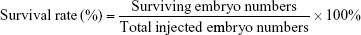Back to Journals » OncoTargets and Therapy » Volume 11
Injection of an SV40 transcriptional terminator causes embryonic lethality: a possible zebrafish model for screening nonhomologous end-joining inhibitors
Authors Yang Z, Chen S, Xue S, Li X, Hu J, Sun Z, Cui H
Received 9 October 2017
Accepted for publication 30 May 2018
Published 17 August 2018 Volume 2018:11 Pages 4945—4953
DOI https://doi.org/10.2147/OTT.S153576
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Jianmin Xu
Zhe Yang,1,2 Shihao Chen,1,2 Songlei Xue,1,2 Xinxiu Li,1,2 Jiang Hu,1,2 Zhen Sun,1,2 Hengmi Cui1–5
1Institute of Epigenetics and Epigenomics, Yangzhou University, Yangzhou, Jiangsu 225009, People’s Republic of China; 2College of Animal Science and Technology, Yangzhou University, Yangzhou, Jiangsu 225009, People’s Republic of China; 3Institute of Comparative Medicine, Yangzhou University, Yangzhou, Jiangsu 225009, People’s Republic of China; 4Jiangsu Co-innovation Center for Prevention and Control of Important Animal Infectious Diseases and Zoonoses, Yangzhou University, Yangzhou, Jiangsu 225009, People’s Republic of China; 5Joint International Research Laboratory of Agricultural & Agri-Product Safety of Educational Ministry of China, Yangzhou University, Yangzhou, Jiangsu 225009, People’s Republic of China
Introduction: DNA repair by the nonhomologous end joining (NHEJ) pathway promotes tumor recurrence after chemotherapy and radiotherapy. Discovery of rapid and high-throughput techniques to screen for an effective NHEJ inhibitor drug is imperative for the suppression of NHEJ during tumor treatment. However, traditional screening methods are too cumbersome to meet the current need. Zebrafish is an ideal model for drug screening due to the specificity of its early embryonic development and similarity of tumor cell generation. By exploiting the high frequency of NHEJ in early embryonic development, we established a model that uses a transcriptional terminator signal fragment from the Simian virus 40 (SV40) to cause embryonic lethality. SV40 fragment-induced embryonic lethality was alleviated by 5,6-bis ((E)-benzylideneamino)-2-mercaptopyrimidin-4-ol or C18H14N4OS (SCR7), an NHEJ inhibitor.
Materials and methods: A 122 bp SV40 terminator fragment (10 ng/µL) was microinjected into zebrafish zygotes. SV40 fragment integration into the zebrafish embryonic genome was detected by Southern blot using a DNA probe for the SV40 terminator. Embryonic lethality rates were observed 24 and 48 h after microinjection. A nonhomologous recombinant inhibitor, SCR7 (5 µM), was used to alleviate embryonic lethality.
Results: Microinjection of zebrafish embryos with the SV40 terminator fragment (10 ng/µL) caused a progressive increase in mortality over time. Using Southern blots, we confirmed that SV40 terminator sequences were integrated into the zebrafish embryonic genome. This phenomenon was effectively alleviated by addition of SCR7.
Conclusion: Injection of an SV40 terminator into zebrafish embryos may cause embryonic lethality due to NHEJ during early zebrafish development. The high mortality of zebrafish embryos could be alleviated by using the NHEJ inhibitor, SCR7. The zebrafish model presented here is simpler and more convenient than traditional methods of screening for NHEJ inhibitors and can be utilized in large-scale drug screens for NHEJ inhibitors and for the development of novel anticancer drugs.
Keywords: nonhomologous end joining, NHEJ, zebrafish embryo, SV40 terminator, SCR7
Introduction
Maintenance of genome integrity is important for the survival of all organisms. Several DNA repair pathways work to protect the genome from genotoxic stresses.1 Typically, genomic integrity is protected by a series of processes, including DNA damage repair, cell cycle checkpoints, and apoptosis. DNA double-strand breaks (DSBs) are considered to be the most lethal type of DNA damage in cells.2 The incorporation of errors during DSB repair may lead to chromosomal rearrangements, such as translocation and deletion, which lead to proto-oncogenic transformations or cell death.3,4 DSBs result from physiological processes, including V(D)J recombination and meiosis, which generate DSBs as intermediates.5 Additionally, DSBs are caused by cancer treatment procedures such as radiotherapy and chemotherapy. These treatments damage tumor cell genomes via physical or chemical methods in order to cause cell death.
