Back to Journals » International Journal of Nanomedicine » Volume 18
Identifying the Potential of miRNAs in Houttuynia cordata-Derived Exosome-Like Nanoparticles Against Respiratory RNA Viruses
Authors Zhu H , Chang M, Wang Q, Chen J, Liu D, He W
Received 14 June 2023
Accepted for publication 10 October 2023
Published 24 October 2023 Volume 2023:18 Pages 5983—6000
DOI https://doi.org/10.2147/IJN.S425173
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Yan Shen
He Zhu,1,2,* Mujun Chang,1,3,* Qiulan Wang,1 Jing Chen,1 Dong Liu,1 Wenxi He1
1Department of Pharmacy, Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, Hubei Province, People’s Republic of China; 2The Center for Biomedical Research, NHC Key Laboratory of Respiratory Diseases, Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, Hubei Province, People’s Republic of China; 3Center for Translational Medicine, Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, Hubei Province, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Dong Liu; Wenxi He, Department of Pharmacy, Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology, No. 1095 Jiefang Avenue, Wuhan, Hubei, People’s Republic of China, Email [email protected]; [email protected]
Introduction: Pathogenic respiratory RNA viruses, including influenza A virus (IAV), respiratory syncytial virus (RSV), and SARS-CoV-2, are major causes of causes of acute respiratory infection globally. Plant-derived exosome-like nanoparticles containing miRNAs have shown substantial cross-kingdom regulatory effects on both viral and human transcripts. Houttuynia cordata (H. cordata), a traditional Chinese medicine frequently used to treat respiratory diseases. However, the role of H. cordata-derived exosome-like nanoparticles (HELNs) and the miRNA they encapsulated are unclear.
Methods: HELNs were isolated from fresh underground roots (uHELNs) and above ground stems and leaves (aHELNs) using differential centrifugation. The HELNs were identified using transmission electron microscopy, nanoparticle tracking analysis, and zeta potential. Small RNA sequencing and RT-PCR were employed to determine the miRNA expression in uHELNs and aHELNs. All genomes were sourced from the NCBI database. Target prediction of viral genomes was performed using RNAhybrid, while human target prediction was conducted using both RNAhybrid and Miranda. Functional enrichment analysis was applied to the predicted human targets to explore the hub targets and their roles in antiviral effects. The accessibility of miRNA target sites was determined through the MFOLD web server, and customized dual-luciferase reporter assays were administered to validate the computational findings.
Results: A total of 12 highly enriched miRNAs were identified in both uHELNs and aHELNs. Upon prediction and verification, miR858a and miR858b were shown to target the NP gene in H1N1, while miR166a-3p targeted the ORF1ab in SARS-CoV-2. However, no valid miRNA targets were found for RSV. Regarding human transcripts, miR168a-3p, miR168b-3p, and miR8175 were found to inhibit MAPK3 expression, and novel_mir2 could suppress both AKT1 and MAPK3 expression.
Discussion: This study sheds light on the collaborative antiviral mechanism of miRNAs in HELNs across two species and explores the potential antiviral scopes of both H. cordata miRNAs and HELNs.
Keywords: plant exosome, nanoparticles, Houttuynia cordata, traditional Chinese medicine, respiratory virus, miRNAs
Introduction
RNA viruses, such as influenza A virus (IAV), respiratory syncytial virus (RSV), and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), employ RNA as their genomic nucleic acids. Owing to their adaptability and high variability, these RNA viruses frequently undergo viral evolution, which enables them to develop resistance to various prophylactic measures, including vaccines and antibodies.1 Currently, IAV, RSV, and SARS-CoV-2 represent the leading causes of acute respiratory infection worldwide, especially in infants, the elderly, and immunocompromised individuals.2–4 IAV is a zoonotic virus with 8 single-stranded negative-sense RNA segments in its genome and periodically causes pandemics.2 Human RSV, with a single negative-sense RNA linear genome, triggers severe respiratory infections in young children with annual winter epidemics.3 SARS-CoV-2, possessing a non-segmented, positive-sense genome, has been responsible for a global pandemic since 2019, posing a severe threat to human health and social development.4 Unfortunately, reliable long-term preventive measures or treatments for these RNA virus-induced acute respiratory infections are lacking.
