Back to Journals » Infection and Drug Resistance » Volume 15
Inducible Resistance to Amikacin in Mycobacterium abscessus Isolated in Beijing, China
Authors Zhang Z, Wang W, Wang Y, Xue Z, Li S , Pang Y
Received 20 January 2022
Accepted for publication 1 April 2022
Published 28 April 2022 Volume 2022:15 Pages 2287—2291
DOI https://doi.org/10.2147/IDR.S357887
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Suresh Antony
Zhijian Zhang,1,* Wei Wang,2,* Yufeng Wang,3 Zhongtan Xue,3 Shanshan Li,2 Yu Pang2
1Department of Respiratory and Critical Care Medicine, the Second Medical Center of Chinese PLA General Hospital, Beijing, 100036, People’s Republic of China; 2Department of Bacteriology and Immunology, Beijing Key Laboratory on Drug-Resistant Tuberculosis Research, Beijing Tuberculosis and Thoracic Tumor Research Institute/Beijing Chest Hospital, Capital Medical University, Beijing, 101149, People’s Republic of China; 3Innovation Alliance on Tuberculosis Diagnosis and Treatment, Beijing, 101149, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Shanshan Li; Yu Pang, Department of Bacteriology and Immunology, Beijing Key Laboratory on Drug-Resistant Tuberculosis Research, Beijing Tuberculosis and Thoracic Tumor Research Institute/Beijing Chest Hospital, Capital Medical University, Beijing, 101149, People’s Republic of China, Tel +86-010-89509368 ; +86-010-89509162, Email [email protected]; [email protected]
Abstract: We aimed to determine the prevalence of amikacin (AMK) resistance of clinical Mycobacterium abscessus (MAB) isolates and to investigate if AMK resistance was induced by AMK exposure. A total of 75 MAB isolates underwent susceptibility testing for AMK after 3 and 14 days of incubation, respectively. The partial fragment of the rrs gene conferring AMK resistance was sequenced. The MIC values for AMK ranged from 0.5 to 128 μg/mL, with MIC50 and MIC90 values of 2 and 32 μg/mL, respectively. In addition, 9.3% of isolates (7/75) were resistant to AMK, all of which harbored a mutation within the rrs locus, including six with A1408G mutation and one with a C1409T mutation. Of note, the MICs of three isolates were significantly increased from 2 μg/mL to 64 μg/mL (one isolate) and 2 μg/mL to 128 μg/mL (two isolates), suggesting that three of the MAB isolates had inducible resistance to AMK. In conclusion, our data demonstrate that approximately one-tenth of clinical MAB isolates in Beijing harbored AMK resistance due to the acquisition of rrs mutations. Additionally, we firstly identified that intrinsic AMK resistance is inducible in MAB isolates, highlighting the urgent need to establish a proper method for the in vitro detection of AMK susceptibility in MAB.
Keywords: amikacin, intrinsic resistance, inducible, Mycobacterium abscessus
Introduction
Nontuberculous mycobacteria (NTM) are increasingly recognized as important pathogens causing infections in both humans and animals.1 Generally, NTM are classified as either slow or rapid growers based on their growth rate.2 Of the rapidly growing mycobacteria (RGM) species, Mycobacterium abscessus (MAB) has been shown to be the most frequently encountered causative agent in human infections, accounting for approximately 65%-80% of RGM lung diseases.1,3 The leading threat posed by this species is due to its resistance to many antibiotics, thereby resulting in unsatisfactory clinical outcomes.4
Amikacin (AMK), a member of the aminoglycoside family of antibiotics, exhibits antimicrobial effects by targeting 16S rRNA and inhibiting protein synthesis.5 It is a cornerstone of antimicrobial chemotherapy treatment for MAB infections in view of its promising efficacy in vitro and in vivo.6 Multiple molecular mechanisms contribute to AMK resistance in MAB isolates, including target mutations and increased efflux transporters.4 The major mechanism conferring AMK resistance is spontaneous single mutations within the rrs gene encoding 16S rRNA, yielding a high level of AMK resistance in MAB clinical isolates.7 Most acquired amikacin-resistant MAB isolates had A1408G or C1409T mutations and other mutations in rrs.8–10 Previous experimental studies have demonstrated that the aminoglycoside-modifying enzyme gene eis2 and the multidrug efflux transporter gene (tap) are also involved in AMK resistance in mycobacteria.11,12 Another important intrinsic mechanism for macrolides in MAB has been described as inducible resistance conferred by a inducible erythromycin ribosomal methylase erm(41) gene.13 Considering that the 16S and 23S rRNAs are the major RNA components in the ribosomal subunits, we hypothesized whether this mechanism was involved in AMK resistance. Thus, the primary objectives of this study were to determine the prevalence of AMK resistance of clinical MAB isolates and to investigate if AMK resistance was induced by AMK exposure.
