Back to Journals » Pharmacogenomics and Personalized Medicine » Volume 13
IKBKB rs2272736 is Associated with Gastric Cancer Survival
Authors Gong Y, Zhao W, Jia Q, Dai J , Chen N, Chen Y, Gu D, Huo X, Chen J
Received 30 April 2020
Accepted for publication 20 July 2020
Published 19 August 2020 Volume 2020:13 Pages 345—352
DOI https://doi.org/10.2147/PGPM.S258761
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Martin H Bluth
Yang Gong,1,* Wenjing Zhao,1,* Qiong Jia,1 Jiali Dai,1 Nan Chen,1 Yuetong Chen,1 Dongying Gu,1 Xinying Huo,1 Jinfei Chen1– 3
1Department of Oncology, Nanjing First Hospital, Nanjing Medical University, Nanjing, Jiangsu, People’s Republic of China; 2Cancer Center, Taikang Xianlin Drum Tower Hospital, Nanjing University School of Medicine, Nanjing, Jiangsu, People’s Republic of China; 3Jiangsu Key Laboratory of Cancer Biomarkers, Prevention and Treatment, Collaborative Innovation Center for Personalized Cancer Medicine, Nanjing Medical University, Nanjing, Jiangsu, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Jinfei Chen
Department of Oncology, Nanjing First Hospital, Nanjing Medical University, Nanjing, Jiangsu, People’s Republic of China
Tel/ Fax +86 258 535 8120
Email [email protected]
Background: IKBKB/IKKβ, as the core catalytic subunit of IκB kinase complex, participates in mediation of the classical NF-κB pathway, which has been linked to inflammation and tumorigenesis. Previous studies have shown that single nucleotide polymorphisms in IKBKB have been related to gastric cancer, but how they associate to the clinical outcome is not yet clear. In this study, we retrospectively investigated the associations between single nucleotide polymorphisms located in IKBKB and gastric cancer survival.
Materials and Methods: IKBKB rs2272736 was genotyped in 1210 patients with primary gastric cancer in a Han Chinese population, and the relationships between rs2272736 and overall survival were evaluated. We conducted Cox proportional hazards regression, which was performed to estimate the effects of single nucleotide polymorphisms on the overall survival of patients, adjusted for potential confounding variables.
Results: We found that patients with rs2272736 A allele in IKBKB had significantly prolonged overall survival time compared to those with the G allele (HR = 0.83, 95% CI = 0.68– 1.00, P = 0.050). In addition, AA genotype was demonstrated to have reduced risk of death for gastric cancer compared with that associated with the GG/GA genotypes, which was more common in patients with cardiac carcinoma, well-differentiated and moderately differentiated tumors, TNM Ⅰ/Ⅱ stages and intestinal type.
Conclusion: Our findings have shown that single nucleotide polymorphism rs2272736 in IKBKB may be a promising prognostic biomarker which should promote personalized treatment.