MicroRNA (miRNA) is a single-stranded noncoding RNA that has an average length of 18–24 nucleotides (nts) and regulates gene expression at the post-transcriptional level. There is growing evidence that miRNAs are stable and capable of cross-kingdom regulation of host transcripts.5 Earlier research has shown that host miRNAs can influence host-virus interactions, which has driven the adoption of miRNA cross-kingdom regulatory strategies for anti-virus.6 In 2012, Zhang et al first reported that plant miR168a from rice can inhibit mammalian gene expression, making the beginning of plant miRNA cross-kingdom regulation.7 Since then, they further reported that miR2911 from honeysuckle inhibits the replication of IAV and SARS-CoV-2, accelerating the recovery process in infected individuals.8,9 However, due to the sequence similarity between some plant miRNAs and human miRNAs and the lack of reproducibility of the data, these reports were later controversial due to the suspicion that contamination during the detection triggered false positive results.10,11 Nonetheless, later research demonstrated that the miRNAs loaded in plant-derived exosome-like nanoparticles (PELNs) do indeed exhibit cross-kingdom effects in mammalian cells. For example, in ginger PELNs, Yin et al reported that 27 highly expressed miRNAs had anti-inflammatory effects through cross-kingdom targeting of intestinal Caco2 cells.12
Recently, PELNs carrying a variety of bioactive substances, including miRNAs, have been widely explored with the advantages of high and eco-friendly yield, no ethical barriers, and nontoxicity, allowing them to circumvent the technical limitations of mammalian microvesicles.13,14 PELNs also showed multiple health benefits, including anti-inflammatory,15 anti-cancer,16 and antibiotic effects.17 Moreover, stability experiments have demonstrated that PELNs can not only be stored at low temperatures for long periods without affecting their activity but can also withstand different pH environments in the gastrointestinal tract when administered orally.16,18,19 This provides a guarantee for the targeted delivery of plant miRNAs to host cells. Recent research by Kalarikkal et al has provided an in-depth analysis of miRNA expression profiles in 11 types of PLENs, including Grapefruit, Hami melon, Pea, Coconut, Blueberry, Tomato, Pear, Ginger, Kiwifruit, Orange, Soybean. This study identified 22 miRNAs potentially targeting the SARS-CoV-2 genome.20 However, these miRNAs were derived from fruits and vegetables rather than traditional Chinese medicine (TCM). Nevertheless, TCM generally has stronger antiviral pharmacological properties than fruits and vegetables. Consequently, the potential for discovering antiviral miRNAs from TCM remains high.
Houttuynia cordata (H. cordata), also known as Herba houttuyniae, is a herbaceous perennial plant that grows in moist and shady environments.21,22 H. cordata is not only a widely distributed medicinal plant but also a functional food offering multiple health benefits, such as anti-inflammatory,23 anti-oxidative,24 and antibiosis.25 Notably, the application of H. cordata in alignment with traditional Chinese medical theory facilitates its use in treating pneumonia caused by viral infections. Moreover, H. cordata and its products have exhibited extensive antagonistic effects against various respiratory viruses, including H1N1,26 HSV,27 norovirus-1,28 and coronavirus.25 In 2003, H. cordata was listed as one of the drugs for the treatment of severe acute respiratory.29 However, whether H. cordata exerts an antiviral effect through miRNAs remains unclear.
Exogenous miRNAs exhibit two different antiviral mechanisms. First, miRNAs can directly target viral genomes or transcripts.30 Second, miRNAs can target the host mRNAs to regulate gene expression and hinder viral entry or replication.31 Accordingly, we first used small RNA sequencing (sRNA-seq) and real-time PCR (RT-PCR) to define miRNAs in H. cordata derived exosome-like nanoparticles (HELNs) from the underground roots (uHELNs) and above ground stems and leaves (aHELNs). By target predicting analysis, we further identified miRNAs highly enriched in uHELNs and aHELNs that could directly target the genome of respiratory RNA viruses, including IAV, RSV, and SARS-CoV-2. On the other hand, we predicted the target human genes of these highly enriched miRNAs to elucidate the endogenous antiviral mechanism in host cells (Figure 1).
 |
Figure 1 Schematic illustration of experimental design for HELNs isolation, and prediction of respiratory RNA viral genomes and human gene targeted by miRNAs in HELNs. Pictures of all the virus structures and genomes were obtained from ViralZone Available from: https://viralzone.expasy.org/. Creative Commons. |
Materials and Methods
HELNs Isolation
Fresh and dry H. cordata was purchased from Hubei Shengdetang TCM Slices Co., LTD, China. HELNs were isolated with the same standard differential centrifugation method as described in our previous study32 (Figure 1). The obtained HELNs were resuspended in sterile PBS and stored at −80 °C until use.
HELNs Identification
Transmission electron microscopy (TEM), nanoparticle tracking analysis (NTA), and zeta potential were used to identify HELNs as previously described.32 NTA and zeta potential were performed by using Zetasizer Nano S90 (Malvern Panalytical, Malvern, UK). TEM imaging of HELNs was performed by using an electron microscope HT7700 TEM (Hitachi, Japan) at 80 kV.
Small RNA Sequencing Analysis
sRNA-seq services were provided by the Bioyi Biotechnology Co., Ltd. in Wuhan, China. Sequencing reads with lengths of 18–25 bp were mapped to miRNA sequences in the miRBase 21.0 database (https://www.mirbase.org/)33 with the selected species as Arabidopsis, because there is no database available for H. cordata and Arabidopsis is the most well-researched plant. All mapped miRNAs were identified as known miRNAs, and the unmapped sequences were identified as novel miRNAs. All miRNA profiling data have been deposited in the NCBI-SRA (www.ncbi.nlm.nih.gov/sra, SRA accession: PRJNA971115) databases.
The differential expression miRNAs between uHELNs and aHELNs were screened with a threshold p-value < 0.05 and |log2FC| ≥ 1. Volcano Plot was performed using the OmicStudio tools at https://www.omicstudio.cn/tool.
RNA Extraction and Real Time-PCR
RNA extraction and RT-PCR were carried out as in our previous study.32 All miRNA primers were designed and synthesized by RibiBio (Guangzhou, China). Mature miRNAs were reverse-transcripted using the miRNA 1st Strand cDNA Synthesis Kit (by tailing A) (Vazyme, Nanjing, China) in accordance with the manufacturer’s instructions. Finally, RT-PCR was performed with the Step-one Plus system (Applied Biosystems, Zug, Switzerland) and Taq Pro Universal SYBR qPCR Master Mix (Vazyme, Nanjing, China) for miRNA level detection.
