Back to Journals » Clinical, Cosmetic and Investigational Dermatology » Volume 17
Integrated Bioinformatics Analysis Reveals Diagnostic Biomarkers and Immune Cell Infiltration Characteristics of Solar Lentigines
Authors Yang X, Xia Z, Fan Y, Xie Y, Ge G, Lang D, Ao J, Yue D, Wu J, Chen T, Zou Y, Zhang M, Yang R
Received 9 October 2023
Accepted for publication 26 December 2023
Published 12 January 2024 Volume 2024:17 Pages 79—88
DOI https://doi.org/10.2147/CCID.S439655
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Prof. Dr. Rungsima Wanitphakdeedecha
Xin Yang,1,2,* Zhikuan Xia,1,* Yunlong Fan,1 Yitong Xie,1 Ge Ge,1 Dexiu Lang,3 Junhong Ao,1 Danxia Yue,1 Jiamin Wu,1 Tong Chen,1 Yuekun Zou,1 Mingwang Zhang,4 Rongya Yang1,2
1Department of Dermatology, The Seventh Medical Center of PLA General Hospital, Beijing, People’s Republic of China; 2Department of Dermatology, Yanbian University Hospital, Yanji, People’s Republic of China; 3Department of Dermatology, XingYi People’s Hospital, Xingyi, People’s Republic of China; 4Department of Dermatology, Southwest Hospital, Army Medical University, Chongqing, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Mingwang Zhang; Rongya Yang, Email [email protected]; [email protected]
Background: Solar lentigines (SLs), serving as a prevalent characteristic of skin photoaging, present as cutaneous aberrant pigmentation. However, the underlying pathogenesis remains unclear and there is a dearth of reliable diagnostic biomarkers.
Objective: The aim of this study was to identify diagnostic biomarkers for SLs and reveal its immunological features.
Methods: In this study, gene expression profiling datasets (GSE192564 and GSE192565) of SLs were obtained from the GEO database. The GSE192564 was used as the training group for screening of differentially expressed genes (DEGs) and subsequent depth analysis. Gene set enrichment analysis (GSEA) was employed to explore the biological states associated with SLs. The weighted gene co-expression network analysis (WGCNA) was employed to identify the significant modules and hub genes. Then, the feature genes were further screened by the overlapping of hub genes and up-regulated differential genes. Subsequently, an artificial neural network was constructed for identifying SLs samples. The GSE192565 was used as the test group for validation of feature genes expression level and the model’s classification performance. Furthermore, we conducted immune cell infiltration analysis to reveal the immune infiltration landscape of SLs.
Results: The 9 feature genes were identified as diagnostic biomarkers for SLs in this study. And an artificial neural network based on diagnostic biomarkers was successfully constructed for identification of SLs. GSEA highlighted potential role of immune system in pathogenesis of SLs. SLs samples had a higher proportion of several immune cells, including activated CD8 T cell, dendritic cell, myeloid-derived suppressor cell and so on. And diagnostic biomarkers exhibited a strong relationship with the infiltration of most immune cells.
Conclusion: Our study identified diagnostic biomarkers for SLs and explored its immunological features, enhancing the comprehension of its pathogenesis.
Keywords: solar lentigines, photoaging, diagnostic biomarkers, immune infiltration, artificial neural network
Introduction
Solar lentigines (SLs), one of the most common benign hyperpigmented lesions in dermatology, predominantly affect middle‐aged and older individuals of Caucasian and Asian descent. They have always been considered a clinical hallmark of skin aging, specifically photoaging, resulting from cumulative ultraviolet radiation (UVR) and chronic hyperpigmentation.1 However, the pathogenesis of SLs has not been completely understood and objective biomarkers are insufficient in SLs.
In recent years, the rapid advancement of bioinformatics techniques has enabled the identification of potential biomarkers through comprehensive analysis of high-throughput data. Weighted co-expression network analysis (WGCNA) has been widely used to identify hub genes in clinically significant modules by constructing gene co-expression network.2 Additionally, artificial neural network, a widely utilized diagnostic model based on machine learning techniques, has demonstrated substantial potential in medical research with its high diagnostic performance across various diseases.3,4 However, the multi-biomarker based diagnostic model has not been developed for the diagnosis of SLs to date.
