Back to Journals » International Journal of General Medicine » Volume 15
Increased Expression of Homeobox 5 Predicts Poor Prognosis: A Potential Prognostic Biomarker for Glioma
Authors Xu C, Huang J , Yang Y, Li L, Li G
Received 20 November 2021
Accepted for publication 5 April 2022
Published 26 April 2022 Volume 2022:15 Pages 4399—4407
DOI https://doi.org/10.2147/IJGM.S350454
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Scott Fraser
Chengran Xu,1 Jinhai Huang,1 Yi Yang,1 Lun Li,2 Guangyu Li1
1Department of Neurosurgery, First Affiliated Hospital of China Medical University, Shenyang, People’s Republic of China; 2Department of Neurosurgery, Anshan Hospital of the First Hospital of China Medical University, Anshan, People’s Republic of China
Correspondence: Guangyu Li, First Affiliated Hospital of China Medical University, Shenyang, People’s Republic of China, Email [email protected]
Background: The homeobox gene 5 (HOXB5) encodes a transcription factor that regulates the embryonic development of the central nervous system. Notably, its expression pattern and prognostic role in glioma remain unelucidated.
Methods: This study identified the relationship between HOXB5 and glioma by investigating HOXB5 expression data from The Cancer Genome Atlas and the Genotype Tissue Expression databases and validating the obtained data using the Chinese Glioma Genome Atlas database. Western blots were used to identify HOXB5 expression levels in glioma cells and clinical samples. Kaplan–Meier and multivariate Cox regression analyses were performed to assess the prognostic value of HOXB5. The key functions and signaling pathways related to HOXB5 were analyzed using GO, KEGG, and GSEA. Immune infiltration was calculated using the microenvironment cell populations-counter, estimate the proportion of immune and cancer, and ESTIMATE algorithms.
Results: The expression of HOXB5 was upregulated in glioma and generally increased with malignancy. HOXB5 was an independent prognostic factor for glioma patients. A nomogram was further built that integrated HOXB5, and it showed stratifying prediction accuracy and efficiency. HOXB5 was associated with the regulation of cell growth, endothelial cell growth, and the IL-6/JAK-STAT3 pathway, and was determined to possibly promote stomatal specimen enrichment and angiogenesis.
Conclusion: HOXB5 protein is overexpressed in glioma and might serve as a good predictive factor of this disease.
Keywords: HOXB5, nomogram, glioma
Introduction
Gliomas are the most widespread and malignant type of primary brain tumors in human adults and have a mortality rate of approximately 30%.1,2 Although surgical resection followed by combined chemo-radiotherapy represents the standard treatment, the prognosis is not encouraging. Following diagnosis, patients with glioblastoma (GBM, the most aggressive type of glioma) have a median survival rate of 14 months.3,4 Although several studies have investigated the molecular mechanisms of glioma malignancy and aimed to identify therapeutic targets,2 the factors involved in promoting malignant proliferation and metastasis have not been completely elucidated. A comprehensive exploration of the oncology-related molecular mechanism might help to identify novel glioma-predictive markers.
The HOX gene family is a family of homeobox genes that encode transcription factors with key roles in both tumor development and malignancy.5,6 There are 39 HOX genes in mammals, which are divided into four clusters named HOXA, HOXB, HOXC, and HOXD. The HOXB cluster consists of 11 genes (including, HOXB1, HOXB2, HOXB3, HOXB4, HOXB5, HOXB6, HOXB7, HOXB8, HOXB9, HOXB10, and HOXB13), which encode nuclear proteins containing a specific DNA-binding domain. Several researchers have demonstrated that some members of the HOXB cluster are dysregulated in glioma tissue, which contributes to oncogenesis. For example, HOXB2, HOXB3, and HOXB9 have been reported to be upregulated in glioma tissue and were shown to promote the proliferation and migration of glioma cells.7–9 Conversely, HOXB1 expression in glioma tissue is significantly downregulated and might be strongly associated with the degree of malignancy.10 In our previous study, we constructed an endogenous RNA network that might have an effect on the survival rate of glioblastoma patients; HOXB5 is a member of this network and serves as the binding target of miR-7.11 However, the clinical and prognostic value of HOXB5 in glioma remains unknown, and its functional role in glioma has never been investigated.