Collection of Respiratory Virus Genomes
The genome of IAV contains 8 segments encoding PB2, PB1, PA, HA, NP, NA, M1, and NS1 genes. Of which the PB2, PB1, and PA together encode the polymerase protein complex on which viral replication depends.2 Meanwhile, NP encodes a protein that wraps around viral nts and acts as a protector.2 As a result, we chose PB2, PB1, PA, and NP (encoded by segments 1, 2, 3, and 5) as targets to do the miRNA targeting site prediction.
All viral genomes were obtained from the NCBI database (https://www.ncbi.nlm.nih.gov/genome/). Four IAV sub-types implicated in human epidemics, including H1N1, H5N1, H7N7, and H7N9, were selected. For H1N1 (A/New York/3442/2009), the complete genome sequences were Accession: CY050779.1, CY050778.1, CY050777.1, and CY050775.1. For H5N1 (A/duck/Shandong/093/2004), the complete genome sequences were Accession: AY856861.1, AY856862.1, AY856863.1, and AY856864.1. For H7N7 (A/mallard/Sweden/95/2005), the complete genome sequences were Accession: CY077023.1, CY077022.1, CY077021.1, and CY077019.1. For H7N9 (A/duck/Japan/AQ-HE29-22/2017), the complete genome sequences were Accession: LC315928.1, LC315926.1, LC315925.1, and LC315924.1. For human RSV, the complete genome sequences were Accession: KM578843. For SARS-CoV-1, the complete genome sequences were Accession: AY545919.1. For SARS-CoV-2, complete genome sequences from three different branches were obtained with Accession: NC_045512.2 (Wuhan), OK091006.1 (Delta), and OP183454.1 (OMICRON).
Prediction of miRNA Binding Sites in Respiratory Virus Genomes
RNAhybrid (https://bibiserv.cebitec.uni-bielefeld.de/rnahybrid/)34 was used to match potential targets of miRNAs in the viral sequence. Three vital rules were followed: (1) minimum free energy (MFE) ≤ −25 kcal/mol; (2) helix constrains between 2–8 nts; (3) maximum allowed internal bulge 0–2 nts. Multiple sequence alignment of viral genome sequences was performed by DNAMAN software version 6.
Bioinformatics Analysis of Predicted Human Target Genes
RNAhybrid and miRanda (http://www.microRNA.org) were used to match potential targets of miRNAs in human genes with a threshold of MFE≤ −25 in both systems. KEGG enrichment analysis was performed by DAVID (https://david-d.ncifcrf.gov/).35 Venn and KEGG were calculated and visualized using the OmicStudio tools at https://www.omicstudio.cn/tool. Protein-protein interaction (PPI) was constructed from the STRING database (http://string-db.org/). All networks were visualized using Cytoscape (v3.9.1). Target site accessibility of miRNA binding sites was validated by the MFLOD web server (http://www.mfold.org/).36 According to the previous study,37 the local secondary structure between 17 nt upstream and 13 nt downstream of the miRNA binding site was updated to analyze the target site accessibility to miRNAs.
Dual-Luciferase Reporter Assay
All sequences of miRNAs and their target plasmids containing the wild-type (WT) or mutant-type (MU) binding sites were synthesized by Wuhan GeneCreate Biological Engineering Co. Ltd. Sequencing was applied to verify these plasmids.
For transfection, HEK 293 T cells were co-transfected with the corresponding plasmids and miRNA mimics/NC with Lipofectamine 3000 (Invitrogen, Carlsbad, USA). After 48 h of incubation, the activity of firefly luciferase activity was measured using the Dual-Luciferase Reporter Assay System (Promega, Madison, USA). The luciferase assay was performed according to the manufacturer’s instructions. The relative luciferase activities were normalized to that of the WT group.
Statistical Analysis
All experiments were performed in triplicate and the data were presented as the mean ± SEM and evaluated by using the Student’s t-test. All p-values were calculated in GraphPad Prism software 8.1 (GraphPad Software Inc., USA). The p < 0.05 was considered statistically significant.
Result
Isolation and Identification of HELNs
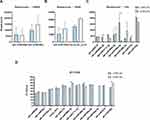
First, HELNs were isolated by using a standard differential centrifugation procedure (Figure 1). The obtained HELNs were identified by TEM and NTA. TEM images of both uHELNs and aHELNs showed a round-shaped membrane-bound structure (Figure 2A and B). However, no HELNs were detected in the sample from dry H. cordata (data not shown). Subsequently, the NTA results showed that the mean diameters of uHELNs and aHELNs are 169.5 and 166.2 nm, respectively (Figure 2C and D), and their surface charges are −23.04 ± 1.22 and −28.73 ± 0.87 mV, respectively (Figure 2E and F). After quantifying, 6.67×1010 particles of uHELNs and 1.5×1010 particles of aHELNs were obtained per gram of fresh tissue. Collectively, these results suggested that HELNs are enriched in fresh H. cordata, rather than dry H. cordata, and have the typical characteristics, which are suitable for subsequent experiments.
Small RNA Sequencing Analysis of HELNs
In total, 326 miRNAs including 323 known and 3 novel miRNAs were annotated through sRNA-seq analysis. Out of these, 18 miRNAs showed differential expression between uHELNs and aHELNs, with 12 being upregulated and 6 being downregulated, as depicted in Supplementary Table 1.