Keywords: gastric cancer, IKBKB, single nucleotide polymorphisms, survival
Introduction
Gastric cancer (GC) is the fifth most common cancer and a leading cause of cancer mortality worldwide.1 Despite the advances in surgery, chemotherapy and radiation therapy, GC is still the most prevalent cancer in East Asia, causing more than 70,000 deaths every year.2 Especially, the 5-year survival of patients with GC is only 30% in China, indicating a very poor prognosis.3 GC is a highly heterogeneous genetic disease and the prognosis of most patients with advanced GC is far from satisfactory. Several studies indicate that NF-κB signaling pathway is associated with biological properties of GC.4–6 NF-κB pathway plays important roles in tumor initiation, promotion and progression.7 NF-κB activation induces DNA damage, oncogenic mutations and genomic instability leading to tumor initiation. Chronic inflammation and NF-κB can also cause chromosomal instability and aneuploidy. NF-κB enhances the proliferation of initiated tumor cells by promoting the production of various cytokines, growth factors and cell cycle proteins.8,9 Most of the altered NF-κB activity discovered in solid tumors is ascribed to enhanced expression of the cytokines activated by the IKK, which comprises the IKBKA and IKBKB catalytic subunits and the IKBKG regulatory subunit.8 As a fundamental regulator of NF-κB activity, not surprisingly, the activity of IKBKB is closely related to tumor development and progression, such as breast cancer,10 skin cancer,11 and GC.12
Genetic variations might be closely associated with the clinical outcome of GC and be potential biomarkers.13,14 SNPs are the most abundant form of DNA variation in the human genome, which can influence the prognosis of GC.15–17 Single nucleotide polymorphisms (SNPs) located within cancer-related genes and non-coding RNAs may affect their expression levels by increasing the promoter activity and specific nuclear protein-binding affinity and other regulatory mechanisms.18–22 So far, reports on the association between IKBKB and GC are rare, and only a few of SNPs in IKBKB were clarified.23
Thus, this study reveals the relationships between genetic variants of IKBKB and the survival of GC in 1210 patients with primary GC in a Han Chinese population.
Materials and Methods
Ethics Statement
All patients gave their written informed consents. The study was approved by the ethical review board of Nanjing medical university.
Study Population
Totally 1210 GC patients were enrolled in this study. All the patients with newly and histologically diagnosed GC between 2005 and 2012 were recruited from Nanjing First Hospital of Nanjing Medical University (Nanjing, Jiangsu Province). Patients with other cancers were excluded from the study. Clinical features on sex, age, alcohol consumption, smoking, tumor site, tumor grade, TNM classification and Lauren classification were collected. Patients were followed up by telephone interviews for survival information from the date of diagnosis to death or the last follow-up in 2014. The median follow-up time was 31 months. Smoking status was assigned as smoker if they smoked at least one cigarette per day for more than 1 year. Drinking status was assigned as drinker if they consumed one or more glasses of alcohol weekly for at least 1 year. All patients provided written informed consent prior to the collection of their information and paraffin-embedded tissue used for research purposes.
SNP Screening
The CHB (Han Chinese in Beijing, China) and CHS (Southern Han Chinese) samples of the 1000 Genomes Project were used to identify the SNPs. Haploview 4.2 was used to select the tag SNPs. RegulomeDB24 and SNPinfo25 were used for SNP function prediction. The cutoff score used for RegulomeDB (old version) was 6. The FuncPred of SNPinfo was used for this study.
SNP Genotyping
Genomic DNA was extracted from the paraffin-embedded tissue using the TGuide FFPE DNA Extraction Kit (TIANGEN, OSR-M405). The SNP genotyping work was accomplished by a custom-by-design 48-Plex SNPscan™ kit. This kit was developed as patented SNP genotyping technology by Genesky Biotechnologies Inc. (Shanghai, China). The kit is based on double ligations and multiplex fluorescence polymerase chain reactions (PCR).26 The genotyping success rate was at least 95% in the samples.
Statistical Analysis
Departures from Hardy–Weinberg equilibrium were tested for each SNPs using the R package SNPassoc 1.9–2. Hazard ratios (HRs) and 95% CIs were calculated to evaluate the relationships between SNPs and GC survival using the unconditional univariate and multivariate Cox proportional hazards model from the R package survival 3.1–8. Kaplan–Meier survival curves were generated by survminer 0.4.6. A P-value equal to or less than 0.05 was considered statistically significant difference. Schoenfeld residuals were used to check the proportional hazards (PH) assumption.27 The results of PH tests are shown in Supplementary Table 1. Given that variable sex is responsible for the seemingly some evidence against the PH assumption (P = 0.022), sex was considered as a stratification variable in the Cox models. All the above statistical analyses were performed using R 3.6.3.
