Back to Journals » Drug Design, Development and Therapy » Volume 16
Integrated Network Pharmacology Analysis and Experimental Validation to Investigate the Molecular Mechanism of Triptolide in the Treatment of Membranous Nephropathy
Authors Zhang PN, Tang JY, Yang KZ, Zheng QY, Dong ZC, Geng YL, Liu YN, Liu WJ
Received 18 August 2022
Accepted for publication 11 November 2022
Published 23 November 2022 Volume 2022:16 Pages 4061—4076
DOI https://doi.org/10.2147/DDDT.S386031
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Anastasios Lymperopoulos
Ping Na Zhang,1,* Jing Yi Tang,1,* Ke Zhen Yang,2,* Qi Yan Zheng,1 Zhao Cheng Dong,1 Yun Ling Geng,1 Yu Ning Liu,1 Wei Jing Liu1
1Renal Research Institution of Beijing University of Chinese Medicine, and Key Laboratory of Chinese Internal Medicine of Ministry of Education and Beijing, Dongzhimen Hospital Affiliated to Beijing University of Chinese Medicine, Beijing, People’s Republic of China; 2School of Acupuncture-Moxibustion and Tuina, Beijing University of Chinese Medicine, Beijing, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Yu Ning Liu; Wei Jing Liu, Renal Research Institution of Beijing University of Chinese Medicine, and Key Laboratory of Chinese Internal Medicine of Ministry of Education and Beijing, Dongzhimen Hospital Affiliated to Beijing University of Chinese Medicine, Beijing, People’s Republic of China, Tel/Fax +86-10-84013190, Email [email protected]; [email protected]
Background: Triptolide, a major active ingredient isolated from Tripterygium wilfordii Hook f., is effective in the treatment of membranous nephropathy (MN); however, its pharmacological mechanism of action has not yet been clarified. We applied an approach that integrated network pharmacology and experimental validation to systemically reveal the molecular mechanism of triptolide in the treatment of MN.
Methods: First, potential targets of triptolide and the MN-related targets were collected from publicly available database. Then, based on a protein–protein interaction network as well as GO and KEGG pathway enrichment analyses, we constructed target-pathway networks to unravel therapeutic targets and pathways. Moreover, molecular docking was applied to validate the interactions between the triptolide and hub targets. Finally, we induced passive Heymann nephritis (PHN) rat models and validated the possible molecular mechanisms of triptolide against MN.
Results: The network pharmacology results showed that 118 intersected targets were identified for triptolide against MN, including mTOR, STAT3, CASP3, EGFR and AKT1. Based on enrichment analysis, signaling pathways such as PI3K/AKT, MAKP, Ras and Rap1 were involved in triptolide treatment of MN. Furthermore, molecular docking confirmed that triptolide could bind with high affinity to the PIK3R1, AKT1 and mTOR, respectively. Then, in vivo experiments indicated that triptolide can reduce 24 h urine protein (P < 0.01) and protect against renal damage in PHN. Serum albumin level was significantly increased and total cholesterol, triglycerides, and low-density lipoprotein levels were decreased by triptolide (P < 0.05). Compared with PHN group, triptolide treatment regulated the PI3K/AKT/mTOR pathway according to Western blot analyses.
Conclusion: Triptolide could exert antiproteinuric and renoprotective effects in PHN. The therapeutic mechanism of triptolide may be associated with the regulation of PI3K/AKT/mTOR signaling pathway. This study demonstrates the pharmacological mechanism of triptolide in the treatment of MN and provides scientific evidence for basic and clinical research.
Keywords: membranous nephropathy, network pharmacology, triptolide, molecular docking, experiment verification
Graphical Abstract:
Introduction
MN is a type of glomerular disease that is characterized by the formation of immune deposits on the outer aspect of the basement membrane.1 Autoantibodies against two major podocyte antigens, phospholipase A2 receptor (PLA2R) and thrombospondin domain-containing 7A, have been identified in 70–80% of patients, which has revolutionized our understanding of the pathophysiology of MN and improved the existing therapies.2,3 Based on clinical and pathological features, MN has been classified as primary MN (pMN) and secondary MN, the latter being caused by lupus, hepatitis B, or malignancies.4 pMN is the most common cause of adult idiopathic nephrotic syndrome and accounts for 30% of cases in non-diabetic patients.4 The prevalence of MN is increasing by 13% every year, while that of the other major glomerulopathies is not increasing.5 The influence of particulate matter 2.5 (PM2.5) should be carefully considered, given that provinces in China with the higher PM2.5 levels show the higher incidence of MN.6 One third of patients may spontaneously remit; however, MN exhibits a highly variable clinical course, and therapeutic means for those with heavy proteinuria and renal failure are limited and may cause renal toxicity.7 In notably, traditional Chinese medicines has been considered as a well-proven strategy for prevention and treatment of MN.8 In clinical trials, natural products exerted therapeutic effects that confirmed by the randomized controlled trials. It has been reported that Shenqi particle were efficacy in patients with IMN and nephrotic syndrome.9 In terms of experimental research, studies provided evidence for the efficacy of traditional Chinese medicines from the perspectives of regulating nuclear factor κB (NF-κB) signaling and decreasing podocyte apoptosis.10,11
Tripterygium wilfordii Hook f. (TWHF) was first recorded by Mao Lan in Dian Nan Ben Cao in 1476. Recently, Tripterygium wilfordii tablets or Tripterygium hypoglaucum hutch tablets were marketed in China that used in the treatment of the autoimmune disorders and inflammation.12 Triptolide, a major active ingredient, was first isolated from TWHF in 1972,13 which has been used in clinical therapy for a long time.14 Owing to its various metabolites and recognized anti-inflammatory and immunosuppressive effects,15 triptolide and its extracts have attracted attention for the treatment of kidney diseases, autoimmune diseases, and rheumatoid arthritis.16 Especially in the treatment of MN, in vitro and in vivo experiments, it was found that triptolide could improve the damage of podocytes mediated by puromycin aminonucleoside and play the role of reducing urinary protein.17 Furthermore, triptolide ameliorated heavy proteinuria and protected against C5b-9-induced podocyte injury in model of PHN.18 In addition, the efficacy of triptolide in reducing proteinuria and protecting podocyte injury has been proved in clinical trials. A clinical study showed that triptolide can reduce proteinuria and increase the complete remission (CR) rate in patients with pMN.19 Moreover, a network meta-analysis of 13 immunosuppressive agents demonstrated that tacrolimus combined with triptolide is more effective than other immunosuppressive agents in terms of CR and proteinuria.20 However, the molecular mechanism whereby triptolide is effective against MN has not been fully elucidated. In this study, unlike in traditional network pharmacology, we combined network pharmacology with molecular docking to reveal the potential targets of triptolide against MN. Furthermore, we used the model of PHN to validate the pharmacological mechanism of triptolide.
Materials and Methods
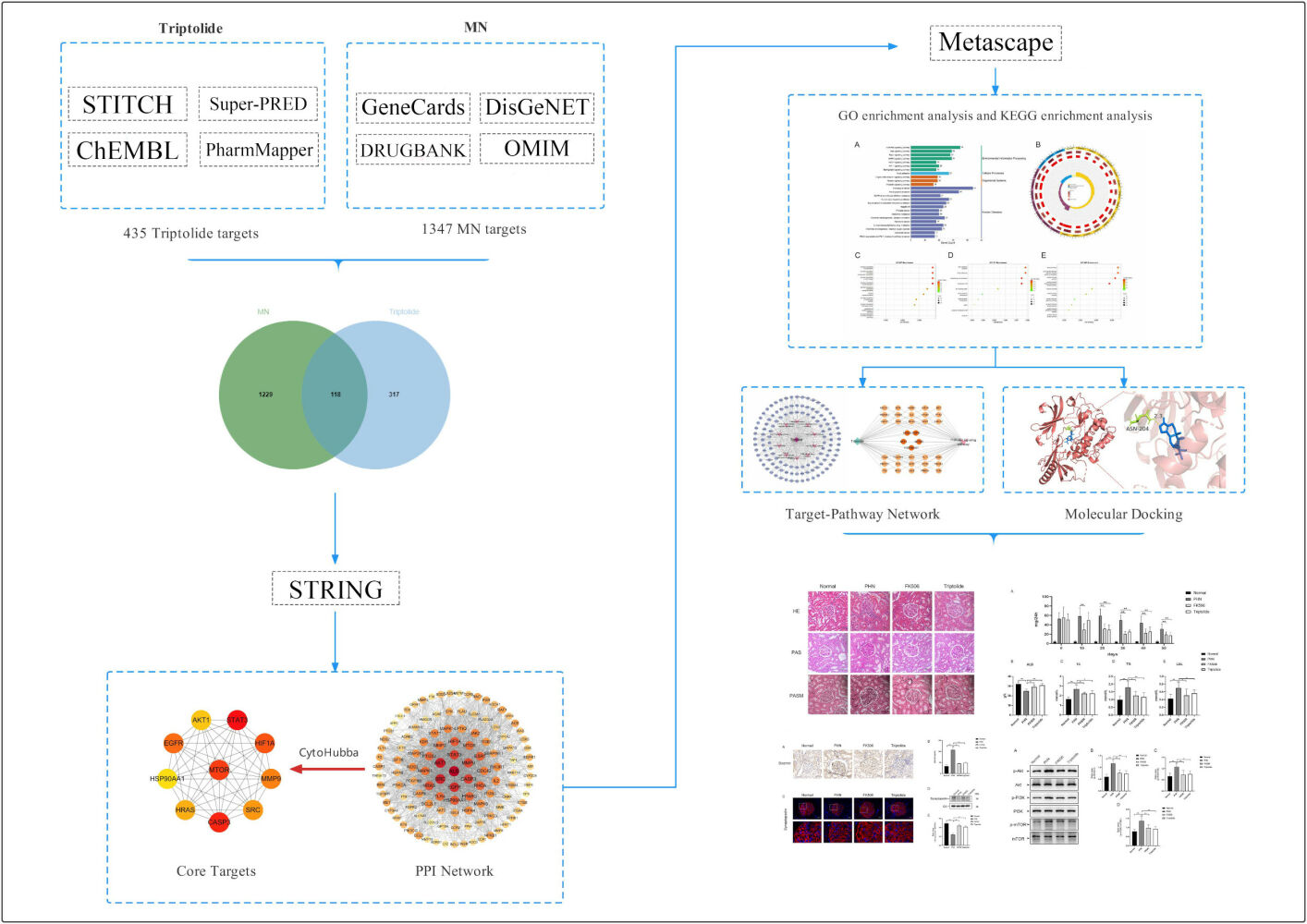
Identification of the Structure and Potential Targets of Triptolide
The 2D structure of triptolide was obtained from PubChem (https://pubchem.ncbi.nlm.nih.gov/). The physicochemical characteristics of triptolide were screened from TCMSP database (http://tcmspw.com/). Potential targets of triptolide were obtained from STITCH (http://stitch.embl.de/), Super-PRED (https://prediction.charite.de/index.php), CHEMBL (https://www.ebi.ac.uk/chembl/) and Pharm Mapper (http://lilab.ecust.edu.cn/). The only species selected was “Homo sapiens” and all obtained targets were converged by the UniProt database (https://www.uniprot.org/) to verify its Gene name and Gene ID.
Screening of MN-Related Targets
The MN-associated targets were obtained from GeneCards (https://www.genecards.org/), DisGeNET (http://www.disgenet.org/home/), the OMIM (http://www.omim.org), and DrugBank (https://go.drugbank.com/). All four databases are public databases, the contents of which are publicly available and allow unrestricted reuse through open licenses, this study would have the need for ethics approval waived (Ethics Committee of Dongzhimen hospital, Beijing University of Chinese Medicine). “Membranous Nephropathy” and “Membranous Glomerulopathy” were imported in the databases as keywords. Meanwhile, the targets from databases were selected by median screening of the relevance score. The MN-related targets were collected by merging all targets from the four disease databases and excluding duplicate targets.
Construction of the Protein–Protein Interaction (PPI) Network
First, we used overlapping circles in a Venn diagram to display the intersection of ingredient-related targets of triptolide and MN-related targets. Then, the core regulated genes were screened by PPI analysis, which was performed by the String database (https://cn.string-db.org/). Meanwhile, an interaction score of 0.4 was used for the extraction of PPI information, and the species were limited to “Homo sapiens”. The topology analysis of the PPI was carried out by Cytoscape 3.9.1 (http://cytoscape.org/.ver.3.9.1). The high-relevant targets were screened by Cytohubba, a plug-in of Cytoscape, and MCC algorithm is used to calculate the score of genes.21
GO and KEGG Enrichment Analyses
To explore the signaling pathways and pharmacological mechanisms involved in the action of triptolide against MN, Metascape (https://metascape.org/) was used to perform KEGG pathway enrichment analyses. The only species selected was “Homo sapiens” with KEGG screening conditions: Min overlap: 3; P: 0.01; min enrichment: 1.5. The heatmap of the top 25 enriched KEGG pathways were also selected and plotted using bioinformatics (http://www.bioinformatics.com.cn/). Go enrichment analysis was carried out through the “clusterProfiler” package of R software, pvalueCutoff = 0.05, padjustmethod = “BH”, qvalueCutoff = 0.05, minGSSize = 3, maxGSSize = 500. The results of the top 10 enriched pathways of the GO terms regarding biological processes (BP), cell composition (CC), and molecular function (MF) were visualized using “ggplot” packages of R software (version 4.0.1).
Construction of a “Target-Pathway” Network
The target-pathway network of triptolide against MN were constructed by Cytoscape to investigate the molecular mechanism of triptolide against MN. The nodes in the network display targets and the signaling pathways in triptolide and MN are represented by the triangle nodes.
Compound-Target Molecular Docking
Molecular docking between the core active compounds of triptolide and proteins selected from the gene targets were used to better understand the interactions from a molecular perspective. The 2D structure of triptolide was obtained in an SDF (.sdf) format from the PubChem database (https://pubchem.ncbi.nlm) and converted into 3D structures, and the minimum free energy of the ligand was optimized through Chem3D software. The protein molecules were downloaded from the PDB database (https://www.rcsb.org/) and saved as PDB files, which were imported into the PyMOL 2.5.2 software (https://pymol.org/2/) to delete water molecules and small molecule ligands. Then, molecular docking was performed by importing the core active compounds and protein molecules into the AutoDockTools 1.5.7 software for dehydration and hydrogenation. Grid boxes were constructed to cover the whole protein receptor. Meanwhile, the protein receptor and small molecule ligand were semi-flexibly docked by evaluating the original ligands and intermolecular interactions, and the lowest binding energy was selected as the docking site. The model was visualized using PyMOL software.
Establishment of the PHN Model and Treatment
Forty male Sprague-Dawley rats with weights of 210 to 230 g were provided by Beijing Vital River Laboratory Animal Technology Co. Ltd (Certificate No. SCXK (JING) 2021—0011). All Sprague-Dawley rats were fed under special pathogen-free conditions and maintained at 25 ± 2 °C, 65% humidity, and a 12 h light/dark cycle by the Experimental Animal Centre of Dongzhimen hospital (Beijing, China, Certificate No. SYXK (JING) 2020—0013). The study was conducted under the national guidelines for laboratory animal welfare and pre-approved by the Animal Ethics Committee, Dongzhimen hospital, Beijing University of Chinese Medicine (Beijing, China, approval No. 21–32).
To induce PHN, rats were injected with 0.5 mL/100 g body weight of sheep anti-rat Fx1A serum (PTX-002S, Probetex, San Antonio, USA) into the tail vein. Then, 24-hour urine collection from individual rats was performed in metabolic cages. Protein in urine was detected using enzymatic methods with specific kits (Nanjing Jiancheng Bioengineering Institute, Nanjing, China). Treatment with triptolide (with 99.6% purity, Batch number 111567-202005, Chinese National Institute for Control of Pharmaceutical and Biological Products, Beijing, China) that dissolved in 0.01% dimethyl sulfoxide started 10 days after sheep anti-rat Fx1A serum injection when proteinuria was already present. Thirty rats with PHN were randomly assigned into three groups. For the PHN model group, rats (n = 10) were provided with water containing 0.01% dimethyl sulfoxide (1 mL/100 g body weight) daily. For the triptolide group, rats (n = 10) were orally administered 200 mg/kg of triptolide daily. In the FK506 group (positive control group), rats (n = 10) were orally treated with 1 mg/kg of FK506. The dose used in rats was converted from patients to animals based on international guidelines. In addition, 10 healthy rats were selected as a normal control group. The medication time was 50 days.
Biochemical Measurements in Urine and Blood
Every 10 days, rats were separately transferred to metabolic cages to collect 24-hour urine samples. During that time, rats had free access to water but no access to food. After 50 days of administration, the rats were fasted for 12 h, and blood samples were then collected from the abdominal aorta after intraperitoneal injection of 35 mg/kg pentobarbital sodium. The samples were centrifuged at 3000 rpm for 10 min, and the serum was collected for the analysis of albumin (ALB), total cholesterol (TC), triglycerides (TG), and low-density lipoprotein (LDL). All biochemical indexes and 24-hour urinary protein were detected according to the manufacturer’s instructions (Nanjing Jiancheng Bioengineering Institute, Nanjing, China).
Pathological Examination of Renal Tissue
Kidney tissues were put into 4% paraformaldehyde (P1110, Solarbio), dehydrated with ethanol, and then embedded in paraffin. The 3 µm paraffin sections were cut for hematoxylin and eosin (H&E), periodic acid-Schiff (PAS), and periodic acid-sliver methenamine (PASM) staining. All stained samples were observed under a light microscope (400× magnification) (BX60, Olympus, Tokyo, Japan).