Subsequently, we identified 12 highly enriched miRNAs in HELNs that had read counts of more than 100 in either uHELNs or aHELNs (Figure 3A–C and Table 1). Among these miRNAs, miR398b-3p, miR168a-5p, and miR167d were found to be significantly upregulated in uHELNs as compared to aHELNs, while miR168a-3p was observed to be significantly downregulated (Figure 3C).
 |
Table 1 Information of the High Abundant miRNAs* |
RT-PCR Validation of the miRNAs in HELNs
To validate the expression of the 12 highly enriched miRNAs identified by sRNA-seq, we performed RT-PCR. As shown in Figure 3D, ath-miR858b and ath-miR858a had the highest expression levels (cycle threshold (Ct) values < 25) without any significant difference between uHELNs and aHELNs. Meanwhile, ath-miR398b-3p and ath-miR159a were significantly overexpressed (with lower Ct values) in uHELNs compared to aHELNs, which was consistent with the sequencing data (Figure 3C). In contrast, ath-miR168b-3p had significantly higher Ct values in uHELNs compared to aHELNs, while sRNA-seq data showed no significant difference. Furthermore, ath-miR168a-3p and ath-miR167d were undetermined in uHELNs and aHELNs, respectively, while both ath-miR398b-3p and ath-miR159a were significantly overexpressed (with lower Ct values) in uHELNs compared to aHELNs. In addition, the remaining five 12 highly enriched miRNAs (ath-miR166a-3p, novel_mir2, ath-miR815, ath-miR171c-3p, and ath-miR168a-5p) expressed equally in both uHELNs and aHELNs, as evidenced by Ct values. Collectively, our findings highlight the substantial abundance of miRNAs in HELNs.
The miRNAs in HELNs Target IAV
H1N1 is a major subtype of IAV and the most frequent cause of influenza in humans. To investigate the potential of the highly enriched miRNAs in HELNs for H1N1 treatment, we used RNAhybrid to predict the miRNA targeted genomic locations. The results showed that 6 miRNAs mightily target the H1N1 genome with MEF ≤ −25 kcal/mol (Table 2). Meanwhile, the secondary structures of the miRNA-target-cRNA duplex for these 6 miRNAs, along with their target site sequences, are shown in Figure 4A–G. Specifically, the PB2 gene in H1N1 segment 1 can be targeted by miR168a-5p, miR398b-3p, and novel_miR2 (Figure 4A, C, and G). Additionally, the overlapped PB1 and PB1-F2 genes in segment 2 can also be targeted by miR168a-5p (Figure 4B). The PA gene in segment 3 was targeted by miR159a (Figure 4D). Together, the above four miRNAs (miR168a-5p, novel_mir2, miR398b-3p, and miR159a) suppress viral polymerase formation by targeting different segment sites. Besides, the NP gene in segment 5 was targeted by miR858a and miR858b at the same locations (Figure 4E and F). Overall, our results indicated that miR858a and miR858b may assist in disrupting H1N1 NP protein synthesis, which can make viral genome exposure.
 |
Table 2 Information of miRNAs Targeting H1N1 |
Subsequently, the other highly pathogenic IAV subtypes H5N1, H7N7, and H7N9 were chosen for validating the broad-spectrum effect of the shortlisted miRNAs in HELNs through multiple sequence alignment. Consequently, four miRNAs (miR159a, miR168a-5p, miR858a, and miR858b) demonstrate full complementarity across all four IAV subtypes, suggesting that these miRNAs can equally suppress the viruses (Figure 4H). MiR398b-3p exhibits complete complementarity with H1N1, H5N1, and H7N7 (Figure 4I). Novel_mir2 displays full complementarity for both H1N1 and H7N9 (Figure 4J). However, miR168a-5p only completely complements H1N1 (Figure 4K).
The miRNAs in HELNs Target RSV
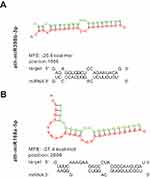
A previous study found that H. cordata has anti-RSV efficacy, and RSV has negative-stranded RNA linear similar to the IAV genome (Figure 1), suggesting a potential therapeutic impact of miRNAs enriched in HELNs against RSV. To corroborate this observation, we used a binding strategy similar to H1N1. Results showed that the N gene can be targeted by miR398b-3p, whereas the P gene can be targeted by miR168a-5p with a meaningful MEF less than −25 kcal/mol (Figure 5 and Table 3).
 |
Table 3 Information of miRNAs Targeting Respiratory Syncytial Virus |
The miRNAs in HELNs Target SARS-CoV-2
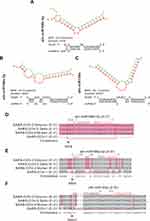
SARS-CoV-2 is a non-segmented, positive-strand RNA virus similar to SARS-COV that attacks the respiratory system severely (Figure 1). To extrapolate the anti-SARS-CoV-2 effect of HELNs, all 12 highly enriched miRNAs in HELNs were applied to target SARS-CoV-2 Omicron genome. The results showed that 3 miRNAs can target virus genomic sequence with miRNA seed-sequence being completely complementary (Figure 6A–C). MiR168b-3p and miR166a-3p can target the ORF1ab gene in SARS-CoV-2 with an energy of −25.5 and −27 kcal/mol, respectively. MiR159a can target the E gene with an energy of −25.1 kcal/mol (Table 4). Furthermore, we conducted multiple sequence alignments to assess the specificity of the selected miRNAs against viral mutations, using two other genomic sequences of SARS-CoV-2 strains, Delta and Wuhan, and SARS-CoV. It showed that miR168b-3p, miR166a-3p, and miR159a are generally effective against SARS-Cov-2; however, miR159a may also have some inhibitory effect on SARS-CoV-1, while miR168b-3p and miR166a-3p are ineffective (Figure 6D–F).