Materials and Methods
For this study, a set of clinical MAB isolates and the reference MAB strain ATCC 19977 were included in our analysis; the AMK susceptibilities of these organisms were tested by using the broth microdilution assay following the guidelines from the Clinical and Laboratory Standards Institute (CLSI).14 Briefly, freshly cultured colonies were harvested from the surface of Löwenstein–Jensen (L-J) medium and then a bacteria suspension equal to the turbidity of a 0.5 McFarland standard was prepared in normal saline. This suspension was diluted 1:20 in cation-adjusted Mueller–Hinton broth (CAMHB) and incubated in a range of AMK concentrations (0.13 to 128 μg/mL). The inoculated plates were incubated at 30 °C for 72 hours. To assess phenotypic induction, the plates were submitted to an extended incubation with reading after 14 days of incubation, respectively. The MIC was defined as the lowest concentration of antibiotics that inhibited visible growth of mycobacteria.
Mycobacterial DNA was extracted using the MasterPureTM Complete DNA and RNA Purification Kit (Lucigen corporation, Wisconsin, USA) and was used as the template for PCR amplification of the rrs gene conferring acquired resistance to AMK in mycobacteria. The 351 bp fragment of the rrs gene was amplified using the primers rrs-F (5′-ATGACGTCAAGTCATCATGCC-3′) and rrs-R (5′-AGGTGATCCAGCCGCACCTTC-3′) as previously reported.10 The PCR products were sent to the Tsingke Company (Beijing, China) and sequenced with the rrs-F primer by Sanger sequencing method. DNA sequences were aligned with the homologous sequences of the reference MAB strain ATCC 19977 using multiple sequence alignments (http://www.ncbi.nlm.nih.gov/BLAST). The mutations were described corresponding to the positions found in the 16S RNA of Escherichia coli.
Results
All 75 MAB isolates underwent susceptibility testing for AMK, the results of which are summarized in Table 1. The MIC values for AMK ranged from 0.5 to 128 μg/mL, with MIC50 and MIC90 values of 2 and 32 μg/mL, respectively. Using the CLSI-endorsed 64 μg/mL as the breakpoint, 9.3% (7/75) were resistant to AMK, comprising of one with an MIC value of 64 μg/mL and six with MIC values of 128 μg/mL. Out of 68 AMK-susceptible MAB isolates on day 3, eight showed at least an 8-fold increase in MIC of AMK on day 14. Of note, the MICs of three isolates were significantly increased from 2 μg/mL to 64 μg/mL (one isolate) and 2 μg/mL to 128 μg/mL (two isolates), suggesting that three of the MAB isolates had inducible resistance to AMK (Table 2).
 |
Table 1 AMK Susceptibility Results and Mutations Within rrs Locus of MAB Isolates |
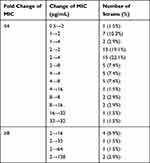 |
Table 2 Fold Changes of MICs of MAB Isolates for 3- and 14-Day Incubation with Amikacin |
We further analyzed the rrs polymorphism conferring AMK resistance. All seven isolates with acquired resistance to AMK harbored a mutation within the rrs locus. Of these isolates, six had a A1408G mutation and one had a C1409T mutation.
Discussion
Due to the intrinsic resistance to a diverse array of antimicrobial agents, clinical management of individuals infected with MAB is challenging since it is difficult to formulate effective therapy regimens;4 however, amikacin holds great promise for the treatment of MAB infections.6 In this study, our data demonstrate that approximately one-tenth of clinical MAB isolates harbored AMK resistance due to the acquisition of rrs mutations. This rate was significantly higher than those from the USA (4.8%),15 South Korea (3.4%),16 France (4.8%),17 and Shanghai (3.9%).18 Several potential explanations could be responsible for the high AMK resistance rate observed in our study. On the one hand, there was a great range in the literature regarding the drug susceptibilities of MAB and M. massiliense. A recent study by Guo and colleagues demonstrated that the rate of AMK resistance for MAB (4.1%) was dramatically higher than that for M. massiliense in Shanghai (0.0%).18 In contrast, a systematic analysis of a global genome dataset found that a slightly higher proportion of M. massiliense genomes contained the canonical rrs mutation conferring high-level AMK resistance than MAB.19 Hence, we speculate that the diverse results across studies may be due to the differences in subspecies compositions. Secondly, the majority of microbiology laboratories do not have the capability to differentiate Mycobacterium tuberculosis from NTM. Therefore, patients infected with NTM could be misdiagnosed as drug-resistant TB, resulting in exposure to inappropriate medications. As the former cornerstone drug for drug-resistant TB, AMK is widely used for the treatment of multidrug-resistant and extensively drug-resistant TB. Although it provides promising efficacy against MAB, the ineffective background regimens would facilitate the emergence of AMK-resistant mycobacteria, which could partly explain the high ratio of AMK resistance in MAB observed in the present study. In view of this potential threat, species identification of mycobacteria is essential to initiate treatment promptly for patients with symptoms suggestive of tuberculosis in this country with a high burden of TB.