In the 1980s, most transgenic animals had been produced by directly injecting DNA into the embryo pronuclei immediately following fertilization. Microinjection of foreign genes into the host genome randomly integrated into the host genome. Many studies have shown that transgenes can be stably transmitted into the reproductive line of fish.6 Studies of transgene integration in mammals suggest that integration seems to be a stochastic process; although sequences in the integration site have some common structural features, there is no so-called integration hotspot.7–10 The integration of exogenous DNA is a mainly random end-to-end tandem process.8,11–15 Studies on transgenic fish have also shown that the integration of foreign genes into the host fish genome is consistent with nonhomologous recombination.15–18 Interestingly, in the early stages of the preparation of transgenic fish, the genome of the embryos was not edited and formed DSBs, and the random integration of foreign genes was entirely through a nonhomologous recombination mechanism.
Higher eukaryotes possess 2 major DSB repair pathways: homologous recombination (HR) and nonhomologous end joining (NHEJ).19 NHEJ is a pathway that repairs DSBs in genomic DNA, which usually arise from ultraviolet (UV) exposure, ionizing radiation (IR), or extreme damage from alkylating agents. NHEJ plays a major role in promoting cellular resistance to radio- and chemotherapy cancer treatment agents.5 NHEJ repair is relatively inaccurate but efficient. DNA ends are recognized by the Ku70 and Ku80 complex, which recruits repair-associated proteins to join DNA ends without the requirement for a homologous template.19 Error-prone or deregulated DNA repair can lead to chromosomal translocations, genome rearrangements, and higher mutation rates, which may provide cancer cells with a survival advantage.20 Although NHEJ is the major DNA DSB repair pathway in mammalian cells,21,22 the inhibitors of NHEJ proteins, including the KU70/80 complex, Artemis, ligase IV/XRCC4, Pol μ, and Pol λ, have been studied as promising targets for tumor therapy.23–25 NHEJ inhibitor drugs, used as sensitizers in the course of tumor therapy, have gradually emerged as direct therapeutic or adjuvant drugs for cancer therapy. In addition, the combination of genotoxic agents (such as radiation) and repair inhibitors can effectively sensitize cancer cells. Importantly, NHEJ inhibitors also reduce the effective dose of radiation and chemotherapeutic drugs and prevent the formation of antitumor and treatment-related side effects. However, traditional drug screening is labor intensive. For example, the HR and NHEJ identification systems were built using I-SceI nuclease in mammalian cells and would require considerable material resources to identify new NHEJ inhibitor drugs.26
Zebrafish has been emerging as a commonly used model organism in the field of small-molecule drug screening since 2000. Although there are differences in the pharmacological effects of zebrafish relative to humans, there are hundreds of small molecules with conserved biological activity in fish and humans.27 Therefore, it is reasonable to speculate that bioactive compounds found in zebrafish-based drug screening will have the same activity in humans. Because zebrafish embryonic development has similar features as tumor development, such as rapid division of cells, apoptosis, and angiogenesis,27 zebrafish embryos are an ideal model to characterize responses to different cancer treatment drugs. NHEJ frequency is high during the early development of zebrafish embryos, as evidenced by the obstacles that occur during transgenic experiments in zebrafish.28 Compared with mouse embryos, zebrafish embryos have a much higher NHEJ efficiency because fertilized eggs and embryos of zebrafish in the wild develop in shallow-water environments with high exposure to UV or strong IR,29 which can have a major impact on genome integrity. To prevent genome damage, zebrafish may have evolved a high frequency of NHEJ repair during embryonic development.