Results
Characteristics of Study Patients
The characteristics of the 1210 GC cases are shown in Table 1. The average age of these patients was 63 years, ranging from 19 to 91 years. Tumors in 34.6% of patients originated from the cardia. The most common tumor grade was poorly differentiated (55.8%). The percentages of TNM classification Ⅰ, Ⅱ, III and Ⅳ were 20.7, 17.9, 34.5 and 26.9, respectively. There were 22.5% of patients with diffuse type and 77.5% of patients with intestinal type in GC samples. Follow-up information was available for 1210 (86.7%) patients; 185 patients were lost to follow-up. Within the follow-up period, 466 (38.5%) patients died.
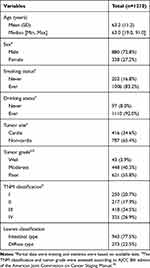 |
Table 1 Clinicopathological Characteristics of GC Patients |
Selection of SNPs in IKBKB
In order to clarify the relationships between genetic variants of IKBKB and the survival of GC, tag SNPs selection in IKBKB was needed. A flow chart of SNPs selection is detailed in Figure 1. First, 72 SNPs were identified from the 1000 Genomes Project. The SNPs will require to meet the following criteria: (1) minor allele frequency (MAF) ≥ 0.05; (2) Hardy–Weinberg equilibrium (HWE) ≥ 0.05; (3) call rate > 95%. Then, tag SNPs were selected with linkage disequilibrium (LD) greater than 0.8 using HaploView. Next, RegulomeDB and SNPinfo were used for SNP function prediction and selected SNPs located in the protein coding and promoter regions. Finally, only one SNP rs2272736 were obtained for genotyping in this study. Hence, the subsequent cohort study focused on this SNP.
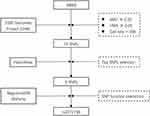 |
Figure 1 Flowchart for selecting SNPs in IKBKB. Abbreviations: MAF, minor allele frequency; HWE, Hardy–Weinberg equilibrium; LD, linkage disequilibrium. |
Association Between Rs2272736 and GC Survival
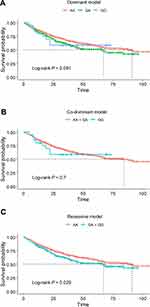
The genotype count of IKBKB rs2272736 and its associations with GC survival according to four genetic models (additive, dominant, codominant and recessive model) are summarized in Table 2. No deviation from HWE (P > 0.05) was detected for IKBKB rs2272736. As is shown in Table 2, the count of the GG, GA, AA genotypes were 20, 261, 836 in GC patients. In the additive model, we found that patients with the A allele showed a significantly increased overall survival time compared to that of those with the G allele (adjusted HR = 0.83, 95% CI = 0.68–1.00, P = 0.050). Compared with GG/GA genotypes, the AA genotype showed better GC survival at a borderline significance level (adjusted HR = 0.81, 95% CI = 0.66–1.01, P = 0.055). Kaplan-Meier survival curves for co-dominant, dominant and recessive models showed consistent results with Cox PH model results (Figure 2A–C). By comparing the results of Akaike information criterion (AIC) in four genetic models (additive, dominant, co-dominant and recessive models), recessive model was chosen for stratification analysis of demographic features. As shown in Supplementary Table 2, we observed a significant association between AA genotype and GC survival in subgroup Age > 63 (adjusted HR = 0.73, 95% CI = 0.55–0.95, P = 0.021); subgroup Smoking Ever (adjusted HR = 0.74, 95% CI = 0.59–0.93, P = 0.011); subgroup Drinking Ever (adjusted HR = 0.73, 95% CI = 0.59–0.91, P = 0.005). In addition, a borderline significance level was obtained (adjusted HR = 0.78, 95% CI = 0.61–1.00, P = 0.052) in male patients.
