Back to Journals » Cancer Management and Research » Volume 13
Identification of Urinary Exosomal miRNAs for the Non-Invasive Diagnosis of Prostate Cancer
Authors Li Z, Li LX, Diao YJ , Wang J, Ye Y, Hao XK
Received 22 August 2020
Accepted for publication 15 December 2020
Published 6 January 2021 Volume 2021:13 Pages 25—35
DOI https://doi.org/10.2147/CMAR.S272140
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Chien-Feng Li
Zhuo Li,1– 3,* La-Xiu Li,3,4,* Yan-Jun Diao,3 Juan Wang,3 Yun Ye,1,3 Xiao-Ke Hao2,3,5
1Department of Laboratory Medicine, The First Affiliated Hospital of Xi’an Medical University, Xi’an, Shaanxi 710077, People’s Republic of China; 2The National Engineering Research Center for Miniaturized Detection Systems, College of Life Science, Northwest University, Xi’an, Shaanxi 710069, People’s Republic of China; 3Department of Laboratory Medicine, Xijing Hospital, The Fourth Military Medical University, Xi’an, Shaanxi 710032, People’s Republic of China; 4Department of Laboratory Medicine, Xi’an Fourth Hospital, Xi’an, Shaanxi 710004, People’s Republic of China; 5School of Medicine, Northwest University, Xi’an, Shaanxi 710069, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Xiao-Ke Hao
School of Medicine, Northwest University, Xi’an, Shaanxi 710069, People’s Republic of China
Email [email protected]
Background: Novel and non-invasive biomarkers with higher sensitivity and specificity for the diagnosis of prostate cancer (PCa) is urgently needed. In this study, we used next-generation sequencing (NGS) to characterize the genome-wide exosomal miRNA expression profiling in urine specimens and explored the diagnostic potential of urinary exosomal miRNAs for PCa.
Methods: Urinary exosomal microRNA expression profiling was performed by next-generation sequencing (NGS) and then validated by quantitative real-time PCR.
Results: Significant downregulation of urinary exosomal miR-375 was observed in PCa patients compared with healthy controls, while the expression levels of urinary exosomal miR-451a, miR-486-3p and miR-486-5p were found to be significantly up-regulated in the PCa patients. Furthermore, the expression level of urinary exosomal miR-375 showed a significant correlation with the clinical T-stage and bone metastasis of patients with PCa (P< 0.05). Receiver operator characteristic curve demonstrated that the urinary exosomal miR-375, miR-451a, miR-486-3p and miR-486-5p levels can be used to differentiate PCa patients from healthy controls, with area under the curves (AUCs) of 0.788, 0.757, 0.704 and 0.796, respectively. The urinary exosomal miR-375 was found to be superior in discriminating between localized and metastatic PCa with an AUC of 0.806. Moreover, PCa patients can be distinguished from patients with benign prostatic hyperplasia by using a panel combining urinary exosomal miR-375 and miR-451a with an AUC of 0.726.
Conclusion: These findings demonstrate that the urinary exosomal miRNAs can serve as novel and non-invasive biomarkers for diagnosing and predicting the progression of PCa.
Keywords: prostate cancer, next-generation sequencing, urinary, exosomal miRNAs
Background
Prostate cancer (PCa) is one of the most common malignant tumours in the male urinary system.1 Although the conventional biopsy is the gold standard for the diagnosis of this tumour, its application is limited due to its invasive procedure. Currently, prostate-specific antigen (PSA) is the most commonly used biomarker for prostate cancer screening. Serum PSA testing for the early detection of PCa, however, strongly depends on the patient’s age and the local prevalence of the disease.2 Most importantly, there are other factors in addition to PCa that can increase the PSA level, such as the presence of benign prostatic hyperplasia (BPH), infection and chronic inflammation,3 leading to a significant over-diagnosis and subsequent overtreatment of indolent PCa. The PSA level, Gleason score, and radiological features are difficult to predict the progression of PCa. Therefore, novel and non-invasive biomarkers with high sensitivity and specificity for the diagnosis of PCa are urgently needed. Exosomal microRNAs (miRNAs or miRs) in circulating body fluids have recently been reported to augment the diagnosis and management of various diseases, including cancer.4–7
Exosomes are small membrane-bound vesicles ranging in size from 30 to 100 nm, which are secreted by numerous cell types into the extracellular environment and can be found in biological fluids such as blood, semen, saliva and urine.8–10 Exosomes contain proteins and various types of RNA molecules including miRNAs. It has been reported that miRNA-rich exosomes can be taken up by neighbouring or distant cells, and subsequently modulate those recipient cells.11 Recent research has revealed that exosomal miRNA profiling can reflect its cell of origin.12,13 In particular, the exosomal miRNAs derived from urine have been identified in recent years.14,15 Recently, extensive attention has been focused on using exosomal miRNAs as a source of biomarkers, because their contents resemble those of the cell of origin. A substantial number of exosomal miRNAs have been reported as diagnostic, prognostic, or even therapeutic biomarkers in cancer patients.16–20 Therefore, exosomal miRNAs may serve as valuable non-invasive biomarkers for the diagnosis and prognosis of certain diseases.