Immunofluorescence on Renal Tissue
Paraffin sections that were 3 µm thick were used for immunofluorescence staining. After baking at 60 °C for 60 min, the sections underwent xylene dewaxing, gradient-alcohol dehydration, and antigen retrieval. Then, the sections were incubated in 0.2% PBST for 20 min for membrane penetration and blocked using 5% donkey serum at 37 °C for 30 min. The sections were then incubated with synaptopodin (sc515842, Santa Cruz, Texas, USA) overnight at 4 °C. After washing with 0.25% PBS, the sections were incubated with Alexa Fluor® 488 donkey anti-rabbit IgG (lot 1927937, Thermo Fisher Scientific) at 37℃ for 60 min. Subsequently, the nuclei were stained with DAPI (ZLI-9557, ZSGB-Bio).
Immunohistochemistry on Renal Tissue
Paraffin sections that were 3 µm thick were used for immunohistochemistry staining (immunochemistry kit: PV-9001, ZSGB-Bio). After baking at 60 °C for 60 min, the sections underwent xylene dewaxing, gradient-alcohol dehydration, and antigen retrieval with a citrate solution at 95 °C for 20 min. Then, the sections were soaked in 3% hydrogen peroxide and blocked with goat serum for 60 min. The sections were then incubated with desmin (ab32362, abcam, Cambridge, UK) overnight at 4 °C. After washing with 0.25% PBS, the sections were incubated with the secondary antibody at 37 °C for 30 min. Subsequently, the sections were colored by DAB and the nuclei were stained with hematoxylin. The integral optical density (IOD) was analyzed using Image-Pro Plus 6.0 software.
Western Blot Analysis
The proteins of the renal cortex were lysed with lysis buffer containing protease inhibitors and quantified with a BCA protein assay kit (BN27109-500T, Biorigin, Beijing, China). Total protein samples were separated on 10% polyacrylamide sodium dodecyl sulfate gels and then transferred to nitrocellulose membranes. Afterwards, the membranes were blocked with 5% skimmed milk powder dissolved in Tris-Buffered Saline and Tween 20 (TBST) for 1 h, followed by incubation with the primary antibodies synaptopodin (ab32127, abcam, Cambridge, UK), AKT (#4685, CST, Boston, USA), p-AKT (#13038, CST, Boston, USA), PI3K (#4257, CST, Boston, USA), p-PI3K (#17366, CST, Boston, USA), mTOR (#2983, CST, Boston, USA) and p-mTOR (#5536, CST, Boston, USA) overnight at 4 °C. After washing with TBST, the membranes were incubated with HRP-conjugated secondary antibodies (donkey anti-mouse, goat anti-rabbit) for 1 h. Finally, an ECL Kit (P1010, Applygen, Beijing, China) was used to detect positive binding under a multi-functional imaging system (Tanon 5200, Shanghai, China). All protein bands were quantified using Image J software.
Statistical Analysis
SPSS 20.0 (IBM, Chicago, IL, United States) was used for statistical analysis. Data were expressed as the mean ± standard deviation. For statistical analysis, the means were compared using one-way ANOVA for multiple comparisons. Statistical significance was set at p < 0.05. Figures were prepared using GraphPad Prism 8.0 software (San Diego, CA, United States).
Results
Targets of Triptolide Against MN
The 2D structure of triptolide was presented in Figure 1A and physicochemical characteristics of triptolide were summarized in Table 1. Applying the STITCH, Super-PRED, CHEMBL and Pharm Mapper databases, 435 potential targets corresponding to the compounds were predicted after deleting duplicate targets, which were normalized to their Gene name and Gene ID by the UniProt database. Based on Gene Cards, the top 25% target genes were taken according to the relevance score and merged with the genes in the databases DisGeNET, OMIM, and DrugBank. A total of 1347 potential MN-associated targets were screened after eliminating duplications. The 435 targets of triptolide (Table S1) were intersected with the 1347 MN-associated targets (Table S2) through a Venn diagram to display 118 intersected targets, which were considered the therapeutic targets of triptolide against MN (Figure 1B).
 |
Table 1 Chemical Information of the Active Compounds of Triptolide |
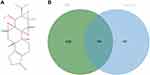 |
Figure 1 (A) The 2D structure of triptolide. (B) Venn diagram of the overlapping genes of triptolide and MN. |
PPI Network of Targets for Triptolide Against MN
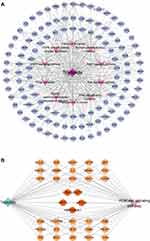
To explore the underlying pharmacological mechanism of triptolide against MN, PPI analysis of the 118 overlapped targets was performed on the String database, and further topology analysis was carried on Cytoscape software (Figure 2A). The top 10 targets were screened according to their MCC score, including mTOR, STAT3, CASP3, EGFR, AKT1, HIF1A, MMP9, SRC, HRAS and HSP90AA1 (Figure 2B). These proteins were the core targets of triptolide in the treatment of MN and therefore used for subsequent molecular docking experiments.
GO and KEGG Pathway Enrichment Analyses
The KEGG pathway enrichment analyses were performed on the 118 intersected targets through Metascape to elucidate the underlying mechanisms of triptolide against MN. KEGG pathway enrichment analysis identified 190 pathways (Table S3). The top 25 enriched KEGG pathways based on the P value were organized and visualized in the bar and circle charts to show the crucial mechanism of triptolide against MN (Figure 3A and B). The main pathways closely related to the role of triptolide against MN were the pathways in cancer, PI3K/AKT, MAKP, Ras and Rap1 signaling pathways. A total of 1604 BPs, 113 CCs, and 152 MFs (Table S4–S6) were screened in the GO analysis through the “clusterProfiler” package of R software (Figure 3C–E). BPs were mostly associated with the positive regulation of cell migration, cellular component movement, and cell motility. The main CC terms were distributed in the cell-substrate junction and focal adhesion. In the MF classification, triptolide may attenuate MN through, kinase activity and protein serine/threonine/tyrosine kinase activity.
