq script (https://hivdb.stanford.edu/page/codfreq/). Then, the codon frequency tables were submitted to HIVdb-NGS beta for genotypic resistance interpretations and quality control analysis. Minimum detection threshold was set to 1% for all samples, because detection below a frequency of 1% may cause failed quality assessment. ShoRAH was applied to convert NGS sequence variants into haplotypes.9
Phylogenetic Analysis
MUSCLE software (v3.8.31)10 was used to align all RT, PR and IN sequences from plasma viral RNA and cellular DNA collected during the ART therapy. Alignments were manually edited and trimmed to 297 nucleotides for PR (HBX2: 2253–2549), 903 nucleotides for RT (HBX2: 2550–3452) and 780 nucleotides for IN (HBX2: 4290–5069) using BioEdit software (v7.0.9). Shorter sequences and sequences with stop codons or gaps larger than a nucleotide triplet were removed from the alignments. The best-fitting nucleotide substitution model was selected with jModeltest software (v2.1.7),11 using the Akaike Information Criterion (AIC). Phylogenetic trees were inferred using PhyML software (v3.0).12 Bootstrap analysis was performed on 1000 replicates.
Demographic Reconstructions
The demographic history of the HIV reservoir in PBMC was estimated using the BEAST software13 and implemented in the Bayesian Markov chain Monte Carlo (MCMC) method. The Bayesian skyline model14 and strict clock model were incorporated in the MCMC method. Multiple independent MCMC runs were performed and assessed for consistency. Convergence of relevant parameters and Bayesian skyline results were assessed by effective sample sizes over 200 in Tracer v1.6 (http://tree.bio.ed.ac.uk/software/tracer/).
The Development of Drug Resistant Mutations
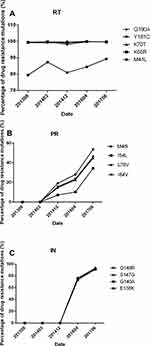
Over the course of three periods of treatment failure, the patient developed a four-class drug resistant virus population, in which we identified thirteen mutations associated with drug resistance. Five were in the RT gene - M41L, K65R, K70T, Y181C and G190A; four in the PR gene - M46I, I54L, L76V, and I84; and four in the IN gene - E138K, G140A, S147G, Q148R.
Sampling for this study was initiated after the patient was first diagnosed with ART failure, five years after ART treatment was first initiated. By that time, In September, 2013, almost 100% of PBMC virus already had mutants resistant to NRTI and NNRTI, and these levels persisted throughout periods of treatment, even during 2014, when plasma viral load was under the limit of detection.
After the introduction of the protease inhibitor Lopinavir/Ritonavir (LPV/r) to the patient’s ART, PI resistant mutants developed slowly in PMBC DNA. After one month, none were found; after sixteen months, less than 20% were mutants. After three years (two months prior to the patient’s death) PI mutants in PMBC DNA were still under 50%. PI resistant mutants in plasma had a different pattern. At sixteen months after the introduction of the PI no sequences could yet be identified because the viral level was too low for amplification. Eventually, substantially higher PI mutant levels were able to be found in plasma - almost 100% two years after a PI drug was switched to ART, by which time viral load had increased to 5.4*104 copies/mL.
Integrase strand transfer inhibitor (INSTI) mutations evolved much more quickly, replacing approximately 75% of the wild genotype in HIV DNA one year after addition of the integrase inhibitor raltegravir to the patient’s ART, and almost 100% after two years.
INSTI-resistant mutations, E138K, G140A, S147G and Q148R, replaced approximately 75% of the wild genotype in HIV DNA one year after addition of RAL to ART, and almost 100% 14 months later, by which time viral load reached 1.8*105 copies/mL. These results are displayed in Table 1 and Figure 2.
Drug Resistance Genotype
In the RT gene, K65R and K70T mutations cause low resistance to 3TC and increased susceptibility to AZT. The Y181C and G190A mutants are associated with high-level resistance to NVP. M41L is a non-polymorphic mutation selected by thymidine analogs AZT. In the PR gene, M46I, L76V and I84V are non-polymorphic mutants selected by protease inhibitors. These mutants reduce susceptibility to LPV/r. In the IN gene, E138K, G140A and Q148R are also non-polymorphic mutants, selected by INSTI (RAL). Q148R is associated with high-level reductions in RAL susceptibility, particularly when it occurs in combination with E138K or G140A mutants (Table 2). All drug resistant mutation associations are based on the Stanford drug resistance database.15
 |
Table 2 Characteristics of Drug-Resistant Mutant Sequences Isolated from PBMC |
Detecting Evolution Over Treatment Time Points
Because there were sufficient sequence data points from PBMC HIV DNA, Bayesian skyline plots were reconstructed to infer the dynamic of the effective population of RT, PR, and IN sequences in the PBMC. The effective population of RT sequences was shown to be stable over the entire testing period. However, the effective population of protease and integrase sequences underwent a significant increase in genetic variation during the period of treatment failure. After the switch of LPV/r to ART, the effective population of PR sequences first decreased and then increased with drug mutants selected by LPV/r. The effective population of IN sequences also decreased after the administration of PI, and stayed low for about six months. After the administration of INSTI, the effective population of IN sequences increased because of the drug resistant mutants selected by RAL. (Figure 3).