Over the last two decades, researchers have emphasized the importance of aberrant miRNA expression, which is a relevant driving force in the initiation and progression of PCa. However, there are few studies reported on the use of exosomal miRNAs in urine specimens for diagnosis and prognosis in patients with PCa. In this study, to explore the diagnostic value of urinary exosomal miRNA for prostate cancer, next-generation sequencing (NGS) technology was used to characterize the genome-wide exosomal miRNA expression profile in urine from PCa patients and healthy controls, and quantitative real-time PCR (qRT-PCR) assay was then performed to further validate the candidate miRNAs. The flow chart of the experimental design was shown in Supplemental Figure S1.
Patients and Methods
Patients and Sample Collection
All urine samples were obtained from the Xijing Hospital of The Fourth Military Medical University (Xi’an, People’s Republic of China). All the patients with PCa or BPH disease enrolled in this study were newly diagnosed cases (no prior treatment). The diagnosis of PCa was confirmed by histological examination or prostate biopsy. Histopathologic Gleason scoring of the tumours was performed according to the WHO criteria. Tumours were staged according to the seventh edition of the American Joint Committee on Cancer tumour-node-metastasis (TNM) system. Clinical T-classification of PCa was divided as organ-confined (T1/T2) or non-organ-confined (T3/T4). First-catch morning urine samples (15–40 mL) were obtained from all participants without prostate rectal exam or massage, and then centrifuged at 10,000× g for 30 mins to remove cell debris and sediment. They were subsequently collected in 50 mL Eppendorf conical tubes and stored at −80°C before exosomal RNA isolation. In this study, six PCa patients and three healthy controls were used as the NGS discovery cohort. Subsequently, another 47 PCa patients, 29 BPH patients and 25 age- and gender-matched healthy individuals were used as the qRT-PCR validation cohort (Table 1). This study was approved by the Medical Ethics Committee of the Xijing Hospital of The Fourth Military Medical University (No. XJYYLL-2,015,129) and was performed in accordance with the regulatory guidelines regarding clinical assays. Informed consent was obtained from all participants or their legal guardians in this non-invasive study. This study was conducted in accordance with the Declaration of Helsinki.
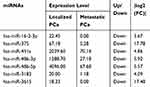 |
Table 1 Expression of seven Significantly Deregulated miRNAs Between Localized PCa Patients and Metastatic PCa Patients Determined by NGS Technology |
Urinary Exosomes Isolation
Exosomes were isolated from urine samples using ExoQuick-TC for tissue culture media and urine (System Biosciences, Mountain View, CA, USA) according to the manufacturer’s protocol. In brief, the urine samples were thawed on ice. Subsequently, 2 mL of ExoQuick-TC reagent was added to 10 mL of urine, and the samples were thoroughly mixed by inverting three times. They were then incubated at 4°C for 12 h, followed by centrifuging at 1500×g for 30 mins. The supernatant was discarded, and the pellets were collected for RNA isolation. The presence of exosomes in the pellets was confirmed by transmission electron microscopy (Supplemental Figure S2).
Transmission Electron Microscopy (TEM)
An aliquot of the exosomal pellet was resuspended in phosphate-buffered saline, and a drop of the suspension was placed on a sheet of parafilm. A copper grid was placed on the drop for 2 min at room temperature and was subsequently placed onto a drop of 2% phosphotungstic acid for a 2 min-staining process. The grid was air-dried for several minutes and was examined by using a Tecnai G2 Spirit electron microscopy (FEI Co., Hillsboro, OR, USA).