 |
Table 2 Association Between rs2272736 in IKBKB and Gastric Cancer Survival |
The associations between IKBKB rs2272736 and clinicopathologic variables related to GC survival stratified by tumor site, tumor grade, TNM classification and Lauren classification are shown in Table 3. We showed a statistically significant correlation between the AA genotype and the GC survival in the cardia; the risk reduction for death was more than 30% (adjusted HR = 0.68, 95% CI = 0.48–0.97, P = 0.032). However, there was no significant association with respect to patients with non-cardia tumors. A subsequent stratification analysis by tumor grade revealed that the AA genotype was correlated with the GC survival in well-differentiated and moderately differentiated GC at a borderline level of significance (adjusted HR = 0.71, 95% CI = 0.49–1.02, P = 0.062), as in TNM Ⅰ and Ⅱ stages (adjusted HR = 0.63, 95% CI = 0.38–1.05, P = 0.076). Further, we found that the AA genotype is significantly associated with better prognosis in the intestinal type of gastric cancer (adjusted HR = 0.65, 95% CI = 0.51–0.84, P = 0.001). No statistically significant correlation was observed in the other subgroups.
 |
Table 3 Stratification Analyses for rs2272736 Genotypes and Gastric Cancer Survival |
Prediction of Functional Effects of IKBKB Rs2272736
Based on the online tool of eQTL analysis from GTEx project, we found that rs2272736 was not an eQTL for IKBKB (data not shown). As IKBKB rs2272736 was a missense variant and located close to an intron-exon boundary, we need to predict whether it has an impact on the biological function of a protein or a possible effect on splicing. This variant was predicted to be “benign” by Polyphen-2,28 “tolerated” by SIFT,29 “splice site changes” by Mutation Taster,30 and “splice-altering” by dbscSNV V1.1.31 The above results suggested that rs2272736 might affect biological functions by altering splicing rather than the structure and function of a protein.
In addition, we checked whether SNPs in LD with the SNP rs2272736 could have potential biological functions. By using LDlink,32 we identified 329 SNPs in LD with rs2272736 (D’ 0.9–1.0) (data not shown). Unfortunately, we did not find any SNP that was predicted functional.
Discussion
This study set out to gain a better understanding of the association between an IKBKB polymorphism and GC survival in a Han Chinese population. We found that A allele of rs2272736 in IKBKB was significantly associated with a reduced risk of death in patients with developing GC. Rs2272736 was not an eQTL for IKBKB and we did not find any SNP in LD with rs2272736 potentially functional. Further, function prediction indicated that rs2272736 might have an effect on splicing, which could be related to the survival differences of GC patients.
IKBKB, inhibitor of nuclear factor kappa B kinase subunit beta, is a key component of the canonical IKK complex in the conventional pathway of NF-κB activation, which is located at 8p11.21.33,34 IKBKB phosphorylates the inhibitors of NF-κB on 2 critical residues, followed by the polyubiquitination of the inhibitors and subsequent degradation by the proteasome.35–37 In addition to involving many diseases associated with chronic or acute inflammation, IKBKB and NF-κB exhibit higher constitutive activity or aberrant regulation reported in multiple types of cancers.10–12,32–39 However, IKBKB function in GC initiation and progression is far from being understood. The role of IKBKB was proposed to be the promotion of the proliferation of the GC cells dependent on the activation of β-catenin and Erk pathways,40 the regulation of gastric carcinogenesis through anti-apoptotic signaling and cell proliferation,12 and the regulation of anoikis resistance by DBC1 in GC cells.41 These studies suggested that IKBKB acts as an important oncogene facilitating GC progression, but further experiments are needed to further confirm the functions of IKBKB.
We evaluated the effect of IKBKB on clinicopathological phenotypes of GC. A previous study showed that IKBKB rs2272733 was associated with gefitinib-induced skin toxicity in East Asian non-small cell lung cancer (NSCLC) patients, which can serve as a potential biomarker for predicting gefitinib-related Adverse Drug Reactions.42 Moreover, Schizandrin A can form key hydrophobic interactions with IKBKB, contributing to its potent IKBKB inhibitory effect, which may enhance the efficacy of gefitinib in NSCLC patients.43 In addition, elevated IKBKB protein expression was considered a be a marker for higher nuclear grade tumors and significantly shorter survival in clear cell renal cell carcinoma patients.44 Accordingly, there is an unambiguous relationship between IKBKB and clinicopathological variables of various forms of cancer. However, no studies have yet been found on the relationship between genetic variants of IKBKB and the GC survival. In our study, we found that the missense polymorphism rs2272736 (allele A) is associated with reduced risk of death for GC. In addition, results showed that compared with the GG/GA genotypes, the AA genotype of rs2272736 in IKBKB was significantly correlated to longer overall survival in cardiac, well-differentiated and moderately differentiated, intestinal type of and early-stage GC. As mentioned in our results, a possible mechanism underlying the effectiveness of rs2272736 is that it may have an influence on splicing, and further studies are needed to confirm and validate these findings.
This study has examined the impact of IKBKB rs2272736 on the prognosis of GC. Although the results of this study support the view that IKBKB has important implications for gastric tumorigenesis in terms of genetic associations, it has certain limitations due to a Han Chinese population of limited size. Therefore, further studies are needed to clarify the relationship between IKBKB and the development and outcome of GC.
Ethics
The study was conducted in accordance with the Helsinki declaration.
Acknowledgments
This work was supported by the Jiangsu Provincial Special Program of Medical Science (BE2017611) and the National Natural Science Foundation of China (81772978, 81972626).
Disclosure
The authors declare no conflicts of interest.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.21492
2. Ferlay J, Shin H-R, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127(12):2893–2917. doi:10.1002/ijc.25516
3. Tian J, Wang X-D, Chen Z-C. Survival of patients with stomach cancer in Changle city of China. World J Gastroenterol. 2004;10(11):1543–1546. doi:10.3748/wjg.v10.i11.1543
4. Fu Z, Lin L, Liu S, et al. Ginkgo biloba extract inhibits metastasis and ERK/nuclear factor kappa B (NF-κB) signaling pathway in gastric cancer. Med Sci Monit. 2019;25:6836–6845. doi:10.12659/MSM.915146
5. Li W, Li Q, Wei L, et al. Rosmarinic acid analogue-11 induces apoptosis of human gastric cancer SGC-7901 cells via the epidermal growth factor receptor (EGFR)/Akt/nuclear factor kappa B (NF-κB) pathway. Med Sci Monit Basic Res. 2019;25:63–75. doi:10.12659/MSMBR.913331
6. Jiang Z, Wu W, Qian M-L. Cellular damage and apoptosis along with changes in NF-kappa B expression were induced with contrast agent enhanced ultrasound in gastric cancer cells and hepatoma cells. Cancer Cell Int. 2012;12(1):8. doi:10.1186/1475-2867-12-8
7. Barcellos-Hoff MH, Lyden D, Wang TC. The evolution of the cancer niche during multistage carcinogenesis. Nat Rev Cancer. 2013;13(7):511–518. doi:10.1038/nrc3536
8. Taniguchi K, Karin M. NF-κB, inflammation, immunity and cancer: coming of age. Nat Rev Immunol. 2018;18(5):309–324. doi:10.1038/nri.2017.142
9. Park MH, Hong JT. Roles of NF-κB in cancer and inflammatory diseases and their therapeutic approaches. Cells. 2016;5(2):15. doi:10.3390/cells5020015
10. Zhang Y, Zhou H, Tao Y, Liu X, Yuan Z, Nie C. ARD1 contributes to IKKβ-mediated breast cancer tumorigenesis. Cell Death Dis. 2018;9(9):860. doi:10.1038/s41419-018-0921-2
11. Page A, Bravo A, Suarez-Cabrera C, et al. IKKβ-mediated resistance to skin cancer development is dependent. Mol Cancer Res. 2017;15(9):1255–1264. doi:10.1158/1541-7786.MCR-17-0157
12. Sakamoto K, Hikiba Y, Nakagawa H, et al. Inhibitor of kappaB kinase beta regulates gastric carcinogenesis via interleukin-1alpha expression. Gastroenterology. 2010;139(1):226–238.e6. doi:10.1053/j.gastro.2010.03.047
13. Shi Y, Hu Z, Wu C, et al. A genome-wide association study identifies new susceptibility loci for non-cardia gastric cancer at 3q13.31 and 5p13.1. Nat Genet. 2011;43(12):1215–1218. doi:10.1038/ng.978
14. Tanikawa C, Kamatani Y, Toyoshima O, et al. Genome-wide association study identifies gastric cancer susceptibility loci at 12q24.11-12 and 20q11.21. Cancer Sci. 2018;109(12):4015–4024. doi:10.1111/cas.13815
15. Wang W, Du M, Li Z, et al. A genetic variant located in promoter region is associated with prognosis of gastric cancer. Cancer Epidemiol Biomarkers Prev. 2018;27(7):822–828. doi:10.1158/1055-9965.EPI-17-1054
16. Gonzalez-Hormazabal P, Romero S, Musleh M, et al. -251T>A (rs4073) polymorphism is associated with prognosis in gastric cancer patients. Anticancer Res. 2018;38(10):5703–5708. doi:10.21873/anticanres.12907
17. Brookes AJ. The essence of SNPs. Gene. 1999;234(2):177–186. doi:10.1016/S0378-1119(99)00219-X
18. Espinosa-Parrilla Y, Muñoz X, Bonet C, et al. Genetic association of gastric cancer with miRNA clusters including the cancer-related genes MIR29, MIR25, MIR93 and MIR106: results from the EPIC-EURGAST study. Int J Cancer. 2014;135(9):2065–2076. doi:10.1002/ijc.28850
19. Pan W, Liu L, Wei J, et al. A functional lncRNA HOTAIR genetic variant contributes to gastric cancer susceptibility. Mol Carcinog. 2016;55(1):90–96. doi:10.1002/mc.22261
20. Sun Q, Gu H, Zeng Y, et al. Hsa-mir-27a genetic variant contributes to gastric cancer susceptibility through affecting miR-27a and target gene expression. Cancer Sci. 2010;101(10):2241–2247. doi:10.1111/j.1349-7006.2010.01667.x
21. Ju H, Lim B, Kim M, et al. A regulatory polymorphism at position −309 in PTPRCAP is associated with susceptibility to diffuse-type gastric cancer and gene expression. Neoplasia. 2009;11(12):1340–1347. doi:10.1593/neo.91132
22. Fan H, Liu D, Qiu X, et al. A functional polymorphism in the DNA methyltransferase-3A promoter modifies the susceptibility in gastric cancer but not in esophageal carcinoma. BMC Med. 2010;8:12. doi:10.1186/1741-7015-8-12
23. Hyland PL, Lin S-W, Hu N, et al. Genetic variants in fas signaling pathway genes and risk of gastric cancer. Int J Cancer. 2014;134(4):822–831. doi:10.1002/ijc.28415
24. Boyle AP, Hong EL, Hariharan M, et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012;22(9):1790–1797. doi:10.1101/gr.137323.112
25. Xu Z, Taylor JA. SNPinfo: integrating GWAS and candidate gene information into functional SNP selection for genetic association studies. Nucleic Acids Res. 2009;37(WebServer issue):W600–W605. doi:10.1093/nar/gkp290