We also firstly identified that intrinsic AMK resistance is inducible in MAB isolates. Indeed, there is increasing evidence that the methylation of 16S rRNA is an important mechanism of resistance against aminoglycosides among gram-negative bacteria and actinomycetes.20,21 In the latter case, multiple actinomycetes are intrinsically resistant to aminoglycosides via the inactivation of aminoglycosides by persistent methylation of 16S rRNA.20 In addition to mutations in the rrs allele, a series of genes, including eis (Rv2416c), tap (Rv1258c), and whiB7 (Rv3197), are reported to confer low-level resistance to AMK in mycobacteria. However, our primary data demonstrate that a proportion of isolates had MICs of 128 μg/mL. Another potential mechanism found in clinical settings is acetylation by the aminoglycoside 6′-N-acetyltransferase type Ib, which prevents the interaction between amikacin and the ribosome; this has been noted in several gram-positive and gram-negative bacteria.22,23 However, the expression of this gene was not inducible under AMK exposure, which may be not directly associated with inducible AMK resistance as observed in MAB isolates. As a consequence, we hypothesize that an inducible 16S ribosome methylase may serve as the underlying mechanism associated with inducible AMK resistance, presenting an efficient means to avoid inhibition of its own protein synthesis without additional fitness cost that makes it untenable in vivo. Further experimental research is required to identify the core 16S rRNA methylase involved in this important phenomenon. Although the exact mechanism remains unclear, our findings provide an important hint for the clinical management of MAB patients. Inducible AMK resistance may manifest in diverse clinical outcomes in MAB cases after treatment with AMK-containing regimens. An extended incubation period is required to accurately identify AMK resistance of MAB isolates.
We also acknowledged several limitations to our study. First, although we observed an increase in MICs after AMK exposure, the mechanism for induced AMK resistance was not clearly illustrated in MAB isolates. Further investigations will be conducted to elucidate its underlying mechanisms. Second, given the fact that all the NTM patients enrolled in our study were outpatients, the lack of treatment outcomes from the outpatients inhibits us from verifying the relationship between induced AMK resistance and treatment outcome. Third, the high proportion of AMK resistance was attributed to previous irregular adherence to treatment as well as antibiotic abuse, but the previous exposure history to aminoglycosides was not clear. Finally, recent studies have declared a significant difference in induced CLA resistance between MAB and Mycobacterium massiliense.24 In the present study, we only included MATB isolates. Thus, it is difficult to clarify whether induced AMK resistance differs significantly among subspecies.
In conclusion, our data demonstrate that approximately one-tenth of clinical MAB isolates in Beijing harbored AMK resistance due to the acquisition of rrs mutations. Additionally, we firstly identified that intrinsic AMK resistance is inducible in MAB isolates, highlighting the urgent need to establish a proper method for the in vitro detection of AMK susceptibility in MAB. Further experimental research is required to identify the core 16S rRNA methylase involved in AMK-inducible resistance in MAB.
Ethics Approval and Consent to Participate
The study was conducted according to the principles found in the Declaration of Helsinki. This study was approved by the Ethics Committee of the Beijing Chest Hospital, affiliated with Capital Medical University. All study participants signed an informed consent that agreed to the anonymous use of clinical samples.
Acknowledgments
This study was supported by the Beijing Hospitals Authority Ascent Plan (DFL20191601), the Beijing Hospitals Authority Clinical Medicine Development of Special Funding (ZYLX202122), and Beijing Key Clinical Specialty Project (20201214).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Johansen MD, Herrmann JL, Kremer L. Non-tuberculous mycobacteria and the rise of Mycobacterium abscessus. Nat Rev Microbiol. 2020;18(7):392–407.
2. Prevots DR, Marras TK. Epidemiology of human pulmonary infection with nontuberculous mycobacteria: a review. Clin Chest Med. 2015;36(1):13–34.
3. Jeon K, Kwon OJ, Lee NY, et al. Antibiotic treatment of Mycobacterium abscessus lung disease: a retrospective analysis of 65 patients. Am J Respir Crit Care Med. 2009;180(9):896–902.
4. Nessar R, Cambau E, Reyrat JM, Murray A, Gicquel B. Mycobacterium abscessus: a new antibiotic nightmare. J Antimicrob Chemother. 2012;67(4):810–818.