Target-Pathway Network
Based on the pathways related to the target genes, a target-pathway network was constructed (Figure 4A). It involved 118 target genes and 10 pathways. To further elucidate the potential pharmacological mechanism of triptolide against MN, a target-pathway network was constructed based on the top 10 signaling pathways and the 118 involved targets. After integrating drug core target, function enrichment and target-pathway network analyses, we identified triptolide mainly acted through the PI3K-AKT signaling pathways. Subsequently, to observe the relationship between triptolide and the PI3K-AKT signaling pathways in more detail, we extracted a subnet to show the mapping path of triptolide at hub genes (Figure 4B). These results confirmed that there were five core targets of triptolide, among which PI3K/AKT signaling pathways showing effects.
Molecular Docking Analysis
Next, we applied molecular docking technology to evaluate the drug-target interaction modes. The binding energy reflects the binding strength between the ligand and receptor. The lower the binding energy, the stronger the interaction and the higher the stability of interaction between the ligand and the receptor. Triptolide and three targets were respectively selected as small molecule ligands and protein receptors (Table 2). Given that the structure of PI3K cannot be obtained from the PDB database, PIK3R1 as a regulatory subunit of PI3K was downloaded to further analyze. The triptolide-AKT1 complex (PDB:7NH5) binds stably to amino acid residue ASN204 via a hydrogen bond with a binding energy of −8.42 (Figure 5A). The triptolide-mTOR complex (PDB: 3JBZ) is stably bound by two hydrogen bonds at amino acid residues VAL2183 and ALA2226, respectively, with a binding energy of −6.16 (Figure 5B). The triptolide-PIK3R1 complex (PDB: 3I5S) binds via two hydrogen bonds at amino acid residues TYR12 and TYR76, respectively, with a binding energy of −5.87 (Figure 5C). The complexes formed by the above three docking can be considered to have good binding effect according to the size of the binding energy. These results on the interaction between triptolide and each target further verified the predictions of the pharmacological network.
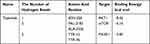 |
Table 2 Virtual Molecular Docking of Triptolide and Its Targets |
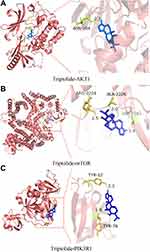 |
Figure 5 Molecular docking of triptolide with AKT1 (A), mTOR (B), and PIK3R1 (C) shown as 3D diagrams. The number indicates the length of the hydrogen bond. |
Triptolide Improved the Biochemical Parameters of PHN Rats
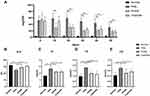
Treatment with triptolide significantly reduced the levels of 24-hour urine total protein quantity (24hUTP) in PHN rats. Compared with the normal control group, PHN rats exhibited high levels of 24hUTP. After treatment with triptolide, 24hUTP was significantly decreased on days 20, 30, 40, and 50 (P < 0.01). Moreover, after 20 days of intervention, triptolide reduced urinary protein to the same extent as FK506 did (Figure 6A).
Consistent with the findings of heavy proteinuria, the level of serum ALB in normal control rats was significantly higher than that in PHN rats (P < 0.01). After treatment with triptolide, serum ALB levels were significantly improved compared with those in untreated PHN rats (P < 0.01), and the effect was similar to that of FK506 (Figure 6B).
Furthermore, blood lipid levels in PHN rats were significantly higher than those in normal control rats (P < 0.01). Triptolide treatment significantly improved blood lipid levels, in which the levels of serum TC, TG, and LDL were decreased compared with those in PHN rats (P < 0.05) (Figure 6C–E).
Triptolide Protects Against Renal Damage in PHN Rats
PHN rats showed glomerular enlargement, thickened basement membrane, protein casts, and some inflammatory infiltration (Figure 7). Compared with PHN rats, rats treated with triptolide or FK506 showed significant improvements in their renal pathological changes.
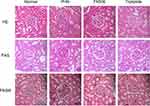 |
Figure 7 The effect of triptolide on renal pathology in PHN rats. Microstructural images of representative renal tissues stained by H&E, PAS, and PASM (400 × magnification) (scale bar = 20 μm). |
Triptolide Reduced Podocyte Injury in PHN Rats
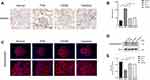
Desmin represents a biomarker of podocyte injury, as it is only expressed in the glomeruli with damaged podocytes. In normal control rats, the expression of desmin in podocytes was barely detected. In PHN rats, the staining of desmin in podocytes was conspicuously enhanced (Figure 8A). Treatment with triptolide significantly decreased the expression of desmin (P < 0.01) (Figure 8B).
Synaptopodin, an actin-associated protein, is responsible for maintaining the motility of podocytes and the function of the glomerular filtration membrane, and is therefore a typical marker of mature podocytes. The results of immunofluorescence staining showed that, compared with normal control rats, PHN rats had decreased expression of synaptopodin. Notably, the expression of synaptopodin was markedly increased after treatment with triptolide (Figure 8C). Meanwhile, Western blotting shows the same results with immunofluorescence staining. The protein expression of synaptopodin was decreased in PHN rats (P < 0.01), but triptolide significantly reversed the expression of synaptopodin (P < 0.01) (Figure 8D and E). (Figures S1 and S2)
Confirmation of PI3K/AKT Signaling Pathway Repressed by Triptolide
In order to further explore the molecular mechanism of triptolide against MN, PI3K/AKT signaling pathway was studied by evaluating the protein expression levels of PI3K and AKT and mTOR by Western blotting (Figure 9A). Western blotting showed that the expression of phosphorylated PI3K, AKT and mTOR were upregulated in PHN rats compared with that in normal rats (P < 0.01). After treatment with triptolide, PI3K/AKT/mTOR expression was markedly downregulated compared with that in untreated PHN rats (P < 0.05) (Figure 9B–D).
