Back to Journals » Journal of Blood Medicine » Volume 8
Identification of patients with congenital hemophilia in a large electronic health record database
Authors Wang M , Cyhaniuk A, Cooper DL , Iyer NN
Received 1 February 2017
Accepted for publication 23 July 2017
Published 30 August 2017 Volume 2017:8 Pages 131—139
DOI https://doi.org/10.2147/JBM.S133616
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Martin Bluth
Michael Wang,1 Anissa Cyhaniuk,2 David L Cooper,3 Neeraj N Iyer3
1Hemophilia and Thrombosis Center, University of Colorado School of Medicine, Aurora, CO, 2AC Analytic Solutions, Barrington, IL, 3Clinical Development, Medical and Regulatory Affairs, Novo Nordisk Inc., Plainsboro, NJ, USA
Background: Electronic health records (EHRs) are an important source of information with regard to diagnosis and treatment of rare health conditions, such as congenital hemophilia, a bleeding disorder characterized by deficiency of factor VIII (FVIII) or factor IX (FIX).
Objective: To identify patients with congenital hemophilia using EHRs.
Design: An EHR database study.
Setting: EHRs were accessed from Humedica between January 1, 2007, and July 31, 2013.
Patients: Selection criteria were applied for an initial ICD-9-CM diagnosis of 286.0 (hemophilia A) or 286.1 (hemophilia B), and confirmation of records 6 months before and 12 months after the first diagnosis. Additional selection criteria included mention of “hemophilia” and “blood” or “bleed” within physician notes identified via natural language processing.
Results: A total of 129 males and 35 females were identified as the analysis population. Of those patients for whom both prothrombin time and activated partial thromboplastin time test results were available, only 56% of males and 7% of females exhibited a pattern of test results consistent with congenital hemophilia (normal prothrombin time and prolonged activated partial thromboplastin time). Few patients had a prescription for a hemophilia treatment; males most commonly received Amicar (10.8%) or FVIII (9.0%), whereas females most commonly received DDAVP (11.0%). The most identifiable sites of pain were the chest and the abdomen; 41% of males and 37% of females had joint pain. To evaluate whether patients had been correctly identified with congenital hemophilia, EHRs of 6 patients were reviewed; detailed assessment of their data was found to be inconsistent with a conclusive diagnosis of congenital hemophilia.
Limitations: Inconsistent coding practices may affect data integrity.
Conclusion: A potentially high number of false positive identifications, particularly among female patients, suggests that ICD-9-CM coding alone may be insufficient to identify patient cohorts. In-depth reviews and multimodal analysis of chart notes may improve data integrity.
Keywords: congenital hemophilia, electronic health record, database, big data
Introduction
Congenital hemophilia is a rare, chronic, inheritable bleeding disorder caused by the deficiency of clotting factors VIII (hemophilia A) or IX (hemophilia B), and over time may cause damage to the joints consequent to recurrent joint bleeding.1 It is typically diagnosed at an early age based on family history or following spontaneous bleeding.1 Males are predominantly affected due to X-linked inheritance, with females serving as carriers; however, females can also have symptomatically low levels of activity of clotting factors in the uncommon instance of lyonization of the normal X-chromosome (~3.2% of the patients with hemophilia).2 The overall prevalence of hemophilia in the United States is estimated to be 20,000,3 with hemophilia A occurring more frequently than hemophilia B (~1 in 5,000 males vs ~1 in 30,000 males, respectively).4 A total of 3,582 females with hemophilia were identified in the Centers for Disease Control and Prevention 2011 Universal Data Collection report.2 Because of the rarity of hemophilia, obtaining data from large numbers of patients is a significant challenge, and therefore, data mining from large electronic health record (EHR) databases may be an effective strategy in obtaining useful information and new insights into the issues related to disease management.5
The importance of data obtained from EHRs has been previously demonstrated in many health conditions, such as diabetes,6 cancer,7 obesity,8 and cardiovascular disease.9 In addition, EHRs have been helpful in extracting information from regular sources of secondary data, such as claims data.10 The systematic structure of information contained in EHRs is highly favorable in the analysis of sequential information with regard to the symptoms and diagnoses of the disease, which can be further refined to stratify patients into subgroups describing patient characteristics, health care utilization, outcomes, prognosis, and interventions.11–13 A common approach used to gather patient data from US or Canadian EHRs is to review the International Classification of Diseases, Ninth Revision (ICD-9) or its clinical modification (ICD-9-CM) codes entered into patient charts, which provides a standardized means of coding structured data about a patient population. This strategy may be especially valuable in cases of rare disorders, such as hemophilia, which is typically studied within the network of federally funded treatment centers through public health surveillance surveys.10,14,15
An important advantage of EHRs over commonly used secondary data sources, such as claims databases, is that EHRs allow access to detailed information contained in chart notes, which is entered by physicians or staff as free text via typing, dictation, or transcription.5,11 Clinicians may prefer entering unstructured data within chart notes rather than in specific coding sections because of the flexibility to document nuances,11 including detailed summaries regarding information of patient admission, diagnostic uncertainties, and treatment.5,13 Valuable information in the unstructured clinical narratives provides context that may not be captured in coded portions,11 and therefore, computer applications based on natural language processing (NLP) have been developed to probe chart notes for relevant data and clinical concepts, by making use of keywords and phrases (or “attributes”).5 To improve the accuracy of information extracted from chart notes, the meaning of key terms may be interpreted using an algorithm that recognizes the context in which the terms appear (or “sentiment”) and also analyzes how the terms relate to each other and to the overall clinical concept.16 This process then generates a list of signs, diseases, and symptoms associated with the study population that is suitable for loading into analytical tools or relational database systems. Use of NLP to analyze structured and unstructured information from EHRs, therefore, offers the potential to examine patient cases in totality, including laboratory testing and imaging studies, and to validate diagnoses within a study population.
The most important aim of studying EHRs is to generate real-world evidence with a practical clinical value13 that may be of particular value in the context of rare and chronic disorders, such as hemophilia. Herein, we report data regarding the use of an EHR database to define a population of patients with congenital hemophilia A or B. Because of the intrinsic issues associated with obtaining data from EHRs, many challenges were encountered, supporting the need for more accurate coding processes.
Materials and methods
Patient data were sourced from Humedica’s EHR database, which is a service offering access to anonymized information of nearly 30 million patients in the United States. Because no individually identifiable data were collected in the database or analyzed in this study, approval by an Institutional Review Board was not required. Data were collected between January 2007 and July 2013, with no specificity for baseline patient age. Patients were included if they received an ICD-9-CM diagnosis code of 286.0 (congenital factor VIII disorder, hemophilia A) or 286.1 (congenital factor IX disorder, hemophilia B), had EHR data extending at least 6 months prior to and 12 months after the first ICD-9-CM hemophilia diagnosis code identified in the database, were identified as receiving care within an integrated delivery network, and had chart notes after the initial diagnosis code that included the words “hemophilia” and “blood” or “bleed”. Clinical meaning was derived from the chart notes using Humedica’s NLP process and extracted as 3 separate categories: term (eg, “bleeding”), location (eg, “nasal”), and attribute (eg, “excessive”) for further evaluation and interpretation. EHR data collected included patient gender, age at the time of receiving access to the database, additional ICD-9-CM 286 diagnosis codes, results of prothrombin time (PT) and activated partial thromboplastin time (aPTT) tests, prescriptions for hemophilia treatments, and the presence and location of pain as identified via NLP. To validate the diagnosis of hemophilia among patients identified using the defined set of inclusion and exclusion criteria, individual signs, diseases, and symptoms obtained via NLP keyword extracts were reviewed for 3 randomly chosen male and female patients each by 2 clinicians; all data regarding bleeding are summarized. Data were analyzed using descriptive statistics; categorical variables are presented as numbers and percentages.
Results
Inclusion criteria were specified to identify a population of individuals with an accurate diagnosis of congenital hemophilia (Figure 1). Initial patient selection based on an ICD-9-CM code of 286.* (indicating the broad category “coagulation defects”) identified approximately equal numbers of males (n=7,913; 50.3%) and females (n=7,824; 49.7%), despite a predominant prevalence of X-linked hemophilia among males. The numbers of males (n=705; 56.3%) and females (n=547; 43.7%) were comparable even when we included only those patients whose first identified diagnosis code was for hemophilia A or hemophilia B.
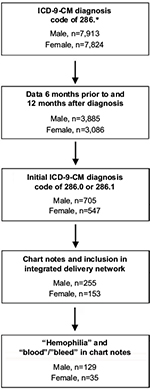  | Figure 1 Patient population. Abbreviation: ICD-9-CM, International Classification of Diseases, Ninth Revision, clinical modification. |
After applying the remaining set of inclusion criteria, an analysis population comprising data of 129 males (78.7%) and 35 females (21.3%) was identified. The mean (±standard deviation (SD)) age at the time of receiving access to the database (2014) was 40 (±25.1) years for males and 48 (±21.9) years for females. Some patients had additional 286.* diagnosis codes, which may reflect a refinement of the initial diagnosis over time or inconsistency of coding from different providers (eg, primary care, hematologist, or hospitalist) (Table 1). More males than females had 1 or more additional 286.* codes (males, 87%; females, 74%). The most common additional 286.* diagnosis codes were for “other and unspecified coagulation defects” (286.9; males, 2%; females, 31%), “von Willebrand’s disease” (286.4; males, 15%; females, 17%), “congenital deficiency of other clotting factors” (286.3; males, 9%; females, 23%), and “acquired coagulation factor deficiency” (286.7; males, 8%; females, 9%). Few patients had non-286 diagnosis codes for bleeding disorders. Glanzmann’s thrombasthenia (qualitative platelet defect, 287.1) was diagnosed in 2% of males and 3% of females. No patients had reported diagnoses of systemic lupus or lupus erythematosus, which might be associated with an artifactual change in aPTT test results.
  | Table 1 Diagnosis codes Note: ^Patients could have multiple diagnosis codes. |
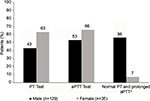
Overall, approximately half of the patients received a coagulation screening test such as PT and aPTT (Figure 2). The availability of results of these tests was higher for females than males. Of the patients who had both PT and aPTT test results, only 56% of the males and 7% of the females had test results consistent with a diagnosis of congenital hemophilia (normal PT and prolonged aPTT).
Few patients had a prescription for hemophilia treatment (Figure 3). The most common prescriptions for males were aminocaproic acid (eg, Amicar®; 10.8%) and coagulation factor VIII (FVIII; 9.0%), whereas the most common prescription for females was desmopressin, or DDAVP (Stimate®; 11.0%). A prescription for an anticoagulant (warfarin, Coumadin®, heparin, Lovenox®, Fragmin®, Eliquis®, Pradaxa®, or Xarelto®), which would typically be expected to be coded as “coagulopathy due to anticoagulants” (289.7) rather than congenital hemophilia, was identified for 36% of males. Dosing information was not obtainable for this study, as few patient records included any data on dosing beyond a single treatment. Nearly all patients’ chart notes included a reference to an NLP term for “pain” (males, 98%; females, 97%); among those that identified a pain location, the most common sites were the chest (male, 88%; female, 83%) and abdomen (male, 85%; female, 91%) (Figure 4). Joint pain was identified in 41% of males and 37% of females.
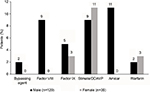  | Figure 3 Hemophilia treatment. Abbreviation: DDAVP, desmopressin. |
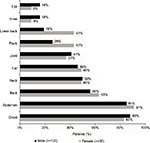  | Figure 4 Sites of pain. |
Case examples
Patient 1 was a 41-year-old female with approximately 2.5 years of chart note data. Her history included multiple instances of bleeding and easy bruising throughout the first 2 years of data, with symptoms including “substantial” and “excessive” bleeding, spontaneous bleeding from the nose, rectum, and bright red blood in the stool. Her last 1.5 years of chart note data included multiple mentions of FVIII deficiency (“mild” and “moderately; decrease”) and hemophilia. Her records did not indicate any use of treatments for hemophilia. She received ICD-9-CM codes of 286.0 and 286.9. Overall, these data seem to indicate a case of symptomatic hemophilia A in a probably heterozygous female (carrier).
Patient 2 was a 33-year-old female with approximately 3 years of chart note data. She experienced at least 3 pregnancies during this period, of which 2 ended in miscarriage and 1 led to “delivery complications”. Recurrent symptoms of bleeding were noted throughout her medical history, which included “unusual” and “abnormal” bleeding, “frequent” nose bleeding, internal bleeding, hematuria, and hematochezia. On the day of her delivery, she experienced “excessive” bleeding and “significant” “intra-abdominal” hemorrhage, and “acquired” FVIII deficiency was noted. Her chart notes included numerous additional mentions of “acquired” FVIII deficiency or “acquired” hemophilia, as well as “rare” blood disorder and “pregnancy-induced” blood disorder. She received ICD-9-CM 286 codes of 286.0, 286.5, and 286.9. Together these data strongly suggest that this was a case of peri-partum acquired hemophilia rather than congenital hemophilia.
Patient 3 was a 78-year-old female with approximately 3.5 years of chart note data. Her history included multiple recurrent bruising and bleeding symptoms, including hematuria, hematochezia, mucosal bleeding, and gastrointestinal bleeding. She also experienced recurrent anemia and thrombocytopenia, potentially as a result of her chronic bleeding symptoms. Although her records did not indicate any use of hemophilia treatments, her chart notes included numerous mentions of “factor VIII inhibitor” associated with FVIII deficiency, coagulopathy, and bruising. She received ICD-9-CM 286 codes of 286.0, 286.5, 286.59, 286.7, and 286.9. Overall, these data seem to suggest that this is a case of acquired hemophilia rather than congenital hemophilia.
Patient 4 was a 69-year-old male with approximately 4 years of chart note data. His history included multiple instances of postoperative bleeding associated with hemarthrosis and a medial meniscus tear, and again with a perforated appendix and acute appendicitis. His chart notes included 4 mentions each of hemophilia A and hemophilia C (an alternate name for factor XI deficiency);17 most often these diagnoses were mentioned on the same day. In most cases, hemophilia A was described as “mild”. His chart notes also included a single mention of FIX deficiency (hemophilia B) and 2 mentions of FVIII deficiency (hemophilia A). He received ICD-9-CM 286 codes of 286.0 and 286.1 and received treatment with FVIII but not FIX. Together, these data are most consistent with a case of congenital hemophilia A due to the specific treatment with FVIII, which would have been ineffective in hemophilia B or C.
Patient 5 was a 57-year-old male with approximately 4 years of chart note data. His history included multiple mentions of hemophilia and FVIII deficiency, as well as chronic hepatitis C, bleeding, “recurrent” and “chronic” hemarthrosis, joint pain, and degenerative joint disease. He received ICD-9-CM 286 codes of 286.0 and 286.2 but had no indications of having received hemophilia treatment. Together these data suggest a case of moderate or severe hemophilia A; however, based upon symptoms and the presence of hepatitis C (implying prior exposure to plasma-derived factor products before viral inactivation steps were added to purification in 1988), the lack of treatment over 4 years is unexpected for an individual with severe hemophilia.
Patient 6 was a 76-year-old male with approximately 6.5 years of chart note data. He had multiple recurrent bleeding symptoms throughout, including frequent mention of “gastro-intestinal” bleeding, hematuria, and hematochezia. His chart notes also included 21 mentions of hemophilia, 16 mentions of factor V deficiency, 5 mentions of factor V Leiden deficiency, and a single mention of factor IV deficiency. He received ICD-9-CM 286 codes of 286.0, 286.3, and 286.9 but had no indications of having received hemophilia treatment. Ten aPTT scores were reported ranging from 83.9 s to 150.01 s; however, no PT scores were reported. Together these data suggest a case of factor VIII deficiency (286.0), V deficiency (coded under 286.3), or a rare case of a FVIII and FV inhibitor, given that factor V Leiden mutation is associated with thrombotic risk (not bleeding).
Discussion
In this study, a large EHR database was used and multiple inclusion/exclusion criteria were specified, in an attempt to identify a population of patients with congenital hemophilia A or B. Criteria required having an ICD-9-CM code of 286.0 (congenital factor VIII disorder) or 286.1 (congenital factor IX disorder) before any other 286 ICD-9-CM codes, as well as mention of “hemophilia” and “blood” or “bleed” within chart notes. In addition, NLP technology was used to derive clinical meaning from the context in which the terms were identified.
Despite stringent inclusion criteria and the incorporation of NLP, the population identified did not consistently reflect the known characteristics of congenital hemophilia. An unexpectedly high percentage of patients were female (21.3%), whereas only approximately 3.2% would be expected based on US epidemiological data.2 In addition, even though the presence of multiple coagulopathies in an individual is uncommon, most patients (87% of males and 74% of females) had multiple “286” codes subsequent to the diagnosis of hemophilia. For patients who had PT and aPTT test results, only 7% of females and approximately half of males showed results consistent with a diagnosis of congenital or acquired hemophilia (normal PT and prolonged aPTT). However, data regarding laboratory testing may be affected by potentially low reporting from patients with congenital hemophilia whose EHRs correspond to ages beyond childhood, as individuals who were diagnosed during infancy or childhood would not be expected to undergo additional PT or aPTT testing in response to bleeding symptoms experienced later in life. Although aPTT results could have been normalized by hemophilia treatment (therapy that replaces the missing clotting factor), such treatment was not reported for any of the females who had normal laboratory values. In addition, 36% of the males had a prescription for an anticoagulant, which would likely be avoided in most people with hemophilia and independently could account for the abnormal aPTT values. Evaluation of the section “Signs, Diseases and Symptoms” accompanying the broad NLP concept of “Pain” identified that the most frequent pain locations indicated were the chest and the abdomen. This contrasts with the typical pain locations among people with hemophilia, ie, a predominant presentation of joint pain;18 however, the observed pattern of pain may have been influenced by patient age, in addition to hemophilia and other identified comorbidities. After a detailed review of physician notes of randomly selected patients from the study cohort (3 females and 3 males), strong evidence of them having congenital hemophilia was found in only 3 out of 6 cases; the remaining cases seemed to more closely resemble acquired hemophilia (2 females) or factor FVIII or V deficiency (1 male).
These findings highlight potential challenges in investigating EHRs and other sources of big data,19 which may be encountered when investigating even relatively common disorders. For example, a review of chart notes from patients with type 2 diabetes found that, of those receiving an ICD-9-CM code, only 16% actually had type 2 diabetes.11,20 Because ICD-9-CM data are typically handled by a coder who enters codes based on the diagnostic labels assigned by the clinician and/or the pharmaceutical products used, the accuracy of information entered is frequently limited by multiple patient- and health care practitioner-related factors. Miscoding may also occur if a patient is not willing to provide information to the clinician or is unable to describe his/her symptoms.19 Potential variability among clinicians in terminology used, as well as shortcomings in compiling information from patient examinations, may also affect data accuracy.19 Additional sources of coding errors may include the coder’s level of experience, the potential for transcription errors, facility-specific coding procedures, and poor legibility in handwritten notes transcribed into the EHR.19 Importantly, even when the process is automated, errors associated with dictation using voice recognition software can occur due to the lack of punctuation, translation inaccuracies, and software editing oversights.21 Specifying sufficiently large vocabularies to recognize relevant terms, abbreviations, and acronyms used in clinical documents can also limit capture and interpretation of complex data.22
In some institutions, coding is required to be complete before the patient is discharged because of insurance billing requirements, and therefore, diagnostic codes may be entered based on incomplete information before a diagnosis is made.11,19 In cases of rare bleeding disorders, the variable language used to describe diagnoses, either during differential diagnosis or when confirmed through testing or consultation, may be confusing to hospital-based or primary-care-based coders or even some health care professionals. In addition, the descriptions in ICD-9-CM are often unclear (eg, extrinsic anticoagulants, circulating inhibitors, congenital hemophilia A-B-C vs acquired or autoimmune hemophilia).
Many physicians are resistant to using EHRs because they feel that technology interferes with their ability to properly care for patients or duplicates other treatment records maintained, and some are concerned that they are spending more time at the computer fulfilling administrative and billing requirements than interacting with patients.23,24 In a study, physicians reported that they spent 44% of their time in front of a computer and only 28% in direct patient care.24 To save time, some physicians may use auto-population features of EHR software to avoid or minimally use chart notes, as evidenced by large blocks of replicated or identical text and long text strings in many notes.23,25 In a survey of physicians at 2 affiliated academic centers, 90% of the participants indicated that they used the copy/paste function in daily progress notes.26 This method can introduce erroneous data or gaps in the medical records, and may have contributed to the over-identification of patients with congenital hemophilia in this study.27
The difficulties encountered with data accuracy in EHRs may be particularly apparent when investigating rare diseases, as missing or inaccurate data can be problematic when sample sizes are small.22 Another study that assessed hemophilia health system costs using records from the Department of Defense also encountered limitations associated with missing data; because of a lack of documentation for homecare management, analyses for treatment patterns yielded little information.28 This lack of accurate home therapy data may be a significant challenge in the context of hemophilia, as home therapy often is either not captured or is recorded retrospectively,29 resulting in an incomplete picture of hemophilia management. Currently, there are no regulatory requirements to evaluate the effectiveness and safety of EHR systems;25 however, the American Medical Informatics Association has proposed guiding principles for clinical data capture and documentation to support efficient, reliable, and high-quality acquisition of information for downstream uses such as policymaking, education, reimbursement, and research.30 Despite these limitations, EHR data are a potentially valuable source of clinical information because of the wide range of patient data that can be obtained from a large population. Further refinement of our analysis strategies will be critical in deriving greater value from this resource.
Study limitations
Multiple limitations of this analysis may have contributed to the difficulties observed in identifying a population of patients with congenital hemophilia. Some limitations are inherent in the analysis of an EHR database, including the potential for ICD-9-CM coding errors, inconsistent coding practices, and other inconsistencies across patient data sources. The incorporation of useful data from chart notes may also be limited by variable chart use among physicians in addition to the potential errors in NLP translation. Finally, the rarity of hemophilia, confusion over descriptions provided to support ICD-9-CM, and lack of consistent diagnosis observed in this study may have contributed to the false identification of patients with hemophilia and limited our ability to characterize the hemophilia patient population.
Conclusion
NLP-based strategies toward incorporating information from chart notes are a potentially valuable approach to obtain clinically important patient data. However, in this study, patient numbers and their gender, ICD-9-CM codes, laboratory test results, prescriptions received, and sites of pain were not consistently aligned with a diagnosis of congenital hemophilia. The seemingly large number of false positive identifications, particularly among female patients, suggests that ICD-9-CM coding alone may be insufficient to identify patient cohorts. A multimodal strategy incorporating a thorough analysis of physician notes in addition to ICD-9-CM codes may be an important approach toward improving data collection, which may be particularly useful in the context of rare diseases. The quality of data from multicenter EHRs is highly dependent on the knowledge of individuals supporting data coding and entry; therefore, the rarer the disorder, the less likely it is for the coding staff to have relevant experience. This information highlights the need for increased focus and training of health care professionals and coders to improve the quality of EHRs to support the “big data” initiatives that hold much promise for helping to improve medical care.
Acknowledgments
Writing assistance was provided by Anna Abt, PhD, and Shawn Keogan, PhD, of ETHOS Health Communications in Yardley, Pennsylvania, and was supported financially by Novo Nordisk Inc., Plainsboro, New Jersey, in compliance with international Good Publication Practice guidelines. We wish to thank Ekaterine Bakhtadze Bagci, Christina Stentoft Hoxer, Anders Krabbe, Snejana Krassova, and Lars Wilkinson for their review and inputs with regard to the preparation of this manuscript. This study was funded and sponsored by Novo Nordisk Inc. An abstract of this study was presented at the ISPOR 21st Annual International Meeting as a poster presentation with interim findings. The poster’s abstract is published in the ISPOR Scientific Presentations Database: https://www.ispor.org/ScientificPresentationsDatabase/Presentation/63564.
Disclosure
Michael Wang is a consultant for Novo Nordisk Inc. Anissa Cyhaniuk is a principal consultant of AC Analytical Solutions, LLC. David L Cooper and Neeraj N Iyer are employees of Novo Nordisk Inc. The authors report no other conflicts of interest in this work.
References
Bolton-Maggs PH, Pasi KJ. Haemophilias A and B. Lancet. 2003;361(9371):1801–1809. | ||
Centers for Disease Control and Prevention [homepage on the Internet]. UDC Data Reports. National Summary Report of UDC Activity: Patient Demographics (Hemophilia); 2011. Available from: https://www2a.cdc.gov/ncbddd/htcweb/UDC_Report/UDC_view1.asp?para1=NATION¶2=ALL¶3=&ScreenWidth=1440&ScreenHeight=900. Accessed October 23, 2015. | ||
Centers for Disease Control and Prevention. Hemophilia: Data and Statistics. Available from: http://www.cdc.gov/ncbddd/hemophilia/data.html. Accessed October 23, 2015. | ||
Mannucci PM, Tuddenham EG. The hemophilias—from royal genes to gene therapy. N Engl J Med. 2001;344(23):1773–1779. | ||
Sujansky WV. The benefits and challenges of an electronic medical record: much more than a “word-processed” patient chart. West J Med. 1998;169(3):176–183. | ||
Crosson JC, Ohman-Strickland PA, Hahn KA, et al. Electronic medical records and diabetes quality of care: results from a sample of family medicine practices. Ann Fam Med. 2007;5(3):209–215. | ||
Houser SH, Colquitt S, Clements K, Hart-Hester S. The impact of electronic health record usage on cancer registry systems in Alabama. Perspect Health Inf Manag. 2012;9:1f. | ||
Wood GC, Chu X, Manney C, et al. An electronic health record-enabled obesity database. BMC Med Inform Decis Mak. 2012;12:45. | ||
Denaxas SC, George J, Herrett E, et al. Data resource profile: cardiovascular disease research using linked bespoke studies and electronic health records (CALIBER). Int J Epidemiol. 2012;41(6):1625–1638. | ||
Tomines A, Readhead H, Readhead A, Teutsch S. Applications of electronic health information in public health: uses, opportunities & barriers. EGEMS (Wash DC). 2013;1(2):1019. | ||
Ramakrishnan N, Hanauer D, Keller B. Mining electronic health records. Computer. 2010;43(10):77–81. | ||
Royer JA, Hardin JW, McDermott S, et al. Use of state administrative data sources to study adolescents and young adults with rare conditions. J Gen Intern Med. 2014;29(Suppl 3):S732–S738. | ||
Jensen PB, Jensen LJ, Brunak S. Mining electronic health records: towards better research applications and clinical care. Nat Rev Genet. 2012;13(6):395–405. | ||
Soucie JM, McAlister S, McClellan A, Oakley M, Su Y. The universal data collection surveillance system for rare bleeding disorders. Am J Prev Med. 2010;38(4 Suppl):S475–S481. | ||
Soucie JM, Miller CH, Kelly FM, et al; Haemophilia Inhibitor Research Study Investigators. A study of prospective surveillance for inhibitors among persons with haemophilia in the United States. Haemophilia. 2014;20(2):230–237. | ||
Liao KP, Ananthakrishnan AN, Kumar V, et al. Methods to develop an electronic medical record phenotype algorithm to compare the risk of coronary artery disease across 3 chronic disease cohorts. PLoS One. 2015;10(8):e0136651. | ||
Gomez K, Bolton-Maggs P. Factor XI deficiency. Haemophilia. 2008;14(6):1183–1189. | ||
Riley RR, Witkop M, Hellman E, Akins S. Assessment and management of pain in haemophilia patients. Haemophilia. 2011;17(6):839–845. | ||
O’Malley KJ, Cook KF, Price MD, Wildes KR, Hurdle JF, Ashton CM. Measuring diagnoses: ICD code accuracy. Health Serv Res. 2005;40(5 Pt 2):1620–1639. | ||
Rhodes ET, Laffel LM, Gonzalez TV, Ludwig DS. Accuracy of administrative coding for type 2 diabetes in children, adolescents, and young adults. Diabetes Care. 2007;30(1):141–143. | ||
American Health Information Management Association. AHIMA e-HIM work group on speech recognition in the EHR; 2003. Available from: http://library.ahima.org/doc?oid=62537#.WZYQflGGPcs. Accessed August 26, 2015. | ||
Sanders CM, Saltzstein SL, Schultzel MM, Nguyen DH, Stafford HS, Sadler GR. Understanