Back to Journals » International Journal of Nanomedicine » Volume 13
Inhibition of H1N1 influenza virus-induced apoptosis by functionalized selenium nanoparticles with amantadine through ROS-mediated AKT signaling pathways
Authors Li Y , Lin Z, Guo M, Zhao M, Xia Y, Wang C, Xu T, Zhu B
Received 3 November 2017
Accepted for publication 26 January 2018
Published 3 April 2018 Volume 2018:13 Pages 2005—2016
DOI https://doi.org/10.2147/IJN.S155994
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Lei Yang
Yinghua Li,* Zhengfang Lin,* Min Guo, Mingqi Zhao, Yu Xia, Changbing Wang, Tiantian Xu, Bing Zhu
Center Laboratory, Guangzhou Women and Children’s Medical Center, Guangzhou Medical University, Guangzhou, China
*These authors contributed equally to this work
Introduction: As a therapeutic antiviral agent, the clinical application of amantadine (AM) is limited by the emergence of drug-resistant viruses. To overcome the drug-resistant viruses and meet the growing demand of clinical diagnosis, the use of biological nanoparticles (NPs) has increased in order to develop novel anti-influenza drugs. The antiviral activity of selenium NPs with low toxicity and excellent activities has attracted increasing attention for biomedical intervention in recent years.
Methods and results: In the present study, surface decoration of selenium NPs by AM (Se@AM) was designed to reverse drug resistance caused by influenza virus infection. Se@AM with less toxicity remarkably inhibited the ability of H1N1 influenza to infect host cells through suppression of the neuraminidase activity. Moreover, Se@AM could prevent H1N1 from infecting Madin Darby Canine Kidney cell line and causing cell apoptosis supported by DNA fragmentation and chromatin condensation. Furthermore, Se@AM obviously inhibited the generation of reactive oxygen species and activation of phosphorylation of AKT.
Conclusion: These results demonstrate that Se@AM is a potentially efficient antiviral pharmaceutical agent for H1N1 influenza virus.
Keywords: selenium nanoparticles, amantadine, influenza virus, apoptosis, nanodrug
Introduction
Influenza virus is a segmented RNA virus which is the most highly contagious pathogen worldwide and affects millions of people with influenza each year in seasonal epidemics.1,2 H1N1 influenza virus, which belongs to influenza A type viruses, is a highly infectious respiratory disease.3 This virus was discovered and identified in Mexico and the USA in 2009 and caused 8,768 deaths in 207 countries.4 The mode of spread of H1N1 influenza is as follows: sneezing, cough, and contaminated materials.5 Owing to the mutation of the genome and antigenic shifts in the emerging cross-species infection by avian influenza virus, the influenza virus has a high variability, which may lead to novel influenza strain among humans.6 The influenza infection cycle comprises of several steps: first, the influenza viruses attach to the host cell surface receptor and fuse with the endosomal membrane; second, uncoating of nucleocapsid and multiplication of the genetic material occurs; and finally, the influenza protein and new viron is expressed and released.7,8 Hemagglutinin (HA) and neuraminidase (NA) found on the surface of influenza virus are the most important glycoproteins. HA is a cell-anchoring viral glycoprotein which plays an important role in viral infection by combining sialic acid-containing receptors on host cells and mediating the entry and fusion of the virus.9,10 NA plays an important role in assisting the virus to cleave the linkage between sialic acid and hemagglutinin.11
Existing antiviral drugs approved by the US Food and Drug Administration are NA inhibitors such as oseltamivir/zanamivir and M2 ion channel inhibitors such as rimantadine/amantadine (AM).12,13 The M2 proton channel is a critical factor in viral replication; the replication cycle is arrested and infection of the host is halted when proton transport through the channel is inhibited.14,15 The M2 protein is a 97-residue integral membrane protein with a TM domain of 19 residues and a 54-residue cytoplasmic tail, with several point mutations in pore-lining residues of the A/M2 TM domain resulting in resistance to AM.16,17 Because of the emergence of such drug resistance, AM and rimantadine are no longer recommended as common clinical anti-influenza treatments.18,19 Therefore, the antiviral therapies must be promoted in an attempt to control the pandemic influenza A virus.
Nanomaterials with unique chemical and physical properties have emerged as a promising alternative for virus control.20 The design of new antiviral nanodrug should consider strategies to effectively control viral infection and also deal with the cytotoxicity related to the exposure of biological materials.21,22 According to Tao et al, consensus M2e peptide was combined with gold nanoparticles (NPs) against H1N1 influenza A viruses.23 According to Ye et al, graphene oxide is a promising antiviral agent due to its unique properties.24 According to Vonnemann et al, different sizes of polyvalent NPs inhibited virus.25 Meanwhile, the potential of NPs against viral infections through immunization was reported by Sokolova et al.26 Wang et al reported that the SiO2-layered double hydroxide NPs enhance the response of hepatitis B virus DNA vaccine.27 According to Wang et al, polyoxometalate with a broad spectrum may be a new type of antiviral agent.28 According to Barras et al, carbon nanodots have high-efficiency functionality as entry inhibitors in the early stage of virus infection.29 According to Khanal et al, phenylboronic-acid-modified NPs possess potential antiviral therapeutic application.30 Among them, selenium NPs (SeNPs) with their unique antimicrobial activities have attracted considerable attention.31,32 Se is an essential nutritional trace element with the ability to regulate cellular redox homeostasis.33–35 The deficiency of Se could increase the susceptibility to infections, including respiratory virus and hepatitis B virus infections.36,37 In this study, we present novel SeNPs that can inhibit the ability of H1N1 influenza virus to infect host cell. We look forward to verifying that AM-modified SeNPs (Se@AM) have excellent antiviral activity. Reactive oxygen species (ROS) plays an important role in many physiological processes; oxidative stress is explicated between consumption of ROS and cellular defense mechanisms.38,39 The imbalance of redox is associated with many pathologies, such as skin disease, diabetes, cancer, Leigh syndrome and other diseases.40 This study was performed to investigate how Se@AM inhibits H1N1 influenza virus-induced host cells apoptosis through ROS-mediated signaling pathways.
Materials and methods
Materials
The Madin–Darby Canine Kidney (MDCK) cells were obtained from ATCC® CCL-34TM. H1N1 influenza virus was provided by the Virus Laboratory, Guangzhou Women and Children’s Medical Center, Guangzhou Medical University. Fetal bovine serum and DMEM were purchased from Thermo Fisher Scientific, Waltham, MA, USA. Na2SeO3, vitamin C, AM, MTT, propidium iodide, 4′,6-diamidino-2-phenylindole (DAPI), and 2′,7′-dichlorofluorescin (DCF)-diacetate were purchased from Sigma-Aldrich Co. (St Louis, MO, USA). AKT and caspase-3 antibodies were purchased from Cell Signaling Technology (Boston, MA, USA).
Preparation of Se@AM
SeNPs were synthesized as follows: 0.25 mL stock solution (0.1 M) of Na2SeO3 was gradually added to 2 mL stock solution (50 mM) of vitamin C. Then, 0.8 mL of 1 μM AM was added to the SeNPs solution. We detected the excess AM, vitamin and Na2SeO3 by dialysis for 24 hours. Se@AM NPs were sonicated and then filtered through 0.2 μm size pores. The concentration of SeNPs and AM was measured by inductively coupled plasma atomic emission spectroscopy.
Characterization of Se@AM
The morphology of Se@AM NPs was characterized by transmission electron microscopy (TEM, H-7650). Elemental composition of Se@AM was determined by energy dispersive X-ray spectroscopy (EDX) (EX-250 system; Horiba, Kyoto, Japan). Zeta potential and size distribution of Se@AM were monitored by Malvern Zetasizer (Malvern Instruments Limited, Manchester, UK) software. Fourier-transform infrared spectroscopy samples were recorded using the potassium bromide-disk method (Equinox 55 IR spectrometer). X-ray photoelectron spectroscopy (XPS) measurement was carried out on ESCALab 250 spectrometer.41
Cell viability by MTT assay
Cytotoxicity of Se@AM NPs was performed as previously described.42 H1N1 influenza virus was incubated for 2 h and then rinsed to remove the viruses that were not internalized. The indicated concentrations of AM with or without SeNPs were added to MDCK cells for 24 h. Then, 20 μL/well MTT was added and incubated for 5 h. The formazan crystals were dissolved by adding dimethyl sulfoxide (150 μL/well) and measured at 570 nm.
TEM of Se@AM-treated H1N1
H1N1 influenza virus was treated with Se@AM, and then attached to the carbon-coated collodion grid. The grids were examined by TEM.43
The NA inhibition assay by Se@AM
MDCK cells were infected with H1N1 influenza virus for 2 h. After 24 h, the cell culture supernatants were harvested. Influenza virus NA activity was determined by quantifying the fluorescent product resulting from the cleavage of the substrate 4-methylumbelliferyl-α-D-N-acetylneur-aminic acid sodium salt hydrate solution by NA.44 The reaction mixture consisted of the tested compounds, after incubation for 30 min at 37°C. The fluorescence of the mixture was recorded at excitation wavelength of 360 nm and emission at 460 nm.
Detection of mitochondrial membrane potential (ΔΨm)
The fluorescence intensity from JC-1 monomers was used to estimate the status of ΔΨm in MDCK cells exposed to Se@oseltamivir (OTV), as previously described.45 Cells were trypsinized and resuspended with 10 μg/mL of JC-1. The MDCK cells were then harvested and analyzed by flow cytometry.
Annexin-V-FLUOS staining assay
Plasma membrane alterations in MDCK cells treated with Se@AM were detected as previously described.46 Briefly, MDCK cells were treated with Se@AM for 24 h and stained with annexin-V-FLUOS, then washed with PBS 3 times, and observed by flow cytometric analysis.
Terminal deoxynucleotidyl transferase-mediated deoxyuridine triphosphate nick-end labeling (TUNEL)-DAPI co-staining assay
MDCK cells were observed under fluorescence microscope with TUNEL and incubated with DAPI for nuclear staining, as previously described.47
Caspase-3 activity
The caspase-3 activity was detected under fluorescence microscope with the excitation at 380 nm and emission at 460 nm wavelengths, as previously described.48
TEM analysis of thin sections of MDCK cells
TEM analysis of MDCK cells was performed in situ as previously described.49
Determination of ROS generation
The ROS level was monitored by analyzing the fluorescence intensity of DCF with excitation at 500 nm and emission at 529 nm wavelengths.50
Western blotting analysis
The total proteins were obtained after MDCK cells were treated with Se@AM and incubated with lysis buffer. BCA assay was used to quantify the protein concentration. The bolts were developed with enhanced chemiluminescence reagent that examines the target proteins on the X-ray film.51
Statistical analysis
All the data are presented as mean ± SD. Differences between the 2 groups were evaluated using 2-tailed Student’s t-test. One-way analysis of variance was used in multiple-group comparisons. Difference with P<0.05 or P<0.01 was considered statistically significant.
Results and discussion
Preparation of Se@AM
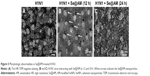
Functionalized Se@AM was synthesized. SeNPs were modified with AM to form more compact and stable NPs (Scheme 1). Light images of AM, SeNPs, and Se@AM are shown in Figure 1A. As shown in Figure 1B and C, the Tyndall effect of Se@AM indicated that Se@AM NPs were synthesized. The morphology of Se@AM presented uniformly spherical particles as shown in Figure 2A. As shown in Figure 2B, EDX indicated the signal of C (20%) and N (8%) from AM, while Se atoms were 72%. The size of Se@AM was decreased from 200 to 70 nm, which indicated much smaller size as shown in Figure 2C and D. The zeta potential of Se@AM (−36.9 mv) was lower than SeNPs (−23.5 mv), demonstrating the higher stability of Se@AM than SeNPs as shown in Figure 2E. The size distribution of Se@AM in Figure 2F revealed that Se@AM was stable for 30 days. Se@AM was further characterized to confirm the chemical binding of AM to the surface of SeNPs. The Fourier transform infrared (FTIR) spectra of unmodified AM, SeNPs, and Se@AM are shown in Figure 3A. The FTIR spectrum of Se@AM resembles that of AM, giving clear evidence that AM ligand forms a part of the nanocomposite. AM displays IR absorbance peaks at 2,903, 1,264, and 739 cm−1 corresponding to −CH2, −NH2, and C−N. The absence of these peaks in Se@AM indicated the formation of Se@AM. The XPS spectra were also recorded to examine the interaction between AM and SeNPs. As shown in Figure 3B, the N 1s peak in the spectrum of Se@AM further confirmed that AM has been successfully conjugated to the SeNPs.
  | Scheme 1 Synthetic route toward Se@AM. |
In vitro antiviral activity of Se@AM
MDCK cells treated with H1N1 influenza virus showed cell viability of 32.34%. AM and SeNPs increased the cell viability to 53.23% and 58.87%, as shown in Figure 4A, but the cell viability of Se@AM was significantly increased to 79.26%. The results indicate that the antiviral effect of Se@AM was superior to that of free SeNPs and AM. As shown in Figure 4B, cells treated with H1N1 influenza virus showed cytoplasmic shrinkage, loss of cell-to-cell contract, and reduction in cells numbers. The MDCK cell morphology changed only slightly by co-treatment with Se@AM and appeared healthy in shape. Synergy was evaluated by calculation of in vitro fractional inhibitory concentration-index values: minimum inhibitory concentration (MIC) of Se in Se@AM (125 μM); MIC of drug B combination concentration of AM in Se@AM (0.1 μM); MIC of drug A alone corresponded to free SeNPs (1 mM); MIC of drug B alone corresponded to free AM (0.4 μM). Fractional inhibitory concentration (FIC) was calculated as (MIC of drug A combination/MIC of drug A alone) + (MIC of drug B combination/MIC of drug B alone) = 125 μM/1 mM + 0.1 μM/0.4 μM = 0.375. FIC was 0.375, below 0.5, indicating synergy. In this study, the FIC index was basically interpreted as follows: FIC <0.5, synergy; FIC between 0.5 and 2, indifference; FIC >2, antagonism. The results suggest that Se@AM effectively inhibited the proliferation of H1N1 influenza virus (Table 1). The titer of H1N1 was detected by Reed-Muench assay (TCID=103.5).
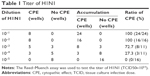  | Table 1 Titer of H1N1 |
H1N1 morphologic changes
The group of H1N1 virus showed typical elliptical or spherically shaped normal H1N1 virus containing virus matrix and capsid (Figure 5A). After interaction of Se@AM with H1N1 virus for 12 and 24 h, the parts of H1N1 viral edges were lost and the viral morphology was destroyed (Figure 5B and C). The results demonstrate that Se@AM could directly interact with virus particles and lead to disruption of viral function.
Identifying NA as the potential target of Se@AM
Surface glycoprotein NA plays an important role in the entry of virus. Untreated H1N1 (100%) or Se@AM-treated H1N1were used to measure the NA enzymatic activity. Compared with AM (75%) and SeNPs (60%), Se@AM (36%) inhibited obvious enzymatic activity of influenza H1N1 viral NA, indicating that the Se@AM NPs had inhibitory effect on NA activity (Figure 6). The anti-influenza mechanism of Se@AM is postulated as: the Se@AM nanoparticles bind tightly to the NA protein, thus forbidding the attachment of H1N1 virus to MDCK cells.
Depletion of mitochondrial membrane potential (ΔΨm) and translocation of phosphatidylserine induced by Se@OTV
MDCK cells were treated with mitochondrial-selective JC-1 dye. As shown in Figure 7A, treatments of MDCK cells with H1N1 influenza virus (30%) resulted in elevation of mitochondrial depolarization and dysfunction. Compared with AM (55%) and SeNPs (65%), when MDCK cells were exposed to Se@AM, the percentage of mitochondrial membrane potential significantly increased to 85%. These results demonstrate that Se@AM inhibited H1N1 influenza virus by apoptosis in MDCK cells by inducing mitochondrial dysfunction. The percentage of living cells treatment with H1N1 influenza virus was 48.80% as shown in Figure 7B. Compared with AM (66.98%) and SeNPs (59.14%), when MDCK cells were exposed to Se@AM, the percentage of MDCK cells treated with Se@AM revealed a decrease in cell number (79.96%). The results demonstrate that Se@AM restrained H1N1 virus infection of MDCK cells mainly through inhibiting apoptosis.
Inhibition of H1N1 infection
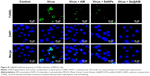
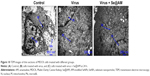
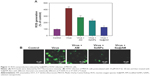
As shown in Figure 8, MDCK cells exhibited apoptotic features by H1N1 influenza virus with DNA fragmentation and nuclear condensation. Co-treatment with Se@AM remarkably prevented the H1N1 influenza virus-induced changes in nuclear morphology. The results indicate that Se@AM rescues the apoptosis of MDCK cells by H1N1 influenza virus.
Inhibition of caspase-3 activation
Caspase-3 activity was used to determine cell apoptosis. As shown in Figure 9, treatment of MDCK cells with H1N1 obviously increased the caspase-3 activity to 519.7%. The caspase-3 activity by AM and SeNPs was 483.6% and 428.3%, respectively. However, Se@AM significantly decreased the caspase-3 activity to 290.2%. The results show that the Se@AM inhibits H1N1 influenza virus activity.
TEM image of thin sections of cells
Mitochondria and microvilli were observed, with no morphological changes in untreated cells as shown in Figure 10A. When MDCK cells were treated with H1N1 influenza virus, TEM image indicated distorted organelles and condensed chromatin, which indicated apoptosis of MDCK cells (Figure 10B). As shown in Figure 10C, the mitochondria of MDCK cells recovered their shape after treatment with Se@AM, which could reveal that H1N1 influenza virus induced apoptosis.
Inhibition of ROS generation by Se@AM
The ROS generation was monitored through DCF assay to indicate the action mechanisms of Se@AM. As shown in Figure 11A, the ROS generation by H1N1 influenza virus was increased to 420%. AM and SeNPs slightly inhibited the ROS generation to 280% and 230%. However, Se@AM remarkably decreased the rate of ROS generation (130%). The fluorescent intensity of DCF was found by H1N1 influenza virus as shown in Figure 11B. The fluorescent intensity of DCF in MDCK cells treated with H1N1 influenza virus was much stronger than in those treated with Se@AM. These results show that Se@AM could downregulate the level of ROS in the antiviral action.
ROS activates apoptotic signaling pathways by Se@AM
The over-generation of ROS could cause DNA damage and affect the regulation of apoptosis signaling pathways. Compared with AM and SeNPs, treatments of MDCK cells with Se@AM remarkably inhibited the expression levels of caspase-3, as shown in Figure 12A. Meanwhile, the level of AKT was significantly increased by Se@AM (Figure 12B). Taken together, Se@AM depressed H1N1 influenza virus-induced host cell apoptosis by ROS-mediated AKT signaling pathways (Figure 12C).
Conclusion
In summary, an efficient chemical method for AM surface-modified SeNPs was described in this study. Se@AM with lower toxicity exhibits enhanced abilities to prevent H1N1 influenza virus infection. The NA activity of H1N1 influenza virus was inhibited by Se@AM. The molecular mechanisms demonstrated that Se@AM inhibited caspase-3 activity and mediated apoptosis through ROS generation. Furthermore, Se@AM inhibited MDCK cell apoptosis by decreasing the level of ROS to trigger AKT pathways. Therefore, Se@AM might provide a promising Se subtype with antiviral properties against H1N1 influenza virus.
Acknowledgments
This work was supported by the Technology Planning Project of Guangzhou (201804010183), Medical Scientific Research Foundation of Guangdong Province (A2018306), Medical Scientific Research Foundation of Guangdong Province (A2018289), The Technology Planning Project of Guangdong Province (2014A020212697), the Technology Planning Project of Guangzhou (201607010120), Guangzhou medical health science and technology project (20181A010022), Science and Technology Project of Guangzhou (201707010014), and National Natural Science Foundation of China (81600399).
Disclosure
The authors report no conflicts of interest in this work.
References
Li Y, Lin Z, Zhao M, et al. Silver nanoparticle based codelivery of oseltamivir to inhibit the activity of the H1N1 influenza virus through ROS-mediated signaling pathways. ACS Appl Mater Interfaces. 2016;8(37):24385–24393. | ||
Wang CZ, Han HH, Tang XY, et al. Sialylglycan-assembled supra-dots for ratiometric probing and blocking of human-infecting influenza viruses. ACS Appl Mater Interfaces. 2017;9(30):25164–25170. | ||
Yang M, Sunderland K, Mao C. Virus-derived peptides for clinical applications. Chem Rev. 2017;117(15):10377–10402. | ||
Li YH, Lin ZF, Zhao MQ, et al. Reversal of H1N1 influenza virus-induced apoptosis by silver nanoparticles functionalized with amantadine. RSC Adv. 2016;6(92):89679–89686. | ||
Li Y, Lin Z, Guo M, et al. Inhibitory activity of selenium nanoparticles functionalized with oseltamivir on H1N1 influenza virus. Int J Nanomedicine. 2017;12:5733–5743. | ||
Jung JH, Park BH, Oh SJ, Choi G, Seo TS. Integration of reverse transcriptase loop-mediated isothermal amplification with an immunochromatographic strip on a centrifugal microdevice for influenza A virus identification. Lab Chip. 2015;15(3):718–725. | ||
Loveday EK, Diederich S, Pasick J, Jean F. Human microRNA-24 modulates highly pathogenic avian-origin H5N1 influenza A virus infection in A549 cells by targeting secretory pathway furin. J Gen Virol. 2015;96(Pt 1):30–39. | ||
Londrigan SL, Short KR, Ma J, et al. Infection of mouse macrophages by seasonal influenza viruses can be restricted at the level of virus entry and at a late stage in the virus life cycle. J Virol. 2015;89(24):12319–12329. | ||
Liu Y, Zhang L, Wei W, et al. Colorimetric detection of influenza A virus using antibody-functionalized gold nanoparticles. Analyst. 2015;140(12):3989–3995. | ||
Hai W, Goda T, Takeuchi H, et al. Specific recognition of human influenza virus with PEDOT bearing sialic acid-terminated trisaccharides. ACS Appl Mater Interfaces. 2017;9(16):14162–14170. | ||
Lin Z, Li Y, Guo M, et al. The inhibition of H1N1 influenza virus-induced apoptosis by silver nanoparticles functionalized with zanamivir. RSC Adv. 2017;7(2):742–750. | ||
Lin Z, Li Y, Guo M, et al. Inhibition of H1N1 influenza virus by selenium nanoparticles loaded with zanamivir through p38 and JNK signaling pathways. RSC Adv. 2017;7(56):35290–35296. | ||
Chen Q, Guo Y. Influenza viral hemagglutinin peptide inhibits influenza viral entry by shielding the host receptor. ACS Infect Dis. 2016;2(3):187–193. | ||
Gleed ML, Busath DD. Why bound amantadine fails to inhibit proton conductance according to simulations of the drug-resistant influenza A M2 (S31N). J Phys Chem B. 2015;119(3):1225–1231. | ||
Gleed ML, Ioannidis H, Kolocouris A, Busath DD. Resistance-mutation (N31) effects on drug orientation and channel hydration in amantadine-bound influenza A M2. J Phys Chem B. 2015;119(35):11548–11559. | ||
Rey-Carrizo M, Barniol-Xicota M, Ma C, et al. Easily accessible polycyclic amines that inhibit the wild-type and amantadine-resistant mutants of the M2 channel of influenza A virus. J Med Chem. 2014;57(13):5738–5747. | ||
Wu Y, Canturk B, Jo H, et al. Flipping in the pore: discovery of dual inhibitors that bind in different orientations to the wild-type versus the amantadine-resistant 531N mutant of the influenza A virus M2 proton channel. J Am Chem Soc. 2014;136(52):17987–17995. | ||