26. Chen X, Li S, Yang Y, et al. Genome-wide association study validation identifies novel loci for atherosclerotic cardiovascular disease. J Thromb Haemost. 2012;10(8):1508–1514. doi:10.1111/j.1538-7836.2012.04815.x
27. Grambsch PM, Therneau TM. Proportional hazards tests and diagnostics based on weighted residuals. Biometrika. 1994;81(3):515–526. doi:10.1093/biomet/81.3.515
28. Adzhubei IA, Schmidt S, Peshkin L, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7(4):248–249. doi:10.1038/nmeth0410-248
29. Sim N-L, Kumar P, Hu J, Henikoff S, Schneider G, Ng PC. SIFT web server: predicting effects of amino acid substitutions on proteins. Nucleic Acids Res. 2012;40(W1):W452–W457. doi:10.1093/nar/gks539
30. Schwarz JM, Cooper DN, Schuelke M, Seelow D. MutationTaster2: mutation prediction for the deep-sequencing age. Nat Methods. 2014;11(4):361–362. doi:10.1038/nmeth.2890
31. Jian X, Boerwinkle E, Liu X. In silico prediction of splice-altering single nucleotide variants in the human genome. Nucleic Acids Res. 2014;42(22):13534–13544. doi:10.1093/nar/gku1206
32. Machiela MJ, Chanock SJ. LDlink: a web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics. 2015;31(21):3555–3557. doi:10.1093/bioinformatics/btv402
33. Shindo M, Nakano H, Sakon S, Yagita H, Mihara M, Okumura K. Assignment of IkappaB kinase beta (IKBKB) to human chromosome band 8p12–>p11 by in situ hybridization. Cytogenet Cell Genet. 1998;82(1–2):32–33. doi:10.1159/000015058
34. Schmid JA, Birbach A. IkappaB kinase beta (IKKbeta/IKK2/IKBKB)–a key molecule in signaling to the transcription factor NF-kappaB. Cytokine Growth Factor Rev. 2008;19(2):157–165. doi:10.1016/j.cytogfr.2008.01.006
35. Häcker H, Karin M. Regulation and function of IKK and IKK-related kinases. Sci STKE. 2006;2006(357):re13. doi:10.1126/stke.3572006re13
36. Perkins ND. Integrating cell-signalling pathways with NF-kappaB and IKK function. Nat Rev Mol Cell Biol. 2007;8(1):49–62. doi:10.1038/nrm2083
37. Scheidereit C. IkappaB kinase complexes: gateways to NF-kappaB activation and transcription. Oncogene. 2006;25(51):6685–6705. doi:10.1038/sj.onc.1209934
38. Hayden MS, Ghosh S. Signaling to NF-kappaB. Genes Dev. 2004;18(18):2195–2224. doi:10.1101/gad.1228704
39. Karin M. NF-kappaB and cancer: mechanisms and targets. Mol Carcinog. 2006;45(6):355–361. doi:10.1002/mc.20217
40. Cao H, Jiang S, Yuan R, et al. Inhibition of IκB kinase 2 attenuated the proliferation and induced apoptosis of gastric cancer. Dig Dis Sci. 2019;64(5):1204–1216. doi:10.1007/s10620-018-5414-8
41. Huan Y, Wu D, Zhou D, Sun B, Li G. DBC1 promotes anoikis resistance of gastric cancer cells by regulating NF-κB activity. Oncol Rep. 2015;34(2):843–849. doi:10.3892/or.2015.4007
42. Xin S, Zhao Y, Wang C, et al. Polymorphisms of NF-κB pathway genes influence adverse drug reactions of gefitinib in NSCLC patients. Pharmacogenomics J. 2020;20(2):285–293. doi:10.1038/s41397-019-0115-z
43. Xian H, Feng W, Zhang J. Schizandrin A enhances the efficacy of gefitinib by suppressing IKKβ/NF-κB signaling in non-small cell lung cancer. Eur J Pharmacol. 2019;855:10–19. doi:10.1016/j.ejphar.2019.04.016
44. Krazinski BE, Kowalczyk AE, Sliwinska-Jewsiewicka A, et al. IKBKB expression in clear cell renal cell carcinoma is associated with tumor grade and patient outcomes. Oncol Rep. 2019;41(2):1189–1197. doi:10.3892/or.2018.6872
45. AJCC 8th edition of the American Joint Commission on cancer staging manual. Available from: https://cancerstaging.org/references-tools/deskreferences/Pages/default.aspx.
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.