Simian virus 40 (SV40) polyA sequences function as transcriptional terminators in many organisms, including human cells30 and zebrafish.28,31,32 In this study, we observed high embryonic lethality after injecting an SV40 transcription terminator into the fertilized eggs of the zebrafish. 5,6-Bis ((E)-benzylideneamino)-2-mercaptopyrimidin-4-ol or C18H14N4OS (SCR7), an NHEJ inhibitor, effectively decreased the embryonic mortality caused by SV40 transcription terminators. Thus, we suggest that this form of NHEJ inhibition in zebrafish can be used as a valuable model to evaluate NHEJ inhibitor anticancer drugs.
Materials and methods
Preparation of zebrafish embryos
Embryos were generated by natural pairwise mating, as described previously.29 For zebrafish mating pairs, 3 females for every 2 males were used and approximately 150–200 embryos per pair were obtained. Embryos were maintained in E3 solution (5 mmol/L NaCl, 0.17 mmol/L KCl, 0.33 mmol/L CaCl2, and 0.33 mmol/L MgSO4; pH =7.2).
Preparation of SV40 terminator
The SV40 transcriptional terminator was amplified using a pIRES2-eGFP plasmid as a template and the following primers: Forward: 5′-ACTTGTTTATTGCAGCTTATAATGGT-3′; and Reverse: 5′-TAAGATACATTGATGAGTTTGGACAAAC-3′. SV40 DNA fragments were prepared and microinjected at 2 concentrations, namely, 5 and 10 ng/μL.
Microinjection of zebrafish embryos with SV40 terminator
The microinjection groups for zebrafish embryos were divided as follows: the experimental group, injected with the SV40 fragment (5 or 10 ng/μL), the first mock group, injected with random 120 bp DNA fragments without terminator function (5 ng/μL), the second mock group, injected with water that was used as a solvent for all DNA fragments, and the uninjected control group. The purpose of the nonterminator fragment group and the water group was to control for embryonic death caused by microinjection, as well as to confirm that all phenotypes were attributable to injection of the terminator DNA fragment. A total of 1 nL of solution was microinjected into each embryo using a microinjector (Tritech Research, Los Angeles, CA, USA; microINJECTOR system contained MINJ-2, MINJ-3, MINJ-4 MINJ-5, and MINJ-6). All embryos were strictly maintained at an ambient temperature of 28°C to avoid temperature fluctuations that affect embryonic development.
Southern blot
Embryos were then placed in 1.5 mL Eppendorf tubes (200–300 per tube) in 500 μL HOM buffer (80 mM EDTA, 100 mM Tris-HCl, 0.5% SDS, pH 8.0) with 10 μL Proteinase K (0.5 mg/mL) and incubated at 55°C for 3 h, vibrating once per hour. Genomic DNA was then extracted with phenol/chloroform. Roche DIG-High Primer DNA Labelling and Detection Starter Kit I (Hoffman-La Roche Ltd., Basel, Switzerland; catalog number 11745832910) was used for DNA fragment detection. Genomic DNA was digested overnight at 37°C with 4 units of EcoRI endonuclease per microgram of DNA, according to manufacturer’s instructions (New England Biolabs, Ipswich, MA, USA; R310S), and then electrophoresed on a 1% agarose gel prepared with Tris-borate-EDTA (TBE). Then, the DNA was stained with ethidium bromide and visualized by UV illumination. Next, the DNA was denatured using NaOH and transferred to nylon membranes (Roche; LOT15569600). The digoxigenin (DIG)-labeled probe was prepared according to the kit instructions. Normal zebrafish embryonic DNA was used as a negative control, and the SV40 sequence was used as a positive control.
Dechorionation of zebrafish embryos for SCR7 treatment test
SCR7 was purchased from MedChemExpress (Monmouth Junction, NJ, USA; HY-127421533426-72-0) and dissolved in DMSO. To exclude the effect of chorion on the treatment of SCR7, the chorion surrounding the embryos was removed enzymatically at 5 min postfertilization (hpf). Dechorionation was performed by exposing 100 embryos to 1.5 mL of 30 mg/mL protease (Sigma; catalog no 9036-06-0) in a 60 mm plastic dish for approximately 5 min until the chorion begins to detach. Then, the embryos were gently rinsed thoroughly with fresh water. Finally, the zebrafish embryos without chorion were treated with SCR7 at concentrations of 0, 2, 4, 6, 8, 10, and 12 μM.