Durrant MG, Eggett DL, Busath DD. Investigation of a recent rise of dual amantadine-resistance mutations in the influenza A M2 sequence. BMC Genet. 2015;16(Suppl 2):S3. | ||
Van Nguyen H, Nguyen HT, Le LT. Investigation of the free energy profiles of amantadine and rimantadine in the AM2 binding pocket. Eur Biophys J. 2016;45(1):63–70. | ||
de Souza E Silva JM, Hanchuk TD, Santos MI, Kobarg J, Bajgelman MC, Cardoso MB. Viral inhibition mechanism mediated by surface-modified silica nanoparticles. ACS Appl Mater Interfaces. 2016;8(26):16564–16572. | ||
Gustafsson S, Mihranyan A. Strategies for tailoring the pore-size distribution of virus retention filter papers. ACS Appl Mater Interfaces. 2016;8(22):13759–13767. | ||
Li YH, Lin ZF, Xu TT, et al. Delivery of VP1 siRNA to inhibit the EV71 virus using functionalized silver nanoparticles through ROS-mediated signaling pathways. RSC Adv. 2017;7(3):1453–1463. | ||
Tao W, Hurst BL, Shakya AK, et al. Consensus M2e peptide conjugated to gold nanoparticles confers protection against H1N1, H3N2 and H5N1 influenza A viruses. Antiviral Res. 2017;141:62–72. | ||
Ye S, Shao K, Li Z, et al. Antiviral activity of graphene oxide: how sharp edged structure and charge matter. ACS Appl Mater Interfaces. 2015;7(38):21571–21579. | ||
Vonnemann J, Sieben C, Wolff C, et al. Virus inhibition induced by polyvalent nanoparticles of different sizes. Nanoscale. 2014;6(4):2353–2360. | ||
Sokolova V, Westendorf AM, Buer J, Uberla K, Epple M. The potential of nanoparticles for the immunization against viral infections. J Mater Chem B. 2015;3(24):4767–4779. | ||
Wang J, Zhu R, Gao B, et al. The enhanced immune response of hepatitis B virus DNA vaccine using SiO2@LDH nanoparticles as an adjuvant. Biomaterials. 2014;35(1):466–478. | ||
Wang J, Liu Y, Xu K, et al. Broad-spectrum antiviral property of polyoxometalate localized on a cell surface. ACS Appl Mater Interfaces. 2014;6(12):9785–9789. | ||
Barras A, Pagneux Q, Sane F, et al. High efficiency of functional carbon nanodots as entry inhibitors of herpes simplex virus type 1. ACS Appl Mater Interfaces. 2016;8(14):9004–9013. | ||
Khanal M, Vausselin T, Barras A, et al. Phenylboronic-acid-modified nanoparticles: potential antiviral therapeutics. ACS Appl Mater Interfaces. 2013;5(23):12488–12498. | ||
Ramya S, Shanmugasundaram T, Balagurunathan R. Biomedical potential of actinobacterially synthesized selenium nanoparticles with special reference to anti-biofilm, anti-oxidant, wound healing, cytotoxic and anti-viral activities. J Trace Elem Med Biol. 2015;32:30–39. | ||
Stevanovic M, Filipovic N, Djurdjevic J, Lukic M, Milenkovic M, Boccaccini A. 45S5Bioglass (R)-based scaffolds coated with selenium nanoparticles or with poly(lactide-co-glycolide)/selenium particles: processing, evaluation and antibacterial activity. Colloids Sur B Biointerfaces. 2015;132:208–215. | ||
Yang Y, Xie Q, Zhao ZN, et al. Functionalized selenium nanosystem as radiation sensitizer of I-125 seeds for precise cancer therapy. ACS Appl Mater Interfaces. 2017;9(31):25857–25869. | ||
Jiang W, Fu Y, Yang F, et al. Gracilaria lemaneiformis polysaccharide as integrin-targeting surface decorator of selenium nanoparticles to achieve enhanced anticancer efficacy. ACS Appl Mater Interfaces. 2014;6(16):13738–13748. | ||