Discussion
In this study, the mutant sequences have emerged during the development of a new four-class drug resistant HIV-1 CRF01_AE variant in a single patient, during several periods of therapy failure. This is a serious and challenging development since PLWHs harboring multi-class drug resistant virus have a high burden of disease, with a worrying incidence of malignancies and poorer survival after treatment failure.3,16
By the study’s first sample collection point, almost 100% of viral sequences already had mutants resistant to NRTI and NNRTI in PBMC, so no significant change in the effective population of these sequences was observed over time. However, PI and INSTI drug resistant mutants gradually replaced the wild genotype, and drove the increase of genetic variability in HIV DNA. Demographic histories of these developments were generated by Bayesian skyline plot analysis, and demonstrate the genetic diversity in viral segment sequences over time, expressed as effective population.
This study’s sequencing data showed significantly reduced genetic variability in both protease and integrase PBMC-derived variants directly following the administration of PI. A study by Besson et al investigated the decay of HIV DNA on ART and showed that the infected cell populations decline initially but then achieve a steady state with the persistence of about 10% of infected cells during effective ART.17 The different phase of decay occurs from the death of infected cells with different half-lives from days to months.18 The effective population increased when the drug resistant mutants were selected.
One previous study reported that the prevalence of INSTI resistance remained low compared with PI and RT resistance in ART-treated populations, but expanded with increased INSTI use between 2009 and 2016.19 The development of INSTI resistance described in this study suggests how that resistant pathway is evolving. Here the CRF01_AE virion developed INSTI resistant mutants by changes at position Q148, the most common mutant pathways previously described in all subtypes.20 There have been numerous reports of the emergence of substitutions involving position Q148 in response to RAL pressure. As substitutions at position Q148 impart a severe fitness cost,20 they are rapidly compensated for by various secondary resistance mutants, and the addition of at least two secondary mutants seems to confer the highest fold changes in resistance to second-generation INSTIs.21 E138K and G140A, identified in our study, are two of these mutants. The prevalence of the INSTI resistance mutants in CRF01_AE needs further investigation through a larger sample.
Drug-resistant mutants in HIV DNA emerged before they appeared in plasma – through the use of next generation sequencing. The difference could be caused by the higher level of cell-associated HIV-1 RNA than in plasma RNA, which may contribute to the generation of new viral genomes, when plasma virus remains below the limit of detection.22 Some mutants could also remain in HIV DNA through persistence and/or proliferation of infected cells. These integrated and unintegrated provirus in latently infected cells may have a delayed contribution to the pool of resistant virus.23
Conclusion
While our study is limited to a single patient and several sampling timepoints, our data set and analysis demonstrated for the first time the evolution of sequences in the development of a four-class drug resistant HIV-1 CRF01_AE virion. It revealed dynamic shifts in the viral population and in drug-resistance mutants, while under the influence of complex ART regimens. This study utilized all samples available for this patient. Collection of baseline samples prior to initiation of ART, and at more sampling time points during treatment, will help us analyze evolutionary change in patient viral population. Our findings also suggested that next generation sequencing can be a very effective tool to detect a low level of drug resistance in HIV DNA, which could be critical for the clinical management of patients, especially those already experiencing virological failure while on particular ART regimens.
Both clinicians and patients need to be aware that a wide pattern of resistance can represent a strong negative prognostic factor for survival. Early detection of the development of drug resistant mutants should become a priority to prevent the further development of resistance through modification of ART regimens and as part of patient education to strengthen adherence to therapy.
Data Sharing Statement
The datasets used in this study are available from the corresponding author on reasonable request.
Ethics Approval and Consent to Participate
The study was approved by the institutional review boards of the First Affiliated Hospital, School of Medicine, Zhejiang University (Reference Number: 2020265). Written informed consent was provided by the patient to allow the case details and any accompanying images to be published.
Acknowledgments
We gratefully thank Susan Joyce Herzog for assistance in editing our manuscript.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work. First co-author: These authors contribute equally to this manuscript: Xiaorong Peng and Yufan Xu.
Funding
This study was supported by National Special Research Program for Important Infectious Diseases (No. 2017ZX10202102-002-002).
Disclosure
The authors declare no conflicts of interest for this work.