 |
Table 4 Information of miRNAs Targeting SARS-CoV-2 |
Human Target Prediction and Functional Analysis
In addition to the direct antiviral effects of highly enriched miRNAs in HELNs, they may activate immune response by targeting host genes, serving as a mechanism of defense against virus infection. We used RNAhybrid and Miranda for the human target prediction, which identified 17,360 and 5484 targets respectively, yielding 5451 common targets after cross-comparison (Figure 7A). KEGG pathway analysis indicated that these cross targets were mostly involved in “Pathways in cancer”, “Herpes simplex virus 1 infection”, and “Human papillomavirus infection”. This suggests that these highly enriched miRNAs in HELNs may regulate gene expression in virus-infected host cells. In addition, these predicted targets are also involved in crucial pathways, the “PI3K-AKT” and “MAPK” signaling pathways (Figure 7B).
To further explore the hub targets in these significant virus-related enrichment pathways, we performed PPI analyses. Intriguingly, the PPI networks showed that AKT1 and MAPK3 were the hub genes based on the degree level (Figure 7C and D).
Validation of miRNAs and Their Target Genes
The ability of a miRNA to successfully suppress a target mRNA is largely determined by the accessibility of the target site to miRNA binding.37 We utilized the MFLOD web server to computationally confirm the associations between miRNA and their targets. This yielded results which established that 8 out of 11 potential miRNAs (namely, miR166a-3p, miR168a-3p, miR168b-3p, miR398b-3p, miR858a, miR858b, miR8175, and novel_mir2) featured at least one predicted secondary structure with an MFE < −15 kcal/mol (Supplementary Table 2).
To affirm the impact of MFLOD predicted miRNAs on cross-targeting genes in organisms, we used the dual-luciferase reporter assay to measure the luciferase activity based on the computational analysis of core sequences. In partial agreement with MFLOD results, the dual-luciferase assay showed a significant reduction in luciferase activity in 7 out of 8 (excluding miR398-3p) WT genes when co-transfecting with miRNA mimic groups, as compared to the other three groups (Figure 8A-G). Specifically, as summarized in Figure 8H, these data showed that miR858a and miR858b may have a suppressive effect on H1N1 by inhibiting the expression of NP (Figure 8A) and that miR166a-3p could inhibit the expression of ORF1ab for resisting SARS-CoV-2 (Figure 8B). Additionally, miR168a-3p, miR168b-3p, and miR8175 can inhibit MAPK3 expression (Figure 8C, E, and F), and novel-mir2 can suppress both AKT1 and MAPK3 expression (Figure 8D and G), suggesting these miRNAs may provide an indirect defense against viral invasion by targeting host genes.
Discussion
PELNs have been isolated from a variety of edible plants, primarily succulent fruits and vegetables; however, TCM has received less attention in this regard.38 This neglect may stem from the fact that TCM is typically subject to high-temperature processing methods such as distillation or drying, which lead to the degradation of most PELNs and encapsulated miRNAs. Although miR2911 has been detected in honeysuckle decoction, its concentration is extremely low, at the femtomolar level, raising doubts about its effectiveness.7 Noteworthy, several recent studies showed that abundant miRNAs can be encapsulated in PELNs to facilitate cross-kingdom targeted transport.39 However, the role of miRNAs in TCM and its PLENs is still unknown.
H. cordata is a widely used TCM for treating respiratory infections,21,22 and both its fresh and dried forms possess medicinal properties, with the fresh product being more potent.39 Notably, H. cordata is consumed directly, following washing and seasoning, in China, Korea, Japan, India, and other Asian countries, particularly throughout many Chinese provinces.21,39 In our previous study, we discovered that ELNs derived from fresh plants effectively retain miRNAs and enable cross-kingdom entry of miRNAs into mammalian cells.32 Interestingly, in this study, we found an abundance of HELNs in fresh H. cordata, while dry H. cordata exhibited none. To this end, we isolated uHELNs and aHELNs in the current study and verified the presence of miRNAs using sRNA-seq and RT-PCR, identifying a wealth of miRNAs in the HELNs. Through target predicting, we further determined that these miRNAs not only can bind viral genomes but also interact with human transcripts (Figure 1).
miRNAs in HELNs Cross-Kingdom Target the Viral Genome Directly
So far, studies on the cross-kingdom regulation of exogenous miRNAs from plants remain limited. In this study, we identified 7 known and 1 novel highly enriched miRNAs in HELNs, including miR159a, miR166a-3p, miR168b-3p, miR168a-5p, miR398b-3p, miR858a, miR858b, and novel_mir2. Although miR398b-3p and miR168a-5p appear to have the potential to target RSV genes (Figure 5), the MFLOD results contradict this expectation (Supplementary Table 2). Upon prediction and verification, 3 of these miRNAs (miR858a, miR858b, and miR166a-3p) were found to significantly suppress gene expression in respiratory viruses (Figure 8H). Notably, miR858a and miR166a-3p have been reported for the first time as demonstrating cross-kingdom regulatory abilities. For miR858b, a previous study identified the anti-HIV properties of miR858b from Moringa oleifera seeds, which were experimentally validated by targeting human VAV1, leading to the enhancement of T cell antigen receptor (TCR) signaling.40 Furthermore, both sRNA-seq and RT-PCR showed high expression levels of miR858a, miR858b, and miR166a-3p in both uHELNs and aHELNs (Figure 3). These findings suggest that HELNs could potentially target NP and ORF1ab genes, thus serving potential therapeutic purposes against H1N1 and SARS-CoV-2 (Figure 8H).