In this study, integrated bioinformatic approaches were employed to identify promising diagnostic biomarkers, construct an artificial neural network (ANN) model with high diagnostic performance, and investigate the immune infiltration landscape in SLs. These findings offer novel insights into the biological mechanisms of SLs. The flowchart of this study is shown in Figure 1.
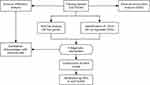 |
Figure 1 Flowchart of the present research. |
Material and Methods
Data Acquisition and Processing
Gene expression profile datasets of SLs (GSE192564, GSE192565) were downloaded from the GEO database (https://www.ncbi.nlm.nih.gov/geo/). The training group for major analysis consisted of 26 SLs lesions and 26 normal samples from GSE192564. The test group for validation, GSE192565, comprised of 12 SLs lesions and 12 normal samples. The probe IDs were matched with gene symbols, and the arithmetic mean of the expression values was calculated for genes that were linked to multiple probe IDs. Subsequently, normalization and log2 transformation were applied to all the data for further analysis.
Identification of the Differentially Expressed Genes (DEGs)
The R package “limma” was employed to identify the DEGs between the control group and the SLs group. The cutoff criteria of |log2FC| ≥ 0.5 and adjusted p value < 0.05 was used. Then, the volcano plot and heatmap of DEGs were plotted by the R package “ggplot2”.
Gene Set Enrichment Analysis (GSEA)
Two pre-defined gene sets (“c2.cp.kegg.v7.5.1.symbols.gmt” and “c5.go.v7.5.1.symbols.gmt”) were downloaded from MSigDB database. GSEA was then conducted using the R package “clusterProfiler”. The biological functions or pathways with |Normalized Enrichment Score (NES)| > 1, adjusted p value < 0.05 and false discovery rate (FDR) < 0.25 were considered to be significantly enriched.
Construction of Weighted Gene Co-Expression Network Analysis
According to descriptions by Zhu et al,2 the R package “WGCNA”was used to construct the gene co-expression network. Data cleaning was firstly performed in training group for removing outliers and genes that ranked in the top 25% of the median absolute deviation were further selected for constructing the correlation matrix. The adjacency matrix was then was converted from the correlation matrix via an appropriate soft threshold power β. Subsequently, the adjacency matrix was turned into a Topological Overlap Matrix (TOM). The genes were then clustered into different gene modules with different colors. Finally, the most significant module associated with SLs was identified through the Pearson’s correlation between modules and clinical traits. Modules exhibiting the highest gene significance (GS) levels were considered as the key module. Furthermore, hub genes were defined as those with Module Membership (MM) > 0.8 and GS > 0.5 in the key module.
Construction and Validation of Artificial Neural Network (ANN) Model
The feature genes for SLs were determined by the overlapping of up-regulated DEGs in the training group and hub genes in the key module. The “Gene Score” of feature genes were firstly evaluated using the R package “limma”. The ANN was then constructed using the R package “neuralnet”. Finally, the classification performance of ANN was evaluated using the R package “pROC”.
Immune Infiltration Analysis
The single-sample gene-set enrichment analysis (ssGSEA) was performed to reveal the difference of immune infiltration between SLs samples and control samples using the R package “GSVA”. Following that, we further evaluated the correlation between feature genes and immune cells by the R package “tidyverse”. P value < 0.05 was considered statistically significant.