This study aimed to identify the relationship between HOXB5 and glioma, as well as its potential prognostic value in glioma. For this, we analyzed the expression level of HOXB5 from glioma and normal brain tissue data from The Cancer Genome Atlas (TCGA) and the Genotype Tissue Expression (GTEx) databases. Moreover, we validated HOXB5 protein expression in glioma cells and clinical samples with Western blotting. Furthermore, we investigated the prognostic value of HOXB5 and built a nomogram for risk quantification. The functional mechanism and tumor microenvironment were explored subsequently.
Materials and Methods
RNA-Sequencing Data and Processing
This study included 1709 glioma samples and 225 non-tumor tissue samples. Samples were summarized from five datasets as follows: TCGA Lower Grade Glioma (LGG); TCGA Glioblastoma (GBM) cohort; The Genotype Tissue Expression (GTEx) datasets; Chinese Glioma Genome Atlas (CGGA) mRNAseq_325 dataset; CGGA mRNA-array_301 dataset and REMBRANDT microarray dataset. We merged five non-tumor samples from TCGA and 200 normal brain cortex samples from GTEx into the normal tissue group of discovery sets for comparison. Moreover, the other three datasets were defined as external validation sets. Furthermore, we used CGGA_mRNA693 dataset as additional validation and put the corresponding results in Figure S1.
The gene expression data (HTSeq-counts and TPM) of TCGA and GTEx datasets were downloaded from the UCSC Xena database (http://xena.ucsc.edu/). The normalized expression matrices of the CGGA microarray cohort, CGGA RNA-seq cohort, and REMBRANDT microarray cohort were obtained from the CGGA database (http://www.cgga.org.cn/). For CGGA RNA-seq data, the normalized count reads from the pre-processed data were log2 transformed after adding a 0.001 pseudo count (to avoid an infinite value upon log transformation). All clinical information was obtained from Gliovis (http://gliovis.bioinfo.cnio.es/). Using the median HOXB5 expression profile, the cases were divided into high and low HOXB5 expression groups. The overall survival (OS) was estimated from data of diagnosis to death or final follow-up. Unavailable or unknown clinical features were regarded as missing values, and the data are summarized in Table S1.
Patient Samples
Patient samples for Western blot were collected at the First Hospital of China Medical University and included seven samples (nine glioma tissues: two cases each for grades II, III, and IV, and one glioma peri-tumor tissue as a control). This study complied with the Declaration of Helsinki and was approved by the Ethics Committee of the First Hospital of China Medical University. All experiments were performed in compliance with the relevant regulations, and all patients provided written informed consent. The patient information are presented in Table S2.
Cell Culture
The human glioma cell line U251 and LN229 cells were purchased from the Chinese Academy of Sciences cell bank (Shanghai, China) in September 2017. U87 was purchased from Gene-Chem (Shanghai, China) in March 2018. HS683 cells were obtained as a gift from Professor Shaowu Ou (Department of Neurosurgery, First Affiliated Hospital of China Medical University). The human neuroblastoma cell line Sh-SY5Y was purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA) in December 2018. Glioma cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM, Bioind, Israel) containing 10% fetal bovine serum (Bioind, Israel) and 1% penicillin/streptomycin (Gibco, USA) at 37°C with 5% CO2. Sh-SY5Y cells were cultured in DMEM/F12 (Gibco, USA).
Protein Extraction and Western Blotting
Total protein was isolated from the samples using whole cell lysis buffer (Beyotime). Next, 20 μg of protein from each sample was loaded into the lanes and subjected to SDS-PAGE, followed by PVDF membrane (Millipore) transfer. Then, the membranes were blocked with 5% skimmed milk for 1 hour at room temperature, followed by primary antibody incubation overnight at 4°C (HOXB5, 1:1000, Bioss, bs-6668R; or GAPDH, 1:2000, Bioss, bs-2188R). Peroxidase-conjugated Affinipure coated anti-rabbit IgG (Bioss; 1:5000) was employed as the secondary antibody. Protein bands were visualized with chemiluminescence ECL reagents (Beyotime) and quantified using Image J software.