Exosomal RNA Isolation
Exosomes were extracted from equal amounts of urine samples collected from healthy controls, BPH and PCa patients. RNA was isolated from the exosomes using miRNeasy Serum/Plasma kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol. To regulate the variability of exosomal miRNA expression levels, at the beginning of RNA isolation, 2 µL of cel-miR-39 (RiboBio, Guangzhou, China) was added to each sample as a spike-in control.21–23
Sequencing Library Preparation and Sequencing Data Analysis
To select miRNAs whose expression levels were altered in PCa patients compared with healthy controls, exosomal microRNA profiling of nine urine samples (6 PCa patients including 3 localized and 3 metastatic PCa patients and 3 age-matched healthy controls) was performed using Illumina NGS technology. Small RNA libraries were constructed using RNA extracted from exosomes. Index libraries were pooled equally for sequencing (Illumina HiSeq2500 platform). The exosomal miRNA RNA-seq data were processed primarily by miRBase21, which indicated miRNA types and expression levels. Exosomal miRNA profiling was normalized using read counts per million mappable miRNA sequences.
Validation of Exosomal miRNAs Using qRT-PCR
The qRT-PCR was performed using a PrimeScript RT Reagent kit and a SYBR Premix Ex Taq kit (Takara Bio, Inc., Shiga, Japan) according to the manufacturer’s protocols. Briefly, 2 µL of total RNA from each sample was reverse transcribed using a PrimeScript RT Reagent kit and specific stem-loop primers that were purchased from RiboBio. The qPCR reactions were performed on an ABI 7500 Fast Detection system (Applied Biosystems, Foster City, CA, USA) using an SYBR Premix Ex Taq kit and sequence-specific primers purchased from RiboBio. Each experiment was carried out in triplicate, and the mean value of the three-cycle threshold was used for further analysis. Cel-miR-39 was used to normalize the exosomal miRNA expression analysis, and the relative quantification analysis of exosomal miRNA was calculated using the 2−ΔΔCt method.24 Exosomal miRNAs with threshold cycle (Ct) values >35 were excluded from the analysis.
Statistical Analysis
Statistical analysis was performed using the Mann–Whitney U-test on log-transformed data between the groups. The box plots represent miRNA levels relative to cel-miR-39, transformed into quantities using the formula 2−ΔΔCt. The box plots and correlation analysis were performed using GraphPad Prism 5. Abnormally distributed data are presented as the median (IQR). P value <0.05 was considered statistically significant.
Receiver operator characteristic (ROC) curves were constructed using SPSS 16.0 to estimate the feasibility of using the specific exosomal miRNAs to differentiate between the PCa patients and the healthy controls. Meanwhile, the area under the curve (AUC) was used to test the power of using the specific exosomal miRNAs and PSA to distinguish PCa patients from BPH patients or to differentiate between localized PCa and metastatic PCa. In brief, the logistic regression was first carried out using miRNA expression levels, and the predicted probabilities were calculated for each single miRNA or all miRNAs together. The AUC was calculated with 95% confidence intervals.
Results
Expression Profiling of Urinary Exosomal miRNAs in Prostate Cancer and the Selection of Candidate Exosomal miRNAs for Validation
Morphological confirmation of exosomes isolated from urine samples was evaluated by TEM (See Supplemental Figure S1). Evaluating aberrant urinary exosomal miRNA expression in PCa can provide a promising tool to identify a specific biomarker for the diagnosis and prediction of PCa. In the initial screening phase using NGS technology, urine samples were collected from six PCa patients (3 localized PCa patients and 3 metastatic PCa patients) and three healthy controls. The standardized count of sequencing reads (total sequencing tags in the library) could be used to compare miRNA expression levels between PCa patients and healthy controls. A genome-wide expression profiling of exosomal miRNAs obtained by Illumina NGS technology showed that urine from PCa patients had 39 down-regulated miRNAs and 14 up-regulated miRNAs compared with urine from the healthy controls (Table 2), and urine from metastatic PCa patients had 7 down-regulated miRNAs compared with the urine of localized PCa patients (Table 1). For validation, four significantly up-regulated miRNAs (miR-16-2-3p, miR-451a, miR-486-3p and miR-486-5p) and one down-regulated miRNA (miR-375) were selected, which showed a mean fold-change >2 between PCa patients and healthy controls, as well as between localized PCa and metastatic PCa.
 |
Table 2 Expression of Significantly Deregulated miRNAs Between PCa Patients and Healthy Controls Determined by NGS Technology |
Validation by qRT-PCR
To further validate the sequencing data, the five differentially expressed urine exosomal miRNAs were examined by using qRT-PCR in a validation sample cohort composed of 47 PCa patients and 25 healthy controls. The clinical and demographic characteristics of the PCa patients are summarized in Table 1. qRT-PCR results demonstrated that the expression of miR-375 was significantly downregulated in urine samples of PCa patients compared with those of healthy controls (Figure 1A). In contrast, the expression levels of miR-451a, miR-486-3p and miR-486-5p were significantly upregulated in PCa samples compared with healthy controls (Figure 1C–E). More importantly, the trend of the alteration of the expression of the four miRNAs was generally concordant between the sequencing data and the validation cohort. In addition, miR-16-2-3p displayed a differential expression tendency between NGS and qRT-PCR, but there was no significant difference in the relative expression of miR-16-2-3p in PCa samples compared with samples from healthy controls (Figure 1B). Therefore, miR-16-2-3p was excluded from further study. This phase generated a panel of four miRNAs (miR-375, miR-451a, miR-486-3p and miR-486-5p) that were significantly altered in PCa patients.