SCR7 treatment and microinjection
The optimal concentration of SCR7 obtained by screening was selected for inhibition of NHEJ-induced embryo death by injection of the SV40 terminator. A total of 1 nL of the SV40 fragment (10 ng/μL) was used for microinjection of each zygote. After the microinjection, the zebrafish survival rate was calculated as follows:
|
|
Results
Microinjection of SV40 transcriptional terminator led to high mortality of zebrafish embryos
Compared with control and mock groups, the experimental group showed a significant decrease in embryonic survival rate after microinjection with the SV40 transcriptional terminator (Figure 1). When comparing the effect of microinjection concentrations on embryo viability, we found that the embryonic survival rate was significantly lower at 10 ng/μL than at 5 ng/μL 48 h after microinjection (12/573, 2.1% vs 219/560; 39.1%). However, no significant difference was found between the survival rates at the 2 concentrations 24 h after microinjection (244/560, 43.5% vs 280/573, 48.87%) (Figure 2). These results indicated that microinjection of the SV40 transcriptional terminator fragment caused high embryonic mortality in zebrafish and that embryonic viability was negatively correlated with SV40 terminator fragment dosage and time after injection.
High embryonic mortality was caused by SV40 terminator integration
We found that the survival rate of zebrafish embryos significantly decreased as the injection dosage of SV40 terminator and time after the injection increased. To confirm whether the injected SV40 terminator fragment was integrated into the zebrafish genome, genomic DNA extracted from the zebrafish injected with the SV40 terminator fragment (10 ng/μL) was analyzed by Southern blot using the SV40 terminator sequences as a probe. We found that the SV40 terminator sequences were detectable in the genomic DNA from SV40-injected zebrafish but not in the control genomic DNA from uninjected embryos (Figure 3). Thus, we speculated that NHEJ activity in the embryos facilitated genomic integration of a large number of SV40 transcriptional terminator fragments and blocked the gene expression required for embryonic development, resulting in high embryonic mortality in the zebrafish.
SCR7 treatment inhibited NHEJ in zebrafish embryos
To confirm that integration of SV40 terminator and embryonic lethality were caused by NHEJ, we treated the embryos with SCR7, an NHEJ inhibitor.33 SCR7 is a tumor treatment drug and has also been used in clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9-targeted transgenic experiments to inhibit NHEJ and avoid HR in transgenic manipulation.34–36 However, the use of SCR7 in zebrafish is not well described. To determine the optimal drug concentration, we treated the dechorionated zebrafish embryos with a series of various SCR7 concentrations. Our results indicated that treatment with several concentrations of SCR7 <10 μM did not cause high mortality in zebrafish embryos. But when SCR7 concentrations were >10 μM, the embryo mortality rate was increased (Table 1). Finally, we selected the concentration of 5 μM SCR7 in this experiment. To determine whether the addition of SCR7 would improve the survival rate of zebrafish embryos, the effects of injection with both SCR7 (5 μM) and the SV40 terminator fragment (10 ng/μL) were examined in zebrafish embryos. The SV40 terminator-injected group without SCR7 had a significantly low embryonic survival rate as low as 2.26% (296/310) at 48 h after injection. In contrast, the survival rate of SCR7-treated embryos was 54.07% (100/213) at 48 h after the microinjection (Figure 4). Our results indicate that using SCR7 significantly increased the survival rate of zebrafish embryos after microinjection with the SV40 transcriptional terminator. Thus, zebrafish embryos microinjected with the SV40 transcriptional terminator may be an ideal model to simplify screening for NHEJ inhibitors or other anticancer drugs.