Discussion
The pathogenesis of MN, an organ-specific autoimmune disease, involves the deposition of subepithelial immune complexes caused by circulating autoantibodies specific to antigens on glomerular podocytes.22 Triptolide has been used for the treatment of kidney diseases, particularly MN, for a long time owing to its prominent immunosuppressive and anti‐inflammatory effects.18,24 Here, we applied an integrated network pharmacology and experimental validation in vivo strategy to systemically reveal the mechanism of triptolide in the treatment of MN. Bioinformatic analysis of the 435 targets of triptolide and the 1347 MN-associated targets revealed a total of 118 intersected targets. According to the MCC score in CytoHubba, the top ten targets were mTOR, STAT3, CASP3, EGFR, AKT1, HIF1A, MMP9, SRC, HRAS and HSP90AA1. Furthermore, combining the network topology parameters with GO and KEGG enrichment analyses revealed that the effect of triptolide against MN involves multiple potential targets and major signaling pathways. Based on KEGG pathway analysis, the PI3K/AKT, MAKP, Ras and Rap1 signaling pathways were among the top 15 significant pathways, and GO enrichment highlighted that the main BPs affected by triptolide treatment are the positive regulation of cell migration and cellular component movement. Meantime, Hydrangea. Paniculata, Sieb exhibited renal protective effect through regulating PI3K/AKT pathways.25 Besides, treatment with Zhen-Wu-Tang ameliorates renal damage in MN, which was associated with inhibition of NF-κB pathway and NLRP3 inflammasome.26 Meanwhile, we induced PHN, whose pathogenic mechanism is similar to that of MN, in rats to investigate the therapeutic effects of triptolide. After treatment with triptolide, the levels of 24hUTP in PHN rats was markedly reduced, and this was accompanied by improvements in biochemical parameters such as serum ALB, TC, TG, and LDL levels. Furthermore, our results showed that triptolide downregulated phosphorylation of PI3K, AKT and mTOR in vivo to attenuate MN.
The normal structure of podocytes is maintained by the actin cytoskeleton, which forms a specialized intercellular junction known as the slit diaphragm that acts as a final barrier, and whose main component is nephrin.23 Triptolide could exert a protective role, reversing PHN-induced actin cytoskeleton disruption and abnormalities in synaptopodin expression, and reducing the expression of desmin. These results indicated that triptolide can effectively restore podocyte injury to reduce proteinuria in MN.
In the current study, triptolide-MN network highlighted 10 promising core targets, and several of those targets and their related pathways had already been reported as being associated with triptolide against MN, which validates the predictive function of the network pharmacology analysis used. The mammalian target of the rapamycin (mTOR) pathway, a central regulator of metabolism, can be inhibited by rapamycin to ameliorate severe proteinuria-induced tubulointerstitial inflammation and interstitial fibrosis in PHN.27 Caspase3 (CASP3) plays a key role in the executive stage of apoptosis, and podocytes apoptosis is a hallmark in the progression of MN.28 The therapeutic effects of triptolide in renal diseases have been confirmed through inhibiting podocytes apoptosis.29,30 STAT3, as master transcription factors, is needed for Th17 cell development. In recent clinical study showed that increased ratio of Th17/Treg cells with transcription factors STAT3 involved in the pathogenesis of IMN.31 Furthermore, matrix metalloproteinase 9 (MMP9) was identified as biomarkers in monitoring IMN, and it was significantly higher in patients with heavy proteinuria.32
To further elucidate the underlying mechanisms whereby triptolide exerts the therapeutic effects in MN, we constructed a target-pathway network. Specifically, based on the analysis results, triptolide may alleviate renal damage in MN via regulating the PI3K/AKT signaling pathways. Then, we extracted triptolide, PI3K/AKT signaling pathways and corresponding targets from the PPI network, and we observed that most targets focused on PI3K/AKT signaling pathway and its downstream pathways (Figure 4). Indeed, the PI3K/AKT signaling pathway is one of the most important signaling pathways involved in attenuating MN. The protein kinase AKT, contains three isoforms (AKT1, AKT2 and AKT3) which are all activated in the same way. AKT can be phosphorylated by 3-phosphoinositide-dependent protein kinase 1, which is activated by PI3K through the Thr308 site to exert biological effects in multiple pathophysiological process.33 The PI3K/AKT signaling pathway is involved in cellular growth, proliferation, cytoskeleton, and metabolism. Combining RNA-sequencing covering mRNA of renal cortex tissues and KEGG analysis, Wang et al25 revealed that PI3K/AKT signaling was significantly higher in the MN model group than in the control group. In their study, over-activated PI3K/AKT and NF-κB pathways were associated with macrophage-induced renal fibrosis, which provided a therapeutic basis for MN. Moreover, PLA2R induced podocytes apoptosis via aberrant activation of PI3K/AKT/mTOR signaling, which could be inhibited by rapamycin.34 Evidence of the critical role of PI3K/AKT in MN is centered on activating autophagy to attenuated MN. Salvianolic acid B, an active ingredient isolated from Danshen,35 could activate renal autophagy to ameliorate cell proliferation and inflammation of MN, which was closely related to inhibition of PI3K/AKT pathway. Another active component of turmeric, curcumin exerted therapeutic effects in MN that suppressed PI3K/AKT/mTOR to improve renal autophagy. In addition, triptolide and its corresponding patent medicines such as Colquhounia root tablets and Kunxian capsules have been applied for the treatment of kidney disease, including MN, diabetic kidney disease and lupus nephritis, which can restore the imbalance of the immune-inflammation system by modulating the PI3K/AKT/NF-κB/TNF-α pathway to exert beneficial effects.36–38 These basic and clinical studies on the relationship between PI3K/AKT/mTOR and MN, and the results of our study support the use of pharmacological network prediction.
In this study, to test whether triptolide can bind stably to PI3K/AKT/mTOR, we predicted that triptolide and the hub target AKT1, PIK3R1 and mTOR were molecularly docked. The results showed that triptolide can bind with relatively strong affinity to AKT1, PIK3R1 and mTOR, respectively, to form complexes with stable structures via hydrogen bonds. These results indicated that triptolide directly inhibited PI3K/AKT/mTOR signaling transduction. Furthermore, our experimental results indicated that triptolide may regulate PI3K/AKT/mTOR pathway to exert its renoprotective effects in MN. In total, the above results support the idea that these active components of triptolide work synergistically to exert antiproteinuric and renoprotective effects. However, this study has several limitations. First, the pharmacological mechanism of triptolide against MN was complex, which cannot be fully revealed in this study. Secondly, although we explore the mechanism of triptolide on MN through network pharmacological analysis and animal experiment, there was still lack of inhibitors of PI3K/AKT pathway to further demonstrate the protective effect of triptolide on podocytes.