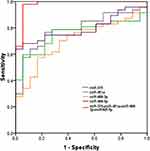
Receiver operator characteristics (ROC) curves were constructed to explore the sensitivity and specificity of urinary exosomal miR-375, miR-451a, miR-486-3p and miR-486-5p in discriminating between PCa patients and healthy controls. The results suggested that the four miRNAs can discriminate between the two groups with high precision: miR-375 (AUC = 0.788; 95% CI = 0.676–0.901), miR-451a (AUC = 0.757; 95% CI = 0.641–0.872), miR-486-3p (AUC = 0.704; 95% CI = 0.576–0.833), miR-486-5p (AUC = 0.796; 95% CI = 0.690–0.901). As a result, we selected a classifier of four miRNAs to be the potential signature for PCa. The diagnostic sensitivity and specificity obtained for the validation cohort were 91% and 89%, respectively, and AUC was 0.979 for the validation cohort (Figure 2).
Correlation of Urinary Exosomal miRNA Levels with Clinical Factors
To determine whether the four miRNA-based biomarkers were influenced by the clinical features of PCa patients, including age, Gleason score, T-stage and serum PSA, the relationships between the four urinary exosomal miRNA expression levels and clinical factors were analyzed using Spearman correlation. The urinary exosomal miR-375 level showed a correlation with the T-stage of PCa patients, while no obvious differences were observed when PCa patients were stratified by age, Gleason score, or serum PSA levels. In addition, the expression level of miR-375 was negatively correlated with the clinical T-stages of PCa, r = - 0.41. The PCa patients with lower levels of urinary exosomal miR-375 expression exhibited advanced clinical T-stages (P < 0.05).
Differentiation Between PCa Patients and BPH Patients by a Biomarker Based on 2 miRNAs
To investigate whether the four urinary exosomal miRNAs could serve as specific biomarkers to distinguish PCa from BPH, their expression levels in 29 BPH patients were measured by using qRT-PCR, and the results were compared with those from the PCa patients in the validation cohort. The clinical and demographic characteristics of the PCa patients are summarized in Table 3. The results clearly indicated that miR-375 and miR-451a could be used to separate PCa from BPH cases (Figure 3A and B). No significant differences were observed in the levels of the other two candidate miRNAs between PCa patients and BPH patients (Figure 3C and D). The ROC curve shows that the AUCs of miR-375 and miR-451a are 0.700 and 0.683, respectively, which suggests their potential efficacy in distinguishing between PCa and BPH. Moreover, miR-375 combined with miR-451a provided a better area under the curve (AUC = 0.726; 95% CI = 0.614–0.842), which means that the biomarker based on those 2 miRNAs is more sensitive in discriminating between PCa samples and BPH samples than PSA (AUC = 0.624; 95% CI = 0.487–0.761; Figure 4).
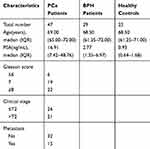 |
Table 3 Clinical Characteristics of Participants in the qRT-PCR Cohort |
Differentiation Between Metastatic PCa Samples and Localized PCa Samples by miR-375
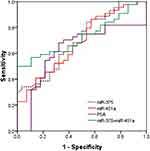
To further analyse whether the 4 miRNAs could serve as a novel biomarker to predict the progression of PCa, the 47 PCa patients in the validation cohort were stratified into 2 groups: localized PCa and metastatic PCa. The comparison shown in Figure 5A clearly suggests that miR-375 could be used to distinguish localized PCa from metastatic PCa. No significant differences were observed in the levels of the other three miRNAs between localized PCa and metastatic PCa (Figure 5B–D). ROC analysis further confirmed that miR-375 (AUC = 0.806; 95% CI = 0.680–0.932) is superior to PSA (AUC = 0.684; 95% CI = 0.510–0.859) in discriminating between localized PCa and metastatic PCa (Figure 6).