Discussion
Our results show that zebrafish embryos injected with an SV40 transcriptional terminator sequence, which leads to embryo lethality, may be a suitable model to screen for NHEJ inhibitor drugs. Many other researchers have previously reported that linearization of foreign genes can be achieved before integration into the host, when used for microinjection.7–12 Studies on transgenic zebrafish have also shown the integration process of exogenous genes characteristics of non-HR.15–18
Therapeutic approaches to cancer are widely classified into 2 types: cytotoxic therapy and molecular targeted drugs.37 Traditional cytotoxic therapies include radiation and chemotherapeutic compounds, such as platinum-based drugs,38–41 which create DSBs in the cell genome. DSBs are considered one of the most lethal types of DNA damages because they affect the integrity and continuity of the genome. Inappropriate repair of DSBs can result in deletions, inversions, duplications, and chromosomal translocations. NHEJ is one of the major DNA DSB repair pathways in cancer cells. Systematic studies of DNA damage and repair using zebrafish have not been extensively reported, despite the advantages of the zebrafish model. Zebrafish are a popular vertebrate model to study embryonic development, and gene function in the zebrafish is conserved across vertebrates, including humans.42 These features make zebrafish a useful system for research on cancer etiology. High-throughput genetic screens for compounds with specific biological activity in a whole organism are feasible using zebrafish embryos.43 The zebrafish genome contains a variety of DNA repair pathways in eukaryotic cells, including direct reversal (DR), mismatch repair (MMR), nucleotide excision repair (NER), base excision repair (BER), HR, NHEJ, and translesion synthesis (TLS). Zebrafish also contain genes in p53-mediated damage recognition pathways. Zebrafish is therefore an ideal model for elucidating DNA damage and repair mechanisms.42
In a natural environment, developing zebrafish embryos are exposed to adverse factors, including UV radiation, IR, and heavy metal ion stimulation. NHEJ can help zebrafish embryos survive the genomic damage caused by these environmental factors. A study of DSB repair in the zebrafish embryos, using the creation of a visual-plus-quantitative analysis system, showed that NHEJ was predominant among the 3 DSB repair pathways (NHEJ, HR, and single-strand annealing [SSA]).44,45 Based on this observation, we explored the idea of building a lethality model using SV40 terminator sequences.
Screening for NHEJ inhibitors is an increasingly important research direction to combat drug resistance and increase radiotherapy sensitivity. Conventional screening methods for NHEJ inhibitors are time consuming. Our findings provide a simpler and more intuitive model to screen for NHEJ inhibitor drugs. Based on the mechanism of NHEJ during zebrafish embryo development, a transcriptional terminator was integrated into the genome. Subsequent embryonic lethality can feasibly be rescued by candidate NHEJ inhibitor drugs.
Compared with the traditional methods of screening for NHEJ inhibitors, this model simplifies the detection process to economically generate more intuitive results. Additionally, this model is more efficient and convenient for large-scale screens for small molecule drugs. However, there are limitations to this model system. Because it is based on the developmental characteristics of zebrafish embryos, several factors, including temperature instability, fertilization rate, and unskilled injections, can affect zebrafish embryonic development and lead to false positives. Additionally, the model can only determine whether a candidate drug is effective but cannot be used to compare which candidate drugs are more effective. Meanwhile, this study also confirmed that SCR7 can really prevent NHEJ in zebrafish. The drug could make the zebrafish genetic modification experiment even more effective, as in mice.
Conclusion
Injection of an SV40 transcriptional terminator into zebrafish embryos may cause embryonic lethality due to NHEJ activity during early zebrafish development. High mortality, seen after the SV40 terminator injection, is alleviated by using the NHEJ inhibitor, SCR7. This model system is simpler and more convenient than traditional NHEJ inhibitor screening methods and may be utilized in large-scale screens for NHEJ inhibitor drugs, as well as in the development of novel anticancer drugs.
Acknowledgments
We appreciate the hard work of the colleagues at the epigenetic lab of our university who helped us reserve and check the relative data. We are also grateful to the people working on zebrafish genetic studies to help us discover this phenomenon.
Author contributions
HC and ZY conceived the study, its design, and coordination. ZY, SC, SX, XL, JH, and ZS conducted the experiments. HC and ZY drafted the manuscript. All authors contributed toward data analysis, drafting and revising the paper and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
Srivastava M, Raghavan SC. DNA double-strand break repair inhibitors as cancer therapeutics. Chem Biol. 2015;22(1):17–29. | ||
Gopalakrishnan V, Raghavan SC. Sequence and structural basis for chromosomal fragility during translocations in cancer. Future Oncol. 2012;8(9):1121–1134. | ||
Nambiar M, Raghavan SC. How does DNA break during chromosomal translocations? Nucleic Acids Res. 2011;39(14):5813–5825. | ||
Zhu C, Mills KD, Ferguson DO, et al. Unrepaired DNA breaks in p53-deficient cells lead to oncogenic gene amplification subsequent to translocations. Cell. 2002;109(7):811–821. | ||
Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem. 2010;79:181–211. | ||
Stuart GW, Mcmurray JV, Westerfield M. Replication, integration and stable germ-line transmission of foreign sequences injected into early zebrafish embryos. Development. 1988;103(2):403–412. | ||
Roth DB, Wilson JH. Nonhomologous recombination in mammalian cells: role for short sequence homologies in the joining reaction. Mol Cell Biol. 1986;6(12):4295–4304. | ||
Hamada T, Sasaki H, Seki R, Sakaki Y. Mechanism of chromosomal integration of transgenes in microinjected mouse eggs: sequence analysis of genome-transgene and transgene-transgene junctions at two loci. Gene. 1993;128(2):197. | ||
Chan AW, Homan EJ, Ballou LU, Burns JC, Bremel RD. Transgenic cattle produced by reverse-transcribed gene transfer in oocytes. Proc Natl Acad Sci U S A. 1998;95(24):14028–14033. | ||
Bill CA, Summers J. Genomic DNA double-strand breaks are targets for hepadnaviral DNA integration. Proc Natl Acad Sci U S A. 2004;101(30):11135. | ||
Houdebine LM, Chourrout D. Transgenesis in fish. Experientia. 1991;47(9):891–897. | ||
Bishop JO, Smith P. Mechanism of chromosomal integration of microinjected DNA. Mol Biol Med. 1989;6(4):283. | ||
Folger KR, Wong EA, Wahl G, Capecchi MR. Patterns of integration of DNA microinjected into cultured mammalian cells: evidence for homologous recombination between injected plasmid DNA molecules. Mol Cell Biol. 1982;2(11):1372. | ||
Dellaire G, Yan J, Little KC, Drouin R, Chartrand P. Evidence that extrachromosomal double-strand break repair can be coupled to the repair of chromosomal double-strand breaks in mammalian cells. Chromosoma. 2002;111(5):304–312. | ||
Penman DJ, Iyengar A, Beeching AJ, Rahman A, Sulaiman Z, Maclean N. Patterns of transgene inheritance in rainbow trout (Oncorhynchus mykiss). Mol Reprod Dev. 1991;30(3):201. | ||
Zhiqiang Z, Zuoyan Z. Transgenes in F4pMThGH-transgenic common carp (Cyprinus carpio L.) are highly polymorphic. Chin Sci Bull. 2001;46(2):143–148. | ||
Bo W, Yong HS, Yan WW, Wang YP, Zuo YZ. Characterization of transgene integration pattern in F4 hGH-transgenic common carp (Cyprinus carpio L.). Cell Res. 2005;15(6):447–454. | ||
Hsiao C-D, Hsieh F-J, Tsai H-J. Enhanced expression and stable transmission of transgenes flanked by inverted terminal repeats from adeno-associated virus in zebrafish. Dev Dyn. 2001;220(4):323. | ||
Pardo B, Gomez-Gonzalez B, Aguilera A. DNA repair in mammalian cells. Cell Mol Life Sci. 2009;66(6):1039–1056. | ||
Lieberman HB. DNA damage repair and response proteins as targets for cancer therapy. Curr Med Chem. 2008;15(4):360–367. | ||
Begg AC, Stewart FA, Vens C. Strategies to improve radiotherapy with targeted drugs. Nat Rev Cancer. 2011;11(4):239–253. | ||
Hefferin ML, Tomkinson AE. Mechanism of DNA double-strand break repair by non-homologous end joining. DNA Repair (Amst). 2005;4(6):639–648. | ||
Chang L, Graham PH, Hao J, et al. PI3K/Akt/mTOR pathway inhibitors enhance radiosensitivity in radioresistant prostate cancer cells through inducing apoptosis, reducing autophagy, suppressing NHEJ and HR repair pathways. Cell Death Dis. 2014;5:e1437. | ||
Beskow C, Skikuniene J, Holgersson A, et al. Radioresistant cervical cancer shows upregulation of the NHEJ proteins DNA-PKcs, Ku70 and Ku86. Br J Cancer. 2009;101(5):816–821. | ||
Bouchaert P, Guerif S, Debiais C, Irani J, Fromont G. DNA-PKcs expression predicts response to radiotherapy in prostate cancer. Int J Radiat Oncol Biol Phys. 2012;84(5):1179–1185. | ||
Rennekamp AJ, Peterson RT. 15 years of zebrafish chemical screening. Curr Opin Chem Biol. 2015;24:58–70. | ||
Amatruda JF, Shepard JL, Stern HM, Zon LI. Zebrafish as a cancer model system. Cancer Cell. 2002;1(3):229–231. | ||
Dai J, Cui XJ, Zhu ZY, Hu W. Non-homologous end joining plays a key role in transgene concatemer formation in transgenic zebrafish embryos. Int J Biol Sci. 2010;6(7):756–768. | ||
Brand M, Granato M, Nüsslein-Volhard C. Keeping and raising zebrafish. Volhard. 2002. | ||
Liu Y, Han X, Yuan J, et al. Biallelic insertion of a transcriptional terminator via the CRISPR/Cas9 system efficiently silences expression of protein-coding and non-coding RNA genes. J Biol Chem. 2017;292(14):5624–5633. | ||
Tiefenbach J, Moll PR, Nelson MR, et al. A live zebrafish-based screening system for human nuclear receptor ligand and cofactor discovery. PLoS One. 2010;5(3):e9797. | ||
Rodrigues AMC, Christen B, Marti M, Belmonte JCI. Skeletal muscle regeneration in Xenopus tadpoles and zebrafish larvae. BMC Dev Biol. 2012;12:9. | ||
Srivastava M, Nambiar M, Sharma S, et al. An inhibitor of nonhomologous end-joining abrogates double-strand break repair and impedes cancer progression. Cell. 2012;151(7):1474–1487. | ||
Chou TY, Tsai PF, Tu YF, Tsai CY. Promoting the efficiency of CRISPR/Cas9-mediated knock-in mice through homology-directed repair via poly(A) tailing and Scr7 treatment. Transgenic Res. 2016;25:226–226. | ||
Vartak SV, Raghavan SC. Inhibition of nonhomologous end joining to increase the specificity of CRISPR/Cas9 genome editing. FEBS J. 2015;282(22):4289–4294. | ||
John F, George J, Srivastava M, et al. Pluronic copolymer encapsulated SCR7 as a potential anticancer agent. Faraday Discuss. 2015;177:155–161. | ||
John F, George J, Vartak SV, et al. Enhanced efficacy of pluronic copolymer micelle encapsulated SCR7 against cancer cell proliferation(a). Macromol Biosci. 2015;15(4):521–534. | ||
Aggarwal S. Targeted cancer therapies. Nat Rev Drug Discov. 2010;9(6):427–428. | ||
Jackson SP. The DNA-damage response: new molecular insights and new approaches to cancer therapy. Biochem Soc Trans. 2009;37(pt 3):483–494. | ||
Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461(7267):1071–1078. | ||
Helleday T, Petermann E, Lundin C, Hodgson B, Sharma RA. DNA repair pathways as targets for cancer therapy. Nat Rev Cancer. 2008;8(3):193–204. | ||
Pei DS, Strauss PR. Zebrafish as a model system to study DNA damage and repair. Mutat Res. 2013;743–744:151–159. | ||
den Hertog J. Chemical genetics: drug screens in zebrafish. Biosci Rep. 2005;25(5–6):289–297. | ||
Bladen CL, Navarre S, Dynan WS, Kozlowski DJ. Expression of the Ku70 subunit (XRCC6) and protection from low dose ionizing radiation during zebrafish embryogenesis. Neurosci Lett. 2007;422(2):97–102. | ||
Bladen CL, Lam WK, Dynan WS, Kozlowski DJ. DNA damage response and Ku80 function in the vertebrate embryo. Nucleic Acids Res. 2005;33(9):3002–3010. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.