Huang Y, Luo Y, Zheng W, Chen T. Rational design of cancer-targeted BSA protein nanoparticles as radiosensitizer to overcome cancer radioresistance. ACS Appl Mater Interfaces. 2014;6(21):19217–19228. | ||
Yu L, Sun L, Nan Y, Zhu LY. Protection from H1N1 influenza virus infections in mice by supplementation with selenium: a comparison with selenium-deficient mice. Biol Trace Elem Res. 2011;141(1–3):254–261. | ||
Cheng Z, Zhi X, Sun G, et al. Sodium selenite suppresses hepatitis B virus transcription and replication in human hepatoma cell lines. J Med Virol. 2016;88(4):653–663. | ||
Li Y, Lin Z, Zhao M, et al. Multifunctional selenium nanoparticles as carriers of HSP70 siRNA to induce apoptosis of HepG2 cells. Int J Nanomedicine. 2016;11:3065–3076. | ||
Li Y, Guo M, Lin Z, et al. Polyethylenimine-functionalized silver nanoparticle-based co-delivery of paclitaxel to induce HepG2 cell apoptosis. Int J Nanomedicine. 2016;11:6693–6702. | ||
Zhu B, Li Y, Lin Z, et al. Silver nanoparticles induce HePG-2 cells apoptosis through ROS-mediated signaling pathways. Nanoscale Res Lett. 2016;11(1):98. | ||
Liu W, Li X, Wong YS, et al. Selenium nanoparticles as a carrier of 5-fluorouracil to achieve anticancer synergism. ACS Nano. 2012;6(8):6578–6591. | ||
Li Y, Li X, Wong YS, et al. The reversal of cisplatin-induced nephrotoxicity by selenium nanoparticles functionalized with 11-mercapto-1-undecanol by inhibition of ROS-mediated apoptosis. Biomaterials. 2011;32(34):9068–9076. | ||
Xiang DX, Chen Q, Pang L, Zheng CL. Inhibitory effects of silver nanoparticles on H1N1 influenza A virus in vitro. J Virol Methods. 2011;178(1–2):137–142. | ||
Xiao S, Si L, Tian Z, et al. Pentacyclic triterpenes grafted on CD cores to interfere with influenza virus entry: a dramatic multivalent effect. Biomaterials. 2016;78:74–85. | ||
Zhang Y, Li X, Huang Z, Zheng W, Fan C, Chen T. Enhancement of cell permeabilization apoptosis-inducing activity of selenium nanoparticles by ATP surface decoration. Nanomedicine. 2013;9(1):74–84. | ||
Wu H, Li X, Liu W, et al. Surface decoration of selenium nanoparticles by mushroom polysaccharides-protein complexes to achieve enhanced cellular uptake and antiproliferative activity. J Mater Chem. 2012;22(19):9602–9610. | ||
Li Y, Li X, Zheng W, Fan C, Zhang Y, Chen T. Functionalized selenium nanoparticles with nephroprotective activity, the important roles of ROS-mediated signaling pathways. J Mater Chem. 2012;22(19):9602–9610. | ||
Li X, Ma L, Zheng W, Chen T. Inhibition of islet amyloid polypeptide fibril formation by selenium-containing phycocyanin and prevention of beta cell apoptosis. Biomaterials. 2014;35(30):8596–8604. | ||
Li JM, Wang YY, Zhao MX, et al. Multifunctional QD-based co-delivery of siRNA and doxorubicin to HeLa cells for reversal of multidrug resistance and real-time tracking. Biomaterials. 2012;33(9):2780–2790. | ||
Fu X, Yang Y, Li X, et al. RGD peptide-conjugated selenium nanoparticles: antiangiogenesis by suppressing VEGF-VEGFR2-ERK/AKT pathway. Nanomedicine. 2016;12(6):1627–1639. | ||
Xia Y, Lin Z, Li Y, et al. Targeted delivery of siRNA using RGDfC-conjugated functionalized selenium nanoparticles for anticancer therapy. J Mater Chem B. 2017;5(33):6941–6952. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.