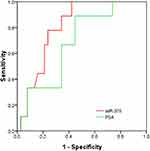 |
Figure 6 Receiver-operating characteristic curve (ROC) for urinary exosomal miR-375 and PSA to differentiate between localized PCa patients and metastatic PCa patients. |
Discussion
PCa has a wide diffusion and represents the second major cause of death worldwide because of its aggressive behaviour.25 MiRNAs have been suggested to be novel diagnostic/therapeutic instruments, as they could play a role as a biomarker in the diagnosis and may also represent possible targets for new therapies.26,27 In the case of PCa, the interest in miRNA is linked to the fact that prostate-specific antigen shows several limitations,28,29 as infection, very common in frail patients.30,31 Early diagnosis, especially in the high risk, as the main cause of death for PCa, is needed.32,33 The challenge nowadays is to better understand the mechanism of cancer development, finding real PCa markers and developing new specific therapies in order to reduce the mortality of PCa.34 Thus, the research on miRNA is of paramount importance.35
A large number of studies have proved that miRNAs play key roles in the pathogenesis and progression of tumours.36,37 Therefore, miRNAs can be used as molecular markers for diagnosis and prognosis. In general, using urine as a specimen for tumour biomarkers is challenging because urine contains a wide variety of biomolecules such as high concentrations of nucleases and RNases. However, exosomes, which contain miRNA, have double-lipid membranes that can protect the miRNA contained within from degradation by RNases in urine.38 In addition, urine samples can be easily collected during the routine examination process. Based on the above advantages, urinary exosomal miRNAs can be used as promising molecular markers for certain diseases. Although miRNA levels have been studied in various diseases, few comprehensive studies have been reported so far for urine exosomal miRNA-based PCa diagnosis.39,40
The aim of this study was to explore whether urine samples contain exosomal miRNAs and whether the expression profiling of these urinary exosomal miRNAs consistently differ between PCa patients and healthy controls. In this study, we investigated hundreds of known exosomal miRNAs to search for diagnostic markers for PCa by using Illumina NGS technology to analyse the urine-based profile of exosomal miRNA expression. After analysis, we selected five significantly deregulated miRNAs, which showed a mean fold-change >2 between PCa patients and healthy controls, as well as between localized PCa and metastatic PCa. The 5 candidate miRNAs were further validated in a larger cohort including 47 PCa patients and 25 healthy controls. qRT-PCR results demonstrated the upregulation of three miRNAs (miR-451a, miR-486-3p, and miR-486-5p) and the downregulation of miR-375 in PCa samples. Furthermore, the expression level of miR-375 was found to be negatively correlated with advanced clinical T-stages of PCa patients.
In our research, we found that the exosomal miR-451a level, which has been shown to be frequently downregulated in hypopharyngeal squamous cell carcinoma,41 papillary thyroid carcinoma,42 melanoma43 and gastric cancer,44 was significantly upregulated in the urine of PCa patients. Deregulation of miR-451a has not been previously reported in PCa. To our knowledge, this is the first study to show that urinary exosomal miR-451a is aberrantly expressed in PCa, which implies that miR-451a could be a non-coding oncogene in PCa, and its function needs further exploration. We found that urinary exosomal miR-486-5p and miR-486-3p levels were upregulated in PCa patients compared with healthy controls. To date, miR-486-5p and miR-486-3p have been rarely reported in PCa patients,45 which still needs further research.
For miR-375, previous studies showed its upregulation in PCa serum, PCa tissue and cell lines in comparison to healthy control serum, adjacent normal tissue and non-tumorigenic epithelial prostatic cells, respectively.46–49 Recently, Foj et al detected miR-375 in urinary exosomes and found it was significantly upregulated in PCa patients.39 However, in the present study based on NGS and qRT-PCR studies, we found an opposite result. Interestingly, in another recent study, the authors found a similar pattern. They indicated that the levels of urinary exosomal miR-196a-5p and miR-501-3p, which had been found to be mainly upregulated in PCa patients in previous studies, were significantly downregulated in PCa patients based on both NGS and qRT-PCR tests. While in our present NGS study, we achieved consistent results showing the downregulation of the two miRNAs in PCa patients. In addition, in the above-mentioned study, the entire list of NGS results showed that miR-375 was reduced in PCa patients compared with the healthy controls, which agreed with our results. It is not clear why there are such different results in urinary exosomal miRNAs studies, but the different patient cohorts using could be a possible explanation. Therefore, to confirm the findings, larger cohorts and more multi-centre studies are needed. In addition, although miR-375 has been mostly considered to be a tumour oncogene in PCa, it is worth mentioning that the recent literature has revealed that miR-375 acts either as an oncomiR or a tumour-suppressor miRNA depending on the cellular context, which seems to agree with the results in our study.
This study has demonstrated the presence of exosomal miRNAs in urine samples as in previous reports. In addition, we have demonstrated that exosomal miR-375, miR-451a, miR-486-3p and miR-486-5p are differentially expressed in urine from PCa patients, and the ROC curve demonstrates that the four miRNAs combined together have good efficacy to distinguish between healthy controls and PCa patients, with a sensitivity of 91% and a specificity of 89%. In addition, our present finding suggests that a combination of miR-375 and miR-451a has the potential to serve as a specific and sensitive molecular marker to distinguish PCa patients from BPH patients, and miR-375 alone can distinguish localized PCa patients from metastatic PCa patients to predict the progression of PCa.
Conclusion
In conclusion, we suggest that certain exosomal miRNAs in urine could serve as powerful and non-invasive tools for the diagnosis and prognosis of PCa. Further studies are needed in a larger cohort of patients to confirm these findings.
Abbreviations
AUC, area under the curve; Ct, threshold cycle; miRNAs, microRNAs; NGS, next-generation sequencing; PCa, prostate cancer; PSA, prostate-specific antigen; ROC, receiver-operating characteristic; TEM, transmission electron microscopy.
Data Sharing Statement
All the datasets in the manuscript are available from the corresponding author on request.
Ethics Approval and Consent to Participate
The procedures were approved by the Ethics Committee of Xijing Hospital, and the written informed consent was signed by each patient.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (General Program, 81872347), the Health Commission of Shaanxi Province (2016D020), the Xi’an Science and Technology Bureau (2017121SF/YX015(3)) and Shaanxi Natural Science Foundation (2018JQ8010).
Author Contributions
All authors contributed to data analysis, drafting or revising the article, have agreed on the journal to which the article will be submitted, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors declare that no competing interests exist.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30. doi:10.3322/caac.21387
2. Grossman DC, Curry SJ, Owens DK, et al. Screening for prostate cancer: US preventive services task force recommendation statement. JAMA. 2018;319(18):1901–1913. doi:10.1001/jama.2018.3710
3. Lilja H, Ulmert D, Vickers AJ. Prostate-specific antigen and prostate cancer: prediction, detection and monitoring. Nat Rev Cancer. 2008;8(4):268–278. doi:10.1038/nrc2351
4. Neylon A, McGorrian C, Blake GJ. miRNA-93-5p and other miRNAs as predictors of coronary artery disease and STEMI. Int J Cardiol. 2016;224:310–316. doi:10.1016/j.ijcard.2016.09.016
5. Giallombardo M, Chacártegui Borrás J, Castiglia M, et al. Exosomal miRNA analysis in non-small cell lung cancer (NSCLC) patients’ plasma through qPCR: a feasible liquid biopsy tool. JoVE. 2016;(111). doi:10.3791/53900
6. Delić D, Eisele C, Schmid R, et al. Urinary exosomal miRNA signature in type II diabetic nephropathy patients. PLoS One. 2016;11(3):e0150154. doi:10.1371/journal.pone.0150154
7. Shi R, Wang PY, Li XY, et al. Exosomal levels of miRNA-21 from cerebrospinal fluids associated with poor prognosis and tumor recurrence of glioma patients. Oncotarget. 2015;6(29):26971–26981. doi:10.18632/oncotarget.4699
8. Cappello F, Logozzi M, Campanella C, et al. Exosome levels in human body fluids: a tumor marker by themselves? Eur J Pharm Sci. 2017;96:93–98. doi:10.1016/j.ejps.2016.09.010
9. Zhang J, Li S, Li L, et al. Exosome and exosomal microRNA: trafficking, sorting, and function. Genomics Proteomics Bioinformatics. 2015;13(1):17–24. doi:10.1016/j.gpb.2015.02.001
10. Colombo M, Raposo G, Théry C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol. 2014;30:255–289. doi:10.1146/annurev-cellbio-101512-122326
11. Franzen CA, Blackwell RH, Foreman KE, Kuo PC, Flanigan RC, Gupta GN. Urinary exosomes: the potential for biomarker utility, intercellular signaling and therapeutics in urological malignancy. J Urol. 2016;195(5):1331–1339. doi:10.1016/j.juro.2015.08.115
12. Iwai K, Minamisawa T, Suga K, Yajima Y, Shiba K. Isolation of human salivary extracellular vesicles by iodixanol density gradient ultracentrifugation and their characterizations. J Extracell Vesicles. 2016;5:30829. doi:10.3402/jev.v5.30829
13. Skog J, Würdinger T, van Rijn S, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10(12):1470–1476. doi:10.1038/ncb1800
14. Cheng L, Sun X, Scicluna BJ, Coleman BM, Hill AF. Characterization and deep sequencing analysis of exosomal and non-exosomal miRNA in human urine. Kidney Int. 2014;86(2):433–444. doi:10.1038/ki.2013.502
15. Gildea JJ, Carlson JM, Schoeffel CD, Carey RM, Felder RA. Urinary exosome miRNome analysis and its applications to salt sensitivity of blood pressure. Clin Biochem. 2013;46(12):1131–1134. doi:10.1016/j.clinbiochem.2013.05.052
16. Huang X, Yuan T, Liang M, et al. Exosomal miR-1290 and miR-375 as prognostic markers in castration-resistant prostate cancer. Eur Urol. 2015;67(1):33–41. doi:10.1016/j.eururo.2014.07.035
17. Haflidadóttir BS, Ceder Y. Exosomal microRNAs as potential biomarkers in castration-resistant prostate cancer. Eur Urol. 2015;67(1):42–43. doi:10.1016/j.eururo.2014.08.067
18. Ogata-Kawata H, Izumiya M, Kurioka D, et al. Circulating exosomal microRNAs as biomarkers of colon cancer. PLoS One. 2014;9(4):e92921. doi:10.1371/journal.pone.0092921
19. Corcoran C, Rani S, O’Driscoll L. miR-34a is an intracellular and exosomal predictive biomarker for response to docetaxel with clinical relevance to prostate cancer progression. Prostate. 2014;74(13):1320–1334. doi:10.1002/pros.22848
20. Hessvik NP, Sandvig K, Llorente A. Exosomal miRNAs as biomarkers for prostate cancer. Front Genet. 2013;4:36. doi:10.3389/fgene.2013.00036
21. Li Z, Ma YY, Wang J, et al. Exosomal microRNA-141 is upregulated in the serum of prostate cancer patients. Onco Targets Ther. 2016;9:139–148. doi:10.2147/OTT.S95565
22. Selth LA, Townley S, Gillis JL, et al. Discovery of circulating microRNAs associated with human prostate cancer using a mouse model of disease. Int J Cancer. 2012;131(3):652–661. doi:10.1002/ijc.26405
23. Mitchell PS, Parkin RK, Kroh EM, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA. 2008;105(30):10513–10518. doi:10.1073/pnas.0804549105
24. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods (San Diego, Calif). 2001;25(4):402–408. doi:10.1006/meth.2001.1262
25. Situ J, Zhang H, Jin Z, Li K, Mao Y, Huang W. MicroRNA-939 directly targets HDGF to inhibit the aggressiveness of prostate cancer via deactivation of the WNT/β-catenin pathway. Onco Targets Ther. 2020;13:4257–4270. doi:10.2147/OTT.S250101
26. Cochetti G, Poli G, Guelfi G, Boni A, Egidi MG, Mearini E. Different levels of serum microRNAs in prostate cancer and benign prostatic hyperplasia: evaluation of potential diagnostic and prognostic role. Onco Targets Ther. 2016;9:7545–7553. doi:10.2147/OTT.S119027
27. Guelfi G, Cochetti G, Stefanetti V, et al. Next generation sequencing of urine exfoliated cells: an approach of prostate cancer microRNAs research. Sci Rep. 2018;8(1):7111. doi:10.1038/s41598-018-24236-y
28. Mearini E, Antognelli C, Del Buono C, et al. The combination of urine DD3(PCA3) mRNA and PSA mRNA as molecular markers of prostate cancer. Biomarkers. 2009;14(4):235–243. doi:10.1080/13547500902807306
29. Talesa VN, Antognelli C, Del Buono C, et al. Diagnostic potential in prostate cancer of a panel of urinary molecular tumor markers. Cancer Biomarkers. 2009;5(6):241–251. doi:10.3233/CBM-2009-0109
30. Errico G, Gagliotti C, Monaco M, et al. Colonization and infection due to carbapenemase-producing Enterobacteriaceae in liver and lung transplant recipients and donor-derived transmission: a prospective cohort study conducted in Italy. Clin Microbiol Infect. 2019;25(2):203–209. doi:10.1016/j.cmi.2018.05.003
31. Gagliotti C, Morsillo F, Moro ML, et al. Infections in liver and lung transplant recipients: a national prospective cohort. Eur J Clin Microbiol Infect Dis. 2018;37(3):399–407. doi:10.1007/s10096-018-3183-0
32. Egidi MG, Cochetti G, Guelfi G, et al. Stability assessment of candidate reference genes in urine sediment of prostate cancer patients for miRNA applications. Dis Markers. 2015;2015:973597. doi:10.1155/2015/973597
33. Rende M, Rambotti MG, Stabile AM, et al. Novel localization of low affinity NGF receptor (p75) in the stroma of prostate cancer and possible implication in neoplastic invasion: an immunohistochemical and ultracytochemical study. Prostate. 2010;70(5):555–561. doi:10.1002/pros.21089
34. Baldassarri M, Fallerini C, Cetta F, et al. Omic approach in non-smoker female with lung squamous cell carcinoma pinpoints to germline susceptibility and personalized medicine. Cancer Research Treat. 2018;50(2):356–365. doi:10.4143/crt.2017.125
35. Cochetti G, Rossi de Vermandois JA, Maulà V, et al. Role of miRNAs in prostate cancer: do we really know everything? Urol Oncol. 2020;38(7):623–635. doi:10.1016/j.urolonc.2020.03.007
36. Hayes J, Peruzzi PP, Lawler S. MicroRNAs in cancer: biomarkers, functions and therapy. Trends Mol Med. 2014;20(8):460–469. doi:10.1016/j.molmed.2014.06.005
37. Cho WC. OncomiRs: the discovery and progress of microRNAs in cancers. Mol Cancer. 2007;6:60. doi:10.1186/1476-4598-6-60
38. Zhang W, Peng P, Shen K. Role of exosome shuttle RNA in cell-to-cell communication. Zhongguo Yi Xue Ke Xue Yuan Xue Bao. 2016;38(4):480–483. doi:10.3881/j.issn.1000-503X.2016.04.020
39. Foj L, Ferrer F, Serra M, et al. Exosomal and non-exosomal urinary miRNAs in prostate cancer detection and prognosis. Prostate. 2017;77(6):573–583. doi:10.1002/pros.23295
40. Filella X, Foj L. Prostate cancer detection and prognosis: from prostate specific antigen (PSA) to exosomal biomarkers. Int J Mol Sci. 2016;17(11):1784. doi:10.3390/ijms17111784
41. Fukumoto I, Kinoshita T, Hanazawa T, et al. Identification of tumour suppressive microRNA-451a in hypopharyngeal squamous cell carcinoma based on microRNA expression signature. Br J Cancer. 2014;111(2):386–394. doi:10.1038/bjc.2014.293
42. Li M, Song Q, Li H, Lou Y, Wang L. Circulating miR-25-3p and miR-451a may be potential biomarkers for the diagnosis of papillary thyroid carcinoma. PLoS One. 2015;10(7):e0132403. doi:10.1371/journal.pone.0132403
43. Babapoor S, Fleming E, Wu R, Dadras SS. A novel miR-451a isomiR, associated with amelanotypic phenotype, acts as a tumor suppressor in melanoma by retarding cell migration and invasion. PLoS One. 2014;9(9):e107502. doi:10.1371/journal.pone.0107502
44. Riquelme I, Tapia O, Leal P, et al. miR-101-2, miR-125b-2 and miR-451a act as potential tumor suppressors in gastric cancer through regulation of the PI3K/AKT/mTOR pathway. Cell Oncol (Dordrecht). 2016;39(1):23–33. doi:10.1007/s13402-015-0247-3
45. Yang Y, Ji C, Guo S, et al. The miR-486-5p plays a causative role in prostate cancer through negative regulation of multiple tumor suppressor pathways. Oncotarget. 2017;8(42):72835–72846. doi:10.18632/oncotarget.20427
46. Pickl JM, Tichy D, Kuryshev VY, et al. Ago-RIP-Seq identifies Polycomb repressive complex I member CBX7 as a major target of miR-375 in prostate cancer progression. Oncotarget. 2016;7(37):59589–59603. doi:10.18632/oncotarget.10729
47. Wach S, Al-Janabi O, Weigelt K, et al. The combined serum levels of miR-375 and urokinase plasminogen activator receptor are suggested as diagnostic and prognostic biomarkers in prostate cancer. Int J Cancer. 2015;137(6):1406–1416. doi:10.1002/ijc.29505
48. Choi N, Park J, Lee JS, et al. miR-93/miR-106b/miR-375-CIC-CRABP1: a novel regulatory axis in prostate cancer progression. Oncotarget. 2015;6(27):23533–23547. doi:10.18632/oncotarget.4372
49. Brase JC, Johannes M, Schlomm T, et al. Circulating miRNAs are correlated with tumor progression in prostate cancer. Int J Cancer. 2011;128(3):608–616. doi:10.1002/ijc.25376
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.