miRNAs in HELNs Cross-Kingdom Target and Regulate Human Genes to Defend Against Respiratory Viruses
Interestingly, plant miRNAs have demonstrated the ability to cross-kingdom regulate human genes to protect against diseases.41 In this context, we found that novel_mir2 can inhibit the expression of AKT1, while miR168a-3p, miR168b-3p, miR8175, and novel_mir2 can inhibit the expression of MAPK3 (Figure 8H).
The AKT1-mediated PI3K-AKT pathway is a promising target against anti-viruses, including IAV, RSV, SARS-CoV, and SARS-CoV-2. During virus infection and inflammatory response, its activation is a key cellular event.42 Previous studies have shown that AKT/mTOR pathway activity is crucial for IAV entry43 and results in the suppression of premature apoptosis during the later stages of infection.42 A recent study using Network Pharmacology and lipidomics claimed that AKT is an ideal drug target for treating RSV-induced lung inflammation.44 Other studies performed in vitro demonstrated that inhibiting PI3K/AKT pathway activation is also applicable to SARS-CoV-2 treatment with an Akt inhibitor.45,46
The mitogen-activated protein kinase (MAPK) p38 is another vital component of the signaling pathways that regulate pro-inflammatory cytokine expression and is associated with virus-supportive functions.47,48 Recent studies have shown that the inhibitory activity of p38 MAPK could suppress RSV and IVA entry and replication during the early stage of infection, as further confirmed using the p38 MAPK inhibitor SB203580.49,50 Another in vitro study using primary human lung explants and lung epithelial organoids showed that p38 inhibitors, PH-797804 and VX-702, could significantly reduce the expression of pro-inflammatory cytokines during SARS-CoV-2 infection.51 Collectively, AKT1 and MAPK3 could serve as ideal targets with broad‐spectrum antiviral effects. The miRNAs identified in HELNs may synergize these two key pathways to achieve antiviral effects.
Limitations of This Study
Despite our promising results, this study has several limitations. Firstly, more in vitro and in vivo experiments are required to validate the miRNAs in HELNs and their targets. The cross-kingdom regulation of plant miRNAs is complex, with the authenticity and regulatory mechanisms remaining unclear, particularly regarding their mode of action on regions beyond the 3’UTR of the viral genome. Moreover, expanding the sequencing library of TCM is crucial for adequately annotating miRNAs and conducting further bioinformatics studies from various plant sources. Finally, additional research on other TCM-derived ELNs is required for assessing therapeutic efficacy in experimental animals and/or clinical settings.
Conclusion
Our study implies that the difference in antiviral ability between fresh and dried TCM may be mediated by the presence of PELNs and encapsulated miRNAs. We investigated the potential antiviral mechanism of fresh H. cordata by identifying and verifying miRNAs in HELNs and predicting their capacity to target the respiratory viral genomes and human genes. This is the first time to reveal the synergistic antiviral mechanism of miRNAs in HELNs across two species.
Funding
This work was supported by the National Natural Science Foundation of China [82104302 and 82100931], and the Nature Science Foundation of Hubei Province [grant number: 2021CFB395].
Disclosure
The authors declare no competing interest in this work.
References
1. Reiss S, Rebhan I, Backes P, et al. Recruitment and activation of a lipid kinase by hepatitis C virus NS5A is essential for integrity of the membranous replication compartment. Cell Host Microbe. 2011;9(1):32–45. doi:10.1016/j.chom.2010.12.002
2. Uyeki TM, Hui DS, Zambon M, Wentworth DE, Monto AS. Influenza. Lancet. 2022;400(10353):693–706. doi:10.1016/S0140-6736(22)00982-5
3. Nam HH, Ison MG. Respiratory syncytial virus infection in adults. BMJ. 2019;366:l5021. doi:10.1136/bmj.l5021
4. Lamers MM, Haagmans BL. SARS-CoV-2 pathogenesis. Nat Rev Microbiol. 2022;20(5):270–284. doi:10.1038/s41579-022-00713-0
5. Saiyed AN, Vasavada AR, Johar SRK. Recent trends in miRNA therapeutics and the application of plant miRNA for prevention and treatment of human diseases. Future J Pharm Sci. 2022;8(1):24. doi:10.1186/s43094-022-00413-9
6. Stern-Ginossar N, Elefant N, Zimmermann A, et al. Host immune system gene targeting by a viral miRNA. Science. 2007;317(5836):376–381. doi:10.1126/science.1140956
7. Zhang L, Hou D, Chen X, et al. Exogenous plant MIR168a specifically targets mammalian LDLRAP1: evidence of cross-kingdom regulation by microRNA. Cell Res. 2012;22(1):107–126. doi:10.1038/cr.2011.158