Survival Analyses and Model Construction
The Kaplan–Meier method followed by a Log rank test was used to assess survival distribution between the high HOXB5 expression group and the low HOXB5 expression group.12 Receiver operating characteristic (ROC) analysis was performed to estimate the accuracy in predicting 1-, 3-, and 5-year survival. Multivariate cox analysis was applied to investigate independent risk factors. Schoenfeld’s Residuals Test analyses were used to test the assumptions of proportionality of the hazard ratios. Variance inflation factors were tested to ensure that all features had no multicollinearity. A nomogram was generated with the R package ‘rms’. Calibration plots and the concordance index (C-index) were used to evaluate the performance of the model.
Bioinformatics Analysis
The ClusterProfiler package was applied to perform GO and KEGG functional enrichment analysis on HOXB5-correlated genes. Gene set enrichment analysis (GSEA) was performed to identify the enrichment of specific gene sets from MSigDB (http://www.gsea-msigdb.org/gsea/msigdb/index.jsp).13 The criteria were |NES > 1.5|, p-value < 0.05, and FDR < 0.05. The stromal score and glioma purity were computed using the ESTIMATE R package.14 Endothelial cell infiltration scores were calculated with Microenvironment Cell Populations (MCP)-counter and estimate the proportion of immune and cancer (EPIC) package.15,16
Statistical Analysis
All statistical analyses and plots were performed using the R v4.02 software. The Wilcoxon rank sum test was used to assess differences in gene expression, and Pearson correlation analysis was used to calculate correlations. Permutation tests were performed with corrections for Western blot results of clinical samples. Other statistical computations and figure generating were performed using R packages (ggplots2, corrplot, coin, and pheatmap). All reported P-values were two-sided and considered significant for values < 0.05.
Results
Expression of HOXB5 in Peri-Tumor and LGG and GBM Tumors
Based on RNA-sequencing (RNA-seq) data from TCGA and GTEx, we discovered that HOXB5 expression was elevated in gliomas compared to that in non-tumor brain tissues (Figure 1A; p < 0.001). Moreover, the expression level gradually increased with WHO grade, with Grade IV GBM having the highest level (Figure 1B and C; p < 0.001). We also confirmed this observation with CGGA_325 data (Figure 1D–F; p < 0.001).
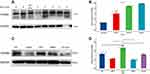
HOXB5 Protein Expression Based on Western Blot Assays
Next, we proved the protein expression pattern of HOXB5 in glioma samples and several central nervous system cancer cell lines. As shown in (Figure 2A and B), six glioma samples showed the upregulation of HOXB5 expression compared with levels in one peri-tumor normal brain tissue (p = 0.02). In addition, we found that GBM U251 presented with the highest expression of HOXB5 protein, whereas HS683 had the least (p < 0.001, Figure 2C and D). These outcomes were consistent with the preceding results. Collectively, these data suggest that HOXB5 protein is overexpressed in aggressive glioma.
Association Between HOXB5 Protein Expression and Glioma Prognosis
As shown (Figure 3A–D), the Kaplan–Meier survival analysis suggested that patients expressing a high level of HOXB5 had a worse prognosis than those expressing a low level of HOXB5 based on four datasets (p < 0.0001). The time-dependent ROC (tROC) curves indicated that the HOXB5 expression profile had moderate accuracy in predicting 1-,3-, and 5- year survival (Figure 3E). Furthermore, multivariate Cox regression analysis indicated that HOXB5 protein expression level was an independent prognostic risk factor in glioma (HR = 1.12, p = 0.01; Table 1), which should arouse the attention of researchers.
 |
Table 1 Multivariate Analysis of Clinical Prognostic Parameters in TCGA |
Construction and Evaluation of a HOXB5-Associated Nomogram
To better quantify the risk assessment and survival probability for glioma patients, we extracted independent prognostic factors and built a nomogram (including age, WHO grade, and HOXB5 expression level (z-score normalized); Figure 3F). Then, we also verified the prediction accuracy of the line chart. The bias-corrected lines of 1-, 3-, and 5-year survival probability in the calibration plot were found to be quite close to the ideal curve (the 45-degree line). The C-index for the nomogram was 0.827 with 1000 bootstrap replicates (Figure 3G). All these results indicated that the nomogram is a promising model of survival prediction for glioma patients.
GO/KEGG Enrichment and GSEA Analysis of HOXB5 in Gliomas
To comprehensively explore the molecular mechanism underlying the effects of HOXB5 in glioma, we investigated 545 genes that were strongly correlated with this marker (Pearson |r| ≥ 0.4; Figure 4A). These genes were enriched in the regulation of cell growth, endothelial cell proliferation, and JAK−STAT signaling pathway, which are involved in tumor progression (Figure 4B–C). GSEA was used to verify the biological function of HOXB5, showing that the high expression group had an activated angiogenesis and IL6/JAK-STAT3 pathway phenotype (Figure 4D–F). These findings indicated that HOXB5 is potentially involved in several tumor-promoting processes.
Glioma Microenvironment Status with HOXB5
Next, we investigated association between HOXB5 and genes involved in angiogenesis among the four datasets. As shown in Figure 5A and Table S3, serval members, including VGEFA, TIPM1, and S100A4, exhibited positive correlations with HOXB5. Furthermore, we examined the microenvironment infiltration status. With ESTIMATE algorithms, we discovered that HOXB5 was positively correlated with the stromal score (r = 0.471) but was negatively correlated with glioma purity (r = −0.428; Figure 5B and C). Additionally, MCP-counter and EPIC algorithms both showed that high expression HOXB5 samples exhibited more endothelial cell infiltration (Figure 5D and E). Together with the preceding results, we indicated that HOXB5 might promote angiogenesis.
Discussion
Glioma represents the most common primary malignancy occurring in the brain, and it has a high mortality rate. Several biomarkers have been reported to be associated with the malignant phenotype of glioma and other disease.17,26,27 However, these are still not adequate. In our study, we comprehensively explored the expression and prognostic role, as well as the functional mechanism, of HOXB5, according to public datasets. Moreover, we validated the protein expression of HOXB5 in glioma cells and clinical tumor samples and respective peri-tumor samples with Western blot experiments. Our analyses proved that HOXB5 shows higher expression in glioma and could serve as a promising prognostic biomarker.
There has been little research on the expression pattern of HOX genes. Under normal conditions, HOX genes regulate the vertebrate central nervous system development at specific time points.18 This pattern is called “spatial and temporal specificity”. An aberrant expression pattern is generally found in poorly differentiated samples and is associated with oncogenic effects.19 However, the expression pattern of HOXB5 in glioma remained to be elucidated. In our study, RNA-seq data from TCGA and CGGA, with protein data from clinical samples, all demonstrated that the expression level of HOXB5 was higher in glioma than in normal brain tissue and that it was increased with increasing grades of glioma.
HOXB5 was reported to be overexpressed and promote poor prognosis in prostate cancer and acute myeloid leukemia.20,21 Furthermore, we comprehensively invested the prognostic value of HOXB5 as a biomarker. Results of four independent cohorts showed that high HOXB5 expression was highly related to an unfavorable OS. Multivariate Cox analysis further proved that HOXB5 is an independent risk factor. Additionally, a nomogram combining HOXB5 and other meaningful clinical features was constructed. Our model showed accuracy and consistency with predicted values for the 1- and 3-year OS. Considering all these results together, we believe that HOXB5 might serve as a promising biomarker for glioma patients.
Angiogenesis is one of the characteristics of malignant glioma, contributing to tumor proliferation and an unfavorable prognosis.22 Activation of the inflammatory response and IL6/JAK-STAT3 signaling pathway has been shown to play important roles in promoting this process.23,24 In the present study, multiple functional analysis of HOXB5-correlated genes revealed a significant association between HOXB5 and the regulation of angiogenesis, inflammatory response, and the IL6/JAK-STAT3 pathway. It is worth noting that Fessner et al reported that the overexpression of HOXB5 can enhance blood vessel remodeling, as well as leukocyte infiltration, in vivo by upregulating IL6.25 In our research, HOXB5 was correlated with endothelial cell proliferation and angiogenesis. Consistently, the population of endothelial cells recruited to the glioma microenvironment was positively associated with HOXB5 expression. Hence, based on these findings, we could conceivably hypothesize that HOXB5 might participate in the regulation of neovascularization. These observations might explain why glioma patients overexpressing HOXB5 have aggressive phenotypes and devastating outcomes. However, further studies are required to better understand the mechanisms through which HOXB5 regulates the glioma microenvironment.
Although our research improved our understanding on the relationship between HOXB5 and glioma, there were a few limitations worth mentioning. First, owing to the limitations of storage conditions, although we validated the expression with cells and clinical samples, there were only seven samples included in the Western blot experiment. Thus, additional glioma samples are required to confirm our results in the future. Furthermore, the study of the direct mechanisms of the involvement of HOXB5 in the development of gliomas requires further functional experimental evaluation.
Conclusions
In summary, the expression level of HOXB5 was evaluated comparing glioma and normal brain samples, and it was found to be generally increased with WHO grade. Moreover, HOXB5 might serve as a promising prognostic biomarker for glioma patients. A nomogram combined with HOXB5 could be beneficial for clinicians in clinical practice. Further, HOXB5 might be associated with endothelial cell growth and could promote angiogenesis.
List of Abbreviations
HOXB5, homeobox gene 5; GTEx, genotype-tissue expression; TCGA, The Cancer Genome Atlas; CGGA, Chinese Glioma Genome Atlas; OS, overall survival; AUC, area under curve; HR, hazard ration; 95% CI, 95% confidence interval; NES, normalized enrichment score; FDR, false discovery rate; GO, gene ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; GSEA, gene set enrichment analysis; MCP-counter, microenvironment cell population counter; EPIC, estimate the proportion of immune and cancer.
Acknowledgments
We thank all the members in Dr. Sun Xun’s lab for helpful advises to our study.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
The project is supported by the Science and Technology Project of Shenyang (18-014-4-03), the Science and Technology Project of the Education Department of Liaoning Province (LFWK201705) and Liaoning BaiQianWan Talents Program.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Weller M, Wick W, Aldape K, et al. Glioma. nature reviews. Dis Primers. 2015;1:15017. doi:10.1038/nrdp.2015.17
2. Barthel F, Wesseling P, Verhaak R. Reconstructing the molecular life history of gliomas. Acta Neuropathol. 2018;135(5):649–670. doi:10.1007/s00401-018-1842-y
3. Wirsching H, Tabatabai G, Roelcke U, et al. Bevacizumab plus hypofractionated radiotherapy versus radiotherapy alone in elderly patients with glioblastoma: the randomized, open-label, phase II ARTE trial. Ann Oncol. 2018;29(6):1423–1430. doi:10.1093/annonc/mdy120
4. Sun L, Yang F, Zhang C, et al. Overexpression of paxillin correlates with tumor progression and predicts poor survival in glioblastoma. CNS Neurosci Ther. 2017;23(1):69–75. doi:10.1111/cns.12606
5. Shah N, Sukumar S. The Hox genes and their roles in oncogenesis. Nat Rev Cancer. 2010;10(5):361–371. doi:10.1038/nrc2826
6. Li B, Huang Q, Wei G. The role of HOX transcription factors in cancer predisposition and progression. Cancers. 2019;11(4):528. doi:10.3390/cancers11040528
7. Pan X, Liu W, Chai Y, Wang J, Zhang Y. Genetic and clinical characterization of HOXB2 in glioma. Onco Targets Ther. 2020;13:10465–10473. doi:10.2147/ott.S268635
8. Li W, Li C, Xiong Q, Tian X, Ru Q. MicroRNA-10b-5p downregulation inhibits the invasion of glioma cells via modulating homeobox B3 expression. Exp Ther Med. 2019;17(6):4577–4585. doi:10.3892/etm.2019.7506
9. Fang L, Xu Y, Zou L. Overexpressed homeobox B9 regulates oncogenic activities by transforming growth factor-β1 in gliomas. Biochem Biophys Res Commun. 2014;446(1):272–279. doi:10.1016/j.bbrc.2014.02.095
10. Han L, Liu D, Li Z, et al. HOXB1 is a tumor suppressor gene regulated by miR-3175 in glioma. PLoS One. 2015;10(11):e0142387. doi:10.1371/journal.pone.0142387
11. Long S, Li G. Comprehensive analysis of a long non-coding RNA-mediated competitive endogenous RNA network in glioblastoma multiforme. Exp Ther Med. 2019;18(2):1081–1090. doi:10.3892/etm.2019.7647
12. Blanche P, Dartigues J, Jacqmin-Gadda H. Estimating and comparing time-dependent areas under receiver operating characteristic curves for censored event times with competing risks. Stat Med. 2013;32(30):5381–5397. doi:10.1002/sim.5958
13. Wu T, Hu E, Xu S, et al. clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innovation. 2021;2(3):100141. doi:10.1016/j.xinn.2021.100141
14. Becht E, Giraldo N, Lacroix L, et al. Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome Biol. 2016;17(1):218. doi:10.1186/s13059-016-1070-5
15. Petitprez F, Levy S, Sun C, et al. The murine microenvironment cell population counter method to estimate abundance of tissue-infiltrating immune and stromal cell populations in murine samples using gene expression. Genome Med. 2020;12(1):86. doi:10.1186/s13073-020-00783-w
16. Racle J, de Jonge K, Baumgaertner P, Speiser D, Gfeller D. Simultaneous enumeration of cancer and immune cell types from bulk tumor gene expression data. eLife. 2017;6. doi:10.7554/eLife.26476
17. Tan Y, Zhang S, Xiao Q, et al. Prognostic significance of ARL9 and its methylation in low-grade glioma. Genomics. 2020;112(6):4808–4816. doi:10.1016/j.ygeno.2020.08.035
18. Wellik D. Hox genes and vertebrate axial pattern. Curr Top Dev Biol. 2009;88:257–278. doi:10.1016/s0070-2153(09)88009-5
19. Guerra S, Maertens O, Kuzmickas R, et al. A deregulated HOX gene axis confers an epigenetic vulnerability in KRAS-mutant lung cancers. Cancer Cell. 2020;37(5):705–19.e6. doi:10.1016/j.ccell.2020.03.004
20. Sekino Y, Pham Q, Kobatake K, et al. HOXB5 overexpression is associated with neuroendocrine differentiation and poor prognosis in prostate cancer. Biomedicines. 2021;9(8):893. doi:10.3390/biomedicines9080893
21. Chen M, Qu Y, Yue P, Yan X. The prognostic value and function of hoxb5 in acute myeloid leukemia. Front Genet. 2021;12:678368. doi:10.3389/fgene.2021.678368
22. Ezhilarasan R, Mohanam I, Govindarajan K, Mohanam S. Glioma cells suppress hypoxia-induced endothelial cell apoptosis and promote the angiogenic process. Int J Oncol. 2007;30(3):701–707.
23. Vandekeere S, Dewerchin M, Carmeliet P. Angiogenesis revisited: an overlooked role of endothelial cell metabolism in vessel sprouting. Microcirculation. 2015;22(7):509–517. doi:10.1111/micc.12229
24. Fan X, He L, Dai Q, et al. Interleukin-1β augments the angiogenesis of endothelial progenitor cells in an NF-κB/CXCR7-dependent manner. J Cell Mol Med. 2020;24(10):5605–5614. doi:10.1111/jcmm.15220
25. Fessner A, Esser J, Bluhm F, et al. The transcription factor HoxB5 stimulates vascular remodelling in a cytokine-dependent manner. Cardiovasc Res. 2014;101(2):247–255. doi:10.1093/cvr/cvt244
26. Cheng J, Zhang J, Wu Z, Sun X. Inferring microenvironmental regulation of gene expression from single-cell RNA sequencing data using scMLnet with an application to COVID-19. Brief Bioinform. 2021;22:988–1005. doi:10.1093/bib/bbaa327
27. Zhang J, Guan M, Wang Q, Zhang J, Zhou T, Sun X. Single-cell transcriptome-based multilayer network biomarker for predicting prognosis and therapeutic response of gliomas. Brief Bioinform. 2020;21:1080–1097. doi:10.1093/bib/bbz040
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.