Results
Identification of DEGs
A total of 200 DEGs were identified in the training group, including 86 up-regulated genes and 114 down-regulated genes (shown in Figure 2; seen in Supplementary Table 1).
 |
Figure 2 Identification of differentially expressed genes following the cutoff criteria of |log2FC| ≥ 0.5 and adjusted p value < 0.05. (a) volcano plot. (b) Heatmap. |
Gene Set Enrichment Analysis
To explore the biological states of SLs, GSEA was conducted using pre-defined gene sets. We found that various cellular components involved in skin development and immune response were significantly enriched in SLs, including intermediate filament cytoskeleton, basement membrane and MHC protein complex. The biological processes and molecular functions related to immune regulation were significant enriched in SLs, involving the activation and migration of immune cells. Furthermore, the KEGG results in SLs group showed that chemokine signaling pathway, cytokine cytokine receptor interaction and leukocyte transendothelial migration were significant enriched pathways. These findings underscore the potential role of the immune system in the pathogenesis of SLs (shown in Figure 3; seen in Supplementary Tables 2 and 3).
 |
Figure 3 Gene set enrichment analysis (GSEA). (a) Top 5 significantly enriched GO terms in SLs. (b) TOP 5 significantly enriched KEGG pathways in SLs. |
Construction of Weighted Gene Co-Expression Network Analysis
In this study, β=8 was set as the optimal soft threshold for constructing the adjacent matrix. The fit index (R2) of scale-free topology was 0.83 when β=8, as well as the matrix exhibited better connectivity (shown in Figure 4a). The adjacent matrix was then transformed into the TOM and the dissTOM (dissTOM =1-TOM) in turn. The similarity genes were split up into various modules via hierarchical clustering (shown in Figure 4b and c). The module–trait relationship analysis showed that the ME pink module had the highest correlation with SLs (R = −0.9, p < 0.001) (shown in Figure 4d). And ME pink module had the highest gene significance (GS) levels among all modules (shown in Figure 4e). Additionally, a total of 36 driver genes with GS > 0.5 and MM > 0.8 were identified as hub genes highly associated with SLs in the ME pink module (shown in Figure 4f; seen in Supplementary Table 4).
Construction and Validation of ANN on 9 Feature Genes
A total of 9 feature genes (SLC1A3, PCDH7, HOXB7, BPGM, HOXD10, HOXD8, HOXD11, PITX2, ZIC2) for SLs were finally identified in this study (shown in Figure 5a). Subsequently, an ANN with 9 neurons in the input layer (9 feature genes) and 2 neurons in the output layer (Control samples and SLs samples) was successfully constructed (shown in Figure 5b). The ANN model demonstrated exceptional classification performance for SLs samples, as evidenced by an AUC score of 1 in the training group (shown in Figure 5d) and an AUC score of 0.976 in the test group (shown in Figure 5e). Significantly, the AUC values of feature genes remained significant (AUC > 0.9; shown in Figure 5c) in the training group, indicating that these 9 feature genes were promising diagnostic biomarkers for SLs. In addition, the significant up-regulated expression levels of all feature genes were also observed in SLs samples of the test group (shown in Figure 5f).
Assessment of Immune Infiltration and Immune Correlation
The present study further investigated immune infiltration of the 28 types of immunocytes in SLs and control samples using ssGSEA. As shown in Figure 6a, in comparison with control samples, SLs samples had a higher proportion of activated CD8 T cell, activated dendritic cell, Eosinophil, immature dendritic cell, MDSC, natural killer T cell, Neutrophil, T follicular helper cell, Effector memory CD4 T cell and Central memory CD4 T cell (all P < 0.05). However, monocyte and memory B cell in SLs were relatively lower than that in control (all P < 0.05). The correlation analysis of the feature genes and immune cells was further investigated, in order to understand the role of feature genes in immune infiltration. The infiltration of most immune cells exhibited a strong relationship with feature genes, with most of these associations being negatively correlated (shown in Figure 6b).
Discussion
Photoaging refers to the cutaneous damage caused by light, particularly exposure to ultraviolet radiation. Clinical manifestations of photoaging encompass dermal thinning, diminished elasticity, and aberrant pigmentation (solar lentigines).5 It is worth noting that SLs are commonly observed on sun-exposed skin areas such as the face and arms, often resulting in a significant psychological impact for individuals, particularly among middle-aged women who prioritize skin aesthetics. However, limited research has been conducted on SLs, and its pathogenesis remains elusive. Recently, Warrick E and his team collected SLs lesions and control samples from Japanese and European volunteers and conducted gene expression analyses, subsequently depositing the datasets into GEO database.6 Based on the GO enrichment analysis of DEGs, they identified several biological functions closely related to SLs, prompting an in-depth discourse. However, their study still has certain limitations. For instance, the conventional GO enrichment analysis tends to overlook genes with minimal changes in expression levels but significant functional importance, and SLs-specific biomarkers have not been identified. Furthermore, the immune microenvironment plays a pivotal role in maintaining skin homeostasis, yet alterations within the immune microenvironment of SLs have not been reported. In order to uncover more potentially valuable information related to SLs, we conducted a comprehensive investigation into the gene expression profiles shared by Warrick et al6 using integrated bioinformatic approaches, including GSEA, WGCNA, ANN model based on machine learning and immune infiltration analysis. The implications of our findings are significant for identifying diagnostic biomarkers of SLs and shedding light on its underlying pathogenic mechanisms.
In this study, a total of 9 feature genes were determined as diagnostic biomarkers for SLs, and their reliability was confirmed by the ANN model and ROC curves. It’s worth noting that these feature genes are not pigmentation-related genes, suggesting that the presence of advanced SLs lesions may not be significantly influenced by an increase in epidermal melanin content and the number of melanocytes. Among the 9 feature genes, four genes belong to the HOX gene family (HOXB7, HOXD10, HOXD8 and HOXD11). HOX genes constitute a family of transcription factors which is part of a developmental regulatory system. Numerous studies have substantiated that hox proteins not only actively participate in modulating normal tissue development but also in the pathogenesis of diverse cancers.7,8 The over-expression of HOX genes, as reported by Warrick et al,6 has been implicated in the SLs morphological changes that can lead to significant deformation of the dermal-epidermal junction. Furthermore, recent research has revealed a novel role of HOX gene family reactivation in the process of aging. For instance, over-expression of Hoxd8 could led to senescence-associated phenotypes in mouse embryonic fibroblasts.9 Thus, we speculated that the HOX genes may potentially involve in development of SLs by regulating the process of cellular senescence in skin cells. Although there is currently no direct evidence linking HOX genes to melanocytes or the production of melanin. However, it has been demonstrated in previous studies that the HOXA10 can induce up-regulation of the Dickkopf 1 gene, which plays a regulatory role in skin pigmentation.10,11 In addition, we identified two genes, PITX2 and SLC1A3 (glutamate transporter), whose potential regulatory effects on keratinocytes differentiation have been reported in previous studies.6,12 The keratinocytic malfunction is widely recognized as a key driver factor of SLs, providing a theoretical basis for PITX2 and SLC1A3 as biomarkers for SLs.13 Besides, Zic2 may involve in melanocyte stem cells (MCSCs) differentiation, based on the regulative role on Wnt signaling pathway.14 In a recent research, Yamada et al reported that the differentiation process of MCSCs into melanoblasts/melanocytes is most likely to be involved in the pathogenesis of SLs.15 However, the molecular function of BPGM gene involved in skin system has not yet been explored. The BPGM gene, encoding enzyme 2,3-bisphosphoglycerate mutase (BPGM), was widely reported in erythrocytosis.16 Further studies are needed to elucidate its potential association with SLs. PCDH7 belongs to protocadherin, which constitutes the largest subfamily of cadherin group. It has been discovered that protocadherin in human skin is intricately linked to the activity of cutaneous sensory neurons and may influence progression of psoriasis by regulating neuroimmune pathways and perception of external stimuli.17 However, further investigations are required to determine whether the up-regulation of PCDH7 expression may potentially enhance the skin’s sensitivity to ultraviolet radiation stimulation, thereby promoting the development of SLs.
In addition, we performed KEGG pathway analysis for 9 feature genes we identified. However, due to too few gene numbers, KEGG pathways containing two or more feature genes can not be significantly enriched (seen in Supplementary Table 5). The GSEA results in this study highlight the significance of immune system in the pathogenesis of SLs, distinguishing it from previous research. Subsequently, through immune infiltration analysis, we identified heterogeneous immune landscape between SLs lesions and normal skin. The proportion of T cells, neutrophils, and dendritic cells in SLs was found to be significantly higher compared to normal skin, indicating their potential role as major effector cells mediating the immune response in SLs. More recently, researchers observed significant increase in T cells contents in photoaged skin through immunohistochemical techniques, further supporting our findings.5 Importantly, UVR may be the key factor resulting in immune homeostasis dysregulation in SLs.18 It has been reported that UVR can stimulate various skin cells, such as keratinocytes, fibroblasts and melanocytes, and promote the secretion of pro-inflammatory cytokines and chemokines, inducing chronic inflammation.19 It is well known that chronic inflammation is an effective inducer of immunosuppression. In this study, we found that immunosuppressive cells MDSC infiltrate significantly in SLs, which may be involved in immunosuppression and preventing excessive inflammatory response. However, it should be noted that prolonged existence of immunosuppressive state could lead to immune senescence and may facilitate the progression of SLs.20 Furthermore, a significant correlation was observed between several feature genes and diverse immune cells, indicating potential influence of these genes on the immune microenvironment of SLs. Overall, these findings suggest that alterations in the immune microenvironment play a pivotal role in the pathogenesis of SLs, highlighting the significance of restoring skin immune homeostasis as a promising avenue for future therapeutic interventions against SLs.
Admittedly, these findings offer novel insights for investigating pathogenesis of SLs; however, there exists some limitations in this study that need to be addressed in future research. First, although an external dataset was used for validation ensuring the credibility of this study, their reproducibility and broad applicability still need further experimental validations. Second, the potential relevance of feature genes and immune cells in SLs remain to be further investigated. In addition, due to the current lack of available in vitro/in vivo models of SLs, it is crucial to procure enough clinical samples for subsequent studies.
Conclusion
In total, this study identified 9 feature genes, which might serve as potential diagnostic biomarkers for SLs. The ANN diagnostic model, developed based on feature genes, demonstrated excellent diagnostic performance for SLs patients. Nevertheless, further investigations into the molecular mechanisms underlying the involvement of these feature genes in SLs are still warranted. Our findings also provide a better understanding of immunologic mechanisms involved in SLs.
Data Sharing Statement
All data generated or analyzed during this study are included in this article and its Supplementary Material Files. Further enquiries can be directed to the corresponding author.
Statement of Ethics
Ethics was approval by the ethics committee of the Seventh Medical Center of PLA General Hospital with ethics number no. 139 in 2023.
Author Contributions
All authors have made significant contributions to this research, encompassing conception, study design, execution, data processing, analysis and interpretation. All authors actively participated in the revision and critical review of this manuscript, ultimately granting their approval for the final manuscript. All authors have reached a consensus on submitting the article to this journal and accept responsibility for all aspects of the research.
Funding
This study was financially supported by the National Natural Science Foundation of China (NO. 82073466, 82373461 and 82373494), and Beijing Natural Science Foundation, China (NO. 7212105 and 7202201).
Disclosure
All authors declare no conflict of interest.
References
1. Praetorius C, Sturm RA, Steingrimsson E. Sun-induced freckling: ephelides and solar lentigines. Pigm Cell Melanoma Res. 2014;27(3):339–350. doi:10.1111/pcmr.12232
2. Zhu Y, Yang X, Zu Y. Integrated analysis of WGCNA and machine learning identified diagnostic biomarkers in dilated cardiomyopathy with heart failure. Front Cell Dev Biol. 2022;10:1089915. doi:10.3389/fcell.2022.1089915
3. Sun D, Peng H, Wu Z. Establishment and analysis of a combined diagnostic model of alzheimer’s disease with random forest and artificial neural network. Front Aging Neurosci. 2022;14:921906. doi:10.3389/fnagi.2022.921906
4. Tian Y, Yang J, Lan M, Zou T. Construction and analysis of a joint diagnosis model of random forest and artificial neural network for heart failure. Aging. 2020;12(24):26221–26235. doi:10.18632/aging.202405
5. Russell-Goldman E, Murphy GF. The pathobiology of skin aging: new insights into an old dilemma. Am J Pathol. 2020;190(7):1356–1369. doi:10.1016/j.ajpath.2020.03.007
6. Warrick E, Duval C, Nouveau S, et al. Actinic lentigines from Japanese and European volunteers share similar impaired biological functions. J Dermatol Sci. 2022;107(1):8–16. doi:10.1016/j.jdermsci.2022.07.001
7. Morgan R, Hunter K, Pandha HS. Downstream of the HOX genes: explaining conflicting tumour suppressor and oncogenic functions in cancer. Int J Cancer. 2022;150(12):1919–1932. doi:10.1002/ijc.33949
8. Mallo M. Reassessing the role of hox genes during vertebrate development and evolution. Trends Genet. 2018;34(3):209–217. doi:10.1016/j.tig.2017.11.007
9. Chen W, Wang X, Wei G, et al. Single-cell transcriptome analysis reveals six subpopulations reflecting distinct cellular fates in senescent mouse embryonic fibroblasts. Front Genet. 2020;11:867. doi:10.3389/fgene.2020.00867
10. Magnusson M, Brun AC, Miyake N, et al. HOXA10 is a critical regulator for hematopoietic stem cells and erythroid/megakaryocyte development. Blood. 2007;109(9):3687–3696. doi:10.1182/blood-2006-10-054676
11. Yamaguchi Y, Passeron T, Hoashi T, et al. Dickkopf 1 (DKK1) regulates skin pigmentation and thickness by affecting Wnt/beta-catenin signaling in keratinocytes. FASEB J. 2008;22(4):1009–1020. doi:10.1096/fj.07-9475com
12. Shi G, Sohn KC, Choi TY, et al. Expression of paired-like homeodomain transcription factor 2c (PITX2c) in epidermal keratinocytes. Exp Cell Res. 2010;316(19):3263–3271. doi:10.1016/j.yexcr.2010.09.013
13. Barysch MJ, Braun RP, Kolm I, et al. Keratinocytic malfunction as a trigger for the development of solar lentigines. Dermatopathology. 2019;6(1):1–11. doi:10.1159/000495404
14. Xu Z, Zheng J, Chen Z, et al. Multilevel regulation of Wnt signaling by Zic2 in colon cancer due to mutation of β-catenin. Cell Death Dis. 2021;12(6):584. doi:10.1038/s41419-021-03863-w
15. Yamada T, Hasegawa S, Inoue Y, et al. Comprehensive analysis of melanogenesis and proliferation potential of melanocyte lineage in solar lentigines. J Dermatol Sci. 2014;73(3):251–257. doi:10.1016/j.jdermsci.2013.11.005
16. Petousi N, Copley RR, Lappin TR, et al. Erythrocytosis associated with a novel missense mutation in the BPGM gene. Haematologica. 2014;99(10):e201–e204. doi:10.3324/haematol.2014.109306
17. Starr I, Seiffert-Sinha K, Sinha AA, Gokcumen O. Evolutionary context of psoriatic immune skin response. Evol Med Public Health. 2021;9(1):474–486. doi:10.1093/emph/eoab042
18. Nakamura M, Morita A, Seité S, Haarmann-Stemmann T, Grether-Beck S, Krutmann J. Environment-induced lentigines: formation of solar lentigines beyond ultraviolet radiation. Exp Dermatol. 2015;24(6):407–411. doi:10.1111/exd.12690
19. Ciążyńska M, Olejniczak-Staruch I, Sobolewska-Sztychny D, Narbutt J, Skibińska M, Lesiak A. Ultraviolet radiation and chronic inflammation-molecules and mechanisms involved in skin carcinogenesis: a narrative review. Life. 2021;11(4). doi:10.3390/life11040326
20. Salminen A, Kaarniranta K, Kauppinen A. Photoaging: UV radiation-induced inflammation and immunosuppression accelerate the aging process in the skin. Inflamm Res. 2022;71(7–8):817–831. doi:10.1007/s00011-022-01598-8
 © 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.