8. Zhou LK, Zhou Z, Jiang XM, et al. Absorbed plant MIR2911 in honeysuckle decoction inhibits SARS-CoV-2 replication and accelerates the negative conversion of infected patients. Cell Discov. 2020;6(1):54. doi:10.1038/s41421-020-00197-3
9. Zhou Z, Li X, Liu J, et al. Honeysuckle-encoded atypical microRNA2911 directly targets influenza A viruses. Cell Res. 2015;25(1):39–49. doi:10.1038/cr.2014.130
10. Kang W, Bang-Berthelsen CH, Holm A, et al. Survey of 800+ data sets from human tissue and body fluid reveals xenomiRs are likely artifacts. Rna. 2017;23(4):433–445. doi:10.1261/rna.059725.116
11. Heintz-Buschart A, Yusuf D, Kaysen A, et al. Small RNA profiling of low biomass samples: identification and removal of contaminants. BMC Biol. 2018;16(1):52. doi:10.1186/s12915-018-0522-7
12. Yin L, Yan L, Yu Q, et al. Characterization of the MicroRNA profile of ginger exosome-like nanoparticles and their anti-inflammatory effects in intestinal Caco-2 cells. J Agric Food Chem. 2022;70(15):4725–4734. doi:10.1021/acs.jafc.1c07306
13. Karamanidou T, Tsouknidas A. Plant-derived extracellular vesicles as therapeutic nanocarriers. Int J Mol Sci. 2021;23(1):191. doi:10.3390/ijms23010191
14. Cui Y, Gao J, He Y, Jiang L. Plant extracellular vesicles. Protoplasma. 2020;257(1):3–12. doi:10.1007/s00709-019-01435-6
15. Chen X, Zhou Y, Yu J. Exosome-like nanoparticles from ginger rhizomes inhibited NLRP3 inflammasome activation. Mol Pharm. 2019;16(6):2690–2699. doi:10.1021/acs.molpharmaceut.9b00246
16. Zhang M, Xiao B, Wang H, et al. Edible ginger-derived nano-lipids loaded with doxorubicin as a novel drug-delivery approach for colon cancer therapy. Mol Ther. 2016;24(10):1783–1796. doi:10.1038/mt.2016.159
17. Teng Y, Xu F, Zhang X, et al. Plant-derived exosomal microRNAs inhibit lung inflammation induced by exosomes SARS-CoV-2 Nsp12. Mol Ther. 2021;29(8):2424–2440. doi:10.1016/j.ymthe.2021.05.005
18. Zhang M, Viennois E, Prasad M, et al. Edible ginger-derived nanoparticles: a novel therapeutic approach for the prevention and treatment of inflammatory bowel disease and colitis-associated cancer. Biomaterials. 2016;101:321–340. doi:10.1016/j.biomaterials.2016.06.018
19. Zhuang X, Deng ZB, Mu J, et al. Ginger-derived nanoparticles protect against alcohol-induced liver damage. J Extracell Vesicles. 2015;4:28713. doi:10.3402/jev.v4.28713
20. Kalarikkal SP, Sundaram GM. Edible plant-derived exosomal microRNAs: exploiting a cross-kingdom regulatory mechanism for targeting SARS-CoV-2. Toxicol Appl Pharmacol. 2021;414:115425. doi:10.1016/j.taap.2021.115425
21. Wu Z, Deng X, Hu Q, et al. Houttuynia cordata Thunb: an Ethnopharmacological Review. Front Pharmacol. 2021;12:714694. doi:10.3389/fphar.2021.714694
22. Rafiq S, Hao H, Ijaz M, Raza A. Pharmacological effects of Houttuynia cordata Thunb (H. cordata): a comprehensive review. Pharmaceuticals. 2022;15(9):1079. doi:10.3390/ph15091079
23. Woranam K, Senawong G, Utaiwat S, Yunchalard S, Sattayasai J, Senawong T. Anti-inflammatory activity of the dietary supplement Houttuynia cordata fermentation product in RAW264.7 cells and Wistar rats. PLoS One. 2020;15(3):e0230645. doi:10.1371/journal.pone.0230645
24. Hsu CC, Yang HT, Ho JJ, Yin MC, Hsu JY. Houttuynia cordata aqueous extract attenuated glycative and oxidative stress in heart and kidney of diabetic mice. Eur J Nutr. 2016;55(2):845–854. doi:10.1007/s00394-015-0994-y
25. Das SK, Mahanta S, Tanti B, Tag H, Hui PK. Identification of phytocompounds from Houttuynia cordata Thunb. as potential inhibitors for SARS-CoV-2 replication proteins through GC-MS/LC-MS characterization, molecular docking and molecular dynamics simulation. Mol Divers. 2022;26(1):365–388. doi:10.1007/s11030-021-10226-2
26. Ling LJ, Lu Y, Zhang YY, et al. Flavonoids from Houttuynia cordata attenuate H1N1-induced acute lung injury in mice via inhibition of influenza virus and toll-like receptor signalling. Phytomedicine. 2020;67:153150. doi:10.1016/j.phymed.2019.153150
27. Hung PY, Ho BC, Lee SY, et al. Houttuynia cordata targets the beginning stage of herpes simplex virus infection. PLoS One. 2015;10(2):e0115475. doi:10.1371/journal.pone.0115475
28. Cheng D, Sun L, Zou S, et al. Antiviral effects of houttuynia cordata polysaccharide extract on murine norovirus-1 (MNV-1)-A human norovirus surrogate. Molecules. 2019;24(9):1835. doi:10.3390/molecules24091835
29. Lau KM, Lee KM, Koon CM, et al. Immunomodulatory and anti-SARS activities of Houttuynia cordata. J Ethnopharmacol. 2008;118(1):79–85.
30. Li L, Gao F, Jiang Y, et al. Cellular miR-130b inhibits replication of porcine reproductive and respiratory syndrome virus in vitro and in vivo. Sci Rep. 2015;5:17010. doi:10.1038/srep17010
31. Li L, Wei Z, Zhou Y, et al. Host miR-26a suppresses replication of porcine reproductive and respiratory syndrome virus by upregulating type I interferons. Virus Res. 2015;195:86–94. doi:10.1016/j.virusres.2014.08.012
32. Wang X, Wu B, Sun G, et al. Selenium biofortification enhanced miR167a expression in broccoli extracellular vesicles inducing apoptosis in human pancreatic cancer cells by targeting IRS1. Int J Nanomedicine. 2023;18:2431–2446. doi:10.2147/IJN.S394133
33. Haunsberger SJ, Connolly NM, Prehn JH. miRNAmeConverter: an R/bioconductor package for translating mature miRNA names to different miRBase versions. Bioinformatics. 2017;33(4):592–593. doi:10.1093/bioinformatics/btw660
34. Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes. Rna. 2004;10(10):1507–1517. doi:10.1261/rna.5248604
35. Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi:10.1038/nprot.2008.211
36. Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31(13):3406–3415. doi:10.1093/nar/gkg595
37. Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. The role of site accessibility in microRNA target recognition. Nat Genet. 2007;39(10):1278–1284. doi:10.1038/ng2135
38. Zhu H, He W. Ginger: a representative material of herb-derived exosome-like nanoparticles. Front Nutr. 2023;10:1223349. doi:10.3389/fnut.2023.1223349
39. Zhu H, Li Y, Li H. Necessity of the development of fresh Chinese medicine decoction pieces by analysis from the quantity of fresh Houttuynia cordata Thunb. Mod Tradit Chin Med Materia Medica--World Sci Technol. 2019;21(12):2753–2758.
40. Minutolo A, Potesta M, Roglia V, et al. Plant microRNAs from Moringa oleifera Regulate Immune Response and HIV Infection. Front Pharmacol. 2020;11:620038. doi:10.3389/fphar.2020.620038
41. Trivedi TS, Patel MP, Nanavaty V, Mankad AU, Rawal RM, Patel SK. MicroRNAs from Holarrhena pubescens stems: identification by small RNA sequencing and their potential contribution to human gene targets. Funct Integr Genomics. 2023;23(2):149. doi:10.1007/s10142-023-01078-0
42. Ehrhardt C, Ludwig S. A new player in a deadly game: influenza viruses and the PI3K/Akt signalling pathway. Cell Microbiol. 2009;11(6):863–871. doi:10.1111/j.1462-5822.2009.01309.x
43. Ranadheera C, Coombs KM, Kobasa D. Comprehending a killer: the Akt/mTOR signaling pathways are temporally high-jacked by the highly pathogenic 1918 influenza virus. EBioMedicine. 2018;32:142–163. doi:10.1016/j.ebiom.2018.05.027
44. Lin L, An L, Chen H, et al. Integrated network pharmacology and lipidomics to reveal the inhibitory effect of qingfei oral liquid on excessive autophagy in RSV-induced lung inflammation. Front Pharmacol. 2021;12:777689. doi:10.3389/fphar.2021.777689
45. Al-Qahtani AA, Pantazi I, Alhamlan FS, et al. SARS-CoV-2 modulates inflammatory responses of alveolar epithelial type II cells via PI3K/AKT pathway. Front Immunol. 2022;13:1020624. doi:10.3389/fimmu.2022.1020624
46. Kyriakopoulos AM, Nigh G, McCullough PA, Seneff S. Mitogen activated protein kinase (MAPK) Activation, p53, and autophagy inhibition characterize the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spike protein induced neurotoxicity. Cureus. 2022;14(12):e32361. doi:10.7759/cureus.32361
47. Cheng Y, Sun F, Wang L, et al. Virus-induced p38 MAPK activation facilitates viral infection. Theranostics. 2020;10(26):12223–12240. doi:10.7150/thno.50992
48. Canovas B, Nebreda AR. Diversity and versatility of p38 kinase signalling in health and disease. Nat Rev Mol Cell Biol. 2021;22(5):346–366. doi:10.1038/s41580-020-00322-w
49. Marchant D, Singhera GK, Utokaparch S, et al. Toll-like receptor 4-mediated activation of p38 mitogen-activated protein kinase is a determinant of respiratory virus entry and tropism. J Virol. 2010;84(21):11359–11373. doi:10.1128/JVI.00804-10
50. Shin HB, Choi MS, Yi CM, Lee J, Kim NJ, Inn KS. Inhibition of respiratory syncytial virus replication and virus-induced p38 kinase activity by berberine. Int Immunopharmacol. 2015;27(1):65–68. doi:10.1016/j.intimp.2015.04.045
51. Faist A, Schloer S, Mecate-Zambrano A, et al. Inhibition of p38 signaling curtails the SARS-CoV-2 induced inflammatory response but retains the IFN-dependent antiviral defense of the lung epithelial barrier. Antiviral Res. 2023;209:105475. doi:10.1016/j.antiviral.2022.105475
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.