References
1. Palella FJ, Delaney KM, Moorman AC, et al. Declining morbidity and mortality among patients with advanced human immunodeficiency virus infection. HIV Outpatient Study Investigators. N Engl J Med. 1998;338:853–860. doi:10.1056/NEJM199803263381301
2. Smith SJ, Zhao XZ, Passos DO, Lyumkis D, Burke TR, Hughes SH. Integrase strand transfer inhibitors are effective anti-HIV drugs. Viruses. 2021;13:205. doi:10.3390/v13020205
3. Zaccarelli M, Tozzi V, Lorenzini P, et al. Multiple drug class-wide resistance associated with poorer survival after treatment failure in a cohort of HIV-infected patients. Aids. 2005;19:1081–1089. doi:10.1097/01.aids.0000174455.01369.ad
4. Armenia D, Di Carlo D, Flandre P, et al. HIV MDR is still a relevant issue despite its dramatic drop over the years. J Antimicrob Chemother. 2020;75:1301–1310. doi:10.1093/jac/dkz554
5. Feng Y, He X, Hsi JH, et al. The rapidly expanding CRF01_AE epidemic in China is driven by multiple lineages of HIV-1 viruses introduced in the 1990s. Aids. 2013;27:1793–1802. doi:10.1097/QAD.0b013e328360db2d
6. Peng X, Wu H, Peng X, Jin C, Wu N. Heterogeneous evolution of HIV-1 CRF01_AE in men who have sex with men (MSM) and other populations in China. PLoS One. 2015;10:e0143699. doi:10.1371/journal.pone.0143699
7. Huang A, Hogan JW, Luo X, et al. Global comparison of drug resistance mutations after first-line antiretroviral therapy across human immunodeficiency virus-1 subtypes. Open Forum Infect Dis. 2016;3:ofv158. doi:10.1093/ofid/ofv158
8. Peng X, Xu Y, Huang Y, Zhu B. Intrapatient evolutionary dynamics in an individual infected with HIV-1 CRF01_AE who experienced periods of treatment failure. AIDS Res Hum Retroviruses. 2021;37:139–146. doi:10.1089/aid.2020.0213
9. Zagordi O, Bhattacharya A, Eriksson N, Beerenwinkel N. ShoRAH: estimating the genetic diversity of a mixed sample from next-generation sequencing data. BMC Bioinform. 2011;12:119. doi:10.1186/1471-2105-12-119
10. Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinform. 2004;5:113. doi:10.1186/1471-2105-5-113
11. Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 2012;9:772. doi:10.1038/nmeth.2109
12. Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–321. doi:10.1093/sysbio/syq010
13. Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 2012;29:1969–1973. doi:10.1093/molbev/mss075
14. Minin VN, Bloomquist EW, Suchard MA. Smooth skyride through a rough skyline: bayesian coalescent-based inference of population dynamics. Mol Biol Evol. 2008;25:1459–1471. doi:10.1093/molbev/msn090
15. Shafer RW. Rationale and uses of a public HIV drug-resistance database. J Infect Dis. 2006;194(Suppl 1):S51–8. doi:10.1086/505356
16. Galli L, Parisi MR, Poli A, et al. Burden of disease in PWH harboring a multidrug-resistant virus: data from the PRESTIGIO registry. Open Forum Infect Dis. 2020;7:ofaa456. doi:10.1093/ofid/ofaa456
17. Besson GJ, Lalama CM, Bosch RJ, et al. HIV-1 DNA decay dynamics in blood during more than a decade of suppressive antiretroviral therapy. Clin Infect Dis. 2014;59:1312–1321. doi:10.1093/cid/ciu585
18. van Zyl G, Bale MJ, Kearney MF. HIV evolution and diversity in ART-treated patients. Retrovirology. 2018;15:14. doi:10.1186/s12977-018-0395-4
19. Kamelian K, Lepik KJ, Chau W, et al. Prevalence of human immunodeficiency virus-1 integrase strand transfer inhibitor resistance in British Columbia, Canada between 2009 and 2016: a longitudinal analysis. Open Forum Infect Dis. 2019;6:ofz060. doi:10.1093/ofid/ofz060
20. Anstett K, Brenner B, Mesplede T, Wainberg MA. HIV drug resistance against strand transfer integrase inhibitors. Retrovirology. 2017;14:36. doi:10.1186/s12977-017-0360-7
21. Tsiang M, Jones GS, Goldsmith J, et al. Antiviral activity of bictegravir (GS-9883), a novel potent HIV-1 integrase strand transfer inhibitor with an improved resistance profile. Antimicrob Agents Chemother. 2016;60:7086–7097. doi:10.1128/AAC.01474-16
22. Scully EP, Gandhi M, Johnston R, et al. Sex-based differences in human immunodeficiency virus type 1 reservoir activity and residual immune activation. J Infect Dis. 2019;219:1084–1094. doi:10.1093/infdis/jiy617
23. Wang YM, Dyer WB, Workman C, Wang B, Sullivan JS, Saksena NK. Molecular evidence for drug-induced compartmentalization of HIV-1 quasispecies in a patient with periodic changes to HAART. Aids. 2000;14:2265–2272. doi:10.1097/00002030-200010200-00007
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.