Conclusion
In this study, the underlying pharmacological mechanisms of triptolide against MN were revealed by applying an integrated strategy that adopted network pharmacological analysis, molecular docking analysis, and experimental validation. Meanwhile, triptolide had excellent stability with the targets PIK3R1, AKT1 and mTOR, respectively. The predictive potential of the identified triptolide-MN network was also validated in PHN. Our analysis revealed that the triptolide therapeutic activity involves in PI3K/AKT/mTOR pathway to alleviate MN. In conclusion, we suggest that triptolide plays multi-target, multi-pathway, and system regulation roles to exerts its therapeutic effects.
Abbreviations
TWHF, Tripterygium wilfordii Hook f; MN, membranous nephropathy; PHN, passive Heymann nephritis; PLA2R, phospholipase A2 receptor; pMN, primary MN; PM2.5, particulate matter 2.5; CR, complete remission; BP, biological processes; CC, cell composition; MF, molecular function; ALB, albumin; TC, total cholesterol; TG, triglycerides; LDL, low density lipoprotein; H&E, hematoxylin and eosin; PAS, periodic acid-Schiff; PASM, periodic acid-sliver methenamine; IOD, integral optical density; TBST, Tris-Buffered Saline and Tween; mTOR, mammalian target of the rapamycin; CASP3, Caspase3; MMP9, metalloproteinase 9.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This study was supported by funds from the Horizontal Subject (HX-DZM-202017) and school-level major project of Beijing University of Traditional Chinese Medicine (2022-JYB-JBZR-038).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Ronco P, Debiec H. Molecular pathogenesis of membranous nephropathy. Annu Rev Pathol. 2020;15(1):287–313. doi:10.1146/annurev-pathol-020117-043811
2. Beck LH, Bonegio RG, Lambeau G, et al. M-type phospholipase A2 receptor as target antigen in idiopathic membranous nephropathy. N Engl J Med. 2009;361(1):11–21. doi:10.1056/NEJMoa0810457
3. Tomas NM, Beck LH, Meyer-Schwesinger C, et al. Thrombospondin type-1 domain-containing 7A in idiopathic membranous nephropathy. N Engl J Med. 2014;371(24):2277–2287. doi:10.1056/NEJMoa1409354
4. Couser WG. Primary membranous nephropathy. Clin J Am Soc Nephrol. 2017;12(6):983–997. doi:10.2215/CJN.11761116
5. Ronco P, Beck L, Debiec H, et al. Membranous nephropathy. Nat Rev Dis Primers. 2021;7(1):69. doi:10.1038/s41572-021-00303-z
6. Xu X, Wang G, Chen N, et al. Long-term exposure to air pollution and increased risk of membranous nephropathy in China. J Am Soc Nephrol. 2016;27(12):3739–3746. doi:10.1681/ASN.2016010093
7. Alsharhan L, Beck LH. Membranous nephropathy: core curriculum 2021. Am J Kidney Dis. 2021;77(3):440–453. doi:10.1053/j.ajkd.2020.10.009
8. Wang YN, Feng HY, Nie X, et al. Recent advances in clinical diagnosis and pharmacotherapy options of membranous nephropathy. Front Pharmacol. 2022;13:907108. doi:10.3389/fphar.2022.907108
9. Chen Y, Deng Y, Ni Z, et al. Efficacy and safety of traditional Chinese medicine (Shenqi particle) for patients with idiopathic membranous nephropathy: a multicenter randomized controlled clinical trial. Am J Kidney Dis. 2013;62(6):1068–1076. doi:10.1053/j.ajkd.2013.05.005
10. Lu H, Luo Y, Su B, et al. Wenyang Lishui decoction ameliorates podocyte injury in membranous nephropathy rat and cell models by regulating p53 and Bcl-2. Evid Based Complement Alternat Med. 2020;2020:6813760. doi:10.1155/2020/6813760
11. Tian R, Wang L, Chen A, et al. Sanqi oral solution ameliorates renal damage and restores podocyte injury in experimental membranous nephropathy via suppression of NFkappaB. Biomed Pharmacother. 2019;115:108904. doi:10.1016/j.biopha.2019.108904
12. Law SK, Simmons MP, Techen N, et al. Molecular analyses of the Chinese herb Leigongteng (Tripterygium wilfordii Hook.f.). Phytochemistry. 2011;72(1):21–26. doi:10.1016/j.phytochem.2010.10.015
13. Kupchan SM, Court WA, Dailey RG, Gilmore CJ, Bryan RF. Triptolide and tripdiolide, novel antileukemic diterpenoid triepoxides from Tripterygium wilfordii. J Am Chem Soc. 1972;94(20):7194–7195. doi:10.1021/ja00775a078
14. Gao J, Zhang Y, Liu X, Wu X, Huang L, Gao W. Triptolide: pharmacological spectrum, biosynthesis, chemical synthesis and derivatives. Theranostics. 2021;11(15):7199–7221. doi:10.7150/thno.57745
15. Bai S, Hu Z, Yang Y, et al. Anti-inflammatory and neuroprotective effects of triptolide via the NF-kappaB signaling pathway in a Rat MCAO model. Anat Rec. 2016;299(2):256–266. doi:10.1002/ar.23293
16. Hou W, Liu B, Triptolide: XH. Medicinal chemistry, chemical biology and clinical progress. Eur J Med Chem. 2019;176:378–392. doi:10.1016/j.ejmech.2019.05.032
17. Zheng CX, Chen ZH, Zeng CH, Qin WS, Li LS, Liu ZH. Triptolide protects podocytes from puromycin aminonucleoside induced injury in vivo and in vitro. Kidney Int. 2008;74(5):596–612. doi:10.1038/ki.2008.203
18. Chen ZH, Qin WS, Zeng CH, et al. Triptolide reduces proteinuria in experimental membranous nephropathy and protects against C5b-9-induced podocyte injury in vitro. Kidney Int. 2010;77(11):974–988. doi:10.1038/ki.2010.41
19. Guo Y, Guo N, Wang J, Wang R, Tang L. Retrospective analysis of Tripterygium wilfordii polyglycoside combined with angiotensin receptor blockers for the treatment of primary membranous nephropathy with sub-nephrotic proteinuria. Ren Fail. 2021;43(1):729–736. doi:10.1080/0886022X.2021.1918555
20. Zheng Q, Yang H, Liu W, et al. Comparative efficacy of 13 immunosuppressive agents for idiopathic membranous nephropathy in adults with nephrotic syndrome: a systematic review and network meta-analysis. BMJ Open. 2019;9(9):e030919. doi:10.1136/bmjopen-2019-030919
21. Chin CH, Chen SH, Wu HH, Ho CW, Ko MT, Lin CY. cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst Biol. 2014;8(Suppl 4):S11. doi:10.1186/1752-0509-8-S4-S11
22. Ronco P, Debiec H. Pathophysiological advances in membranous nephropathy: time for a shift in patient’s care. Lancet. 2015;385(9981):1983–1992. doi:10.1016/s0140-6736(15)60731-0
23. Beck LH, Salant DJ. Membranous nephropathy: from models to man. J Clin Invest. 2014;124(6):2307–2314. doi:10.1172/JCI72270
24. Liu S, Li X, Li H, Liang Q, Chen J, Chen J. Comparison of tripterygium wilfordii multiglycosides and tacrolimus in the treatment of idiopathic membranous nephropathy: a prospective cohort study. BMC Nephrol. 2015;16:200. doi:10.1186/s12882-015-0199-x
25. Wang W, Sheng L, Chen Y, et al. Total coumarin derivates from Hydrangea paniculata attenuate renal injuries in cationized-BSA induced membranous nephropathy by inhibiting complement activation and interleukin 10-mediated interstitial fibrosis. Phytomedicine. 2022;96:153886. doi:10.1016/j.phymed.2021.153886
26. Liu B, Lu R, Li H, et al. Zhen-Wu-Tang ameliorates membranous nephropathy rats through inhibiting NF-kappaB pathway and NLRP3 inflammasome. Phytomedicine. 2019;59:152913. doi:10.1016/j.phymed.2019.152913
27. Bonegio RG, Fuhro R, Wang Z, et al. Rapamycin ameliorates proteinuria-associated tubulointerstitial inflammation and fibrosis in experimental membranous nephropathy. J Am Soc Nephrol. 2005;16(7):2063–2072. doi:10.1681/ASN.2004030180
28. Sun Z, Xu Q, Ma Y, Yang S, Shi J. Circ_0000524/miR-500a-5p/CXCL16 axis promotes podocyte apoptosis in membranous nephropathy. Eur J Clin Invest. 2021;51(3):e13414. doi:10.1111/eci.13414
29. Yang YQ, Liang J, Han XD, et al. Dual-function of triptriolide in podocytes injury: inhibiting of apoptosis and restoring of survival. Biomed Pharmacother. 2019;109:1932–1939. doi:10.1016/j.biopha.2018.11.031
30. Gao J, Liang Z, Zhao F, Liu X, Ma N. Triptolide inhibits oxidative stress and inflammation via the microRNA-155-5p/brain-derived neurotrophic factor to reduce podocyte injury in mice with diabetic nephropathy. Bioengineered. 2022;13(5):12275–12288. doi:10.1080/21655979.2022.2067293
31. Motavalli R, Etemadi J, Soltani-Zangbar MS, et al. Altered Th17/Treg ratio as a possible mechanism in pathogenesis of idiopathic membranous nephropathy. Cytokine. 2021;141:155452. doi:10.1016/j.cyto.2021.155452
32. Gilbert A, Changjuan A, Guixue C, Jianhua L, Xiaosong Q. Urinary matrix metalloproteinase-9 and nephrin in idiopathic membranous nephropathy: a cross-sectional study. Dis Markers. 2021;2021:1620545. doi:10.1155/2021/1620545
33. Qiu D, Kao PN. Immunosuppressive and anti-inflammatory mechanisms of triptolide, the principal active diterpenoid from the Chinese medicinal herb Tripterygium wilfordii Hook. f. Drugs R D. 2003;4(1):1–18. doi:10.2165/00126839-200304010-00001
34. Chiou TT, Chau YY, Chen JB, Hsu HH, Hung SP, Lee WC. Rapamycin attenuates PLA2R activation-mediated podocyte apoptosis via the PI3K/AKT/mTOR pathway. Biomed Pharmacother. 2021;144:112349. doi:10.1016/j.biopha.2021.112349
35. Meim X-D, Cao Y-F, Che -Y-Y, et al. Danshen: a phytochemical and pharmacological overview. Chin J Nat Med. 2019;17(1):59–80. doi:10.1016/s1875-5364(19)30010-x
36. Tang Y, Zhang Y, Li L, Xie Z, Wen C, Huang L. Kunxian capsule for rheumatoid arthritis: inhibition of inflammatory network and reducing adverse reactions through drug matching. Front Pharmacol. 2020;11:485. doi:10.3389/fphar.2020.00485
37. Ma Z, Liu Y, Li C, Zhang Y, Lin N. Repurposing a clinically approved prescription Colquhounia root tablet to treat diabetic kidney disease via suppressing PI3K/AKT/NF-kB activation. Chin Med. 2022;17(1):2. doi:10.1186/s13020-021-00563-7
38. Zhang Y, Jin D, Kang X, et al. Signaling pathways involved in diabetic renal fibrosis. Front Cell Dev Biol. 2021;9:696542. doi:10.3389/fcell.2021.696542
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.