5. Ramirez MS, Tolmasky ME. Amikacin: uses, Resistance, and Prospects for Inhibition. Molecules. 2017;22(12):34.
6. Daley CL, Iaccarino JM, Lange C, et al. Treatment of Nontuberculous Mycobacterial Pulmonary Disease: an Official ATS/ERS/ESCMID/IDSA Clinical Practice Guideline. Clin Infect Dis. 2020;71(4):905–913.
7. Prammananan T, Sander P, Brown BA, et al. A single 16S ribosomal RNA substitution is responsible for resistance to amikacin and other 2-deoxystreptamine aminoglycosides in Mycobacterium abscessus and Mycobacterium chelonae. J Infect Dis. 1998;177(6):1573–1581.
8. Daniel-Wayman S, Shallom S, Azeem N, Olivier KN, Zelazny AM, Prevots DR. Amikacin exposure and susceptibility of macrolide-resistant Mycobacterium abscessus. ERJ Open Res. 2019;5(2):3456.
9. Kim SY, Kim DH, Moon SM, et al. Association between 16S rRNA gene mutations and susceptibility to amikacin in Mycobacterium avium Complex and Mycobacterium abscessus clinical isolates. Sci Rep. 2021;11(1):6108.
10. Nessar R, Reyrat JM, Murray A, Gicquel B. Genetic analysis of new 16S rRNA mutations conferring aminoglycoside resistance in Mycobacterium abscessus. J Antimicrob Chemother. 2011;66(8):1719–1724.
11. Hurst-Hess K, Rudra P, Ghosh P. Mycobacterium abscessus WhiB7 Regulates a Species-Specific Repertoire of Genes To Confer Extreme Antibiotic Resistance. Antimicrob Agents Chemother. 2017;61:11.
12. Ramon-Garcia S, Mick V, Dainese E, et al. Functional and genetic characterization of the tap efflux pump in Mycobacterium bovis BCG. Antimicrob Agents Chemother. 2012;56(4):2074–2083.
13. Nash KA, Brown-Elliott BA, Wallace RJ. A novel gene, erm(41), confers inducible macrolide resistance to clinical isolates of Mycobacterium abscessus but is absent from Mycobacterium chelonae. Antimicrob Agents Chemother. 2009;53(4):1367–1376.
14. CLSI. Performance Standards for Susceptibility Testing of Mycobacteria, Nocardia Spp., And Other Aerobic Actinomycetes.
15. Realegeno S, Mirasol R, Garner OB, Yang S. Clinical Whole Genome Sequencing for Clarithromycin and Amikacin Resistance Prediction and Subspecies Identification of Mycobacterium abscessus. J Mol Diagn. 2021;23(11):1460–1467.
16. Huh HJ, Kim SY, Shim HJ, et al. GenoType NTM-DR Performance Evaluation for Identification of Mycobacterium avium Complex and Mycobacterium abscessus and Determination of Clarithromycin and Amikacin Resistance. J Clin Microbiol. 2019;57(8):32.
17. Mougari F, Amarsy R, Veziris N, et al. Standardized interpretation of antibiotic susceptibility testing and resistance genotyping for Mycobacterium abscessus with regard to subspecies and erm41 sequevar. J Antimicrob Chemother. 2016;71(8):2208–2212.
18. Guo Y, Cao X, Yu J, et al. Antimicrobial Susceptibility of Mycobacterium abscessus Complex Clinical Isolates from a Chinese Tertiary Hospital. Infect Drug Resist. 2020;13:2001–2010.
19. Bronson RA, Gupta C, Manson AL, et al. Global phylogenomic analyses of Mycobacterium abscessus provide context for non cystic fibrosis infections and the evolution of antibiotic resistance. Nat Commun. 2021;12(1):5145.
20. Doi Y, Arakawa Y. 16S ribosomal RNA methylation: emerging resistance mechanism against aminoglycosides. Clin Infect Dis. 2007;45(1):88–94.
21. Wachino J, Shibayama K, Kurokawa H, et al. Novel plasmid-mediated 16S rRNA m1A1408 methyltransferase, NpmA, found in a clinically isolated Escherichia coli strain resistant to structurally diverse aminoglycosides. Antimicrob Agents Chemother. 2007;51(12):4401–4409.
22. Ramirez MS, Tolmasky ME. Aminoglycoside modifying enzymes. Drug Resist Updat. 2010;13(6):151–171.
23. Chiem K, Fuentes BA, Lin DL, et al. Inhibition of aminoglycoside 6’-N-acetyltransferase type Ib-mediated amikacin resistance in Klebsiella pneumoniae by zinc and copper pyrithione. Antimicrob Agents Chemother. 2015;59(9):5851–5853.
24. Koh WJ, Jeon K, Lee NY, et al. Clinical significance of differentiation of Mycobacterium massiliense from Mycobacterium abscessus. Am J Respir Crit Care Med. 2011;183(3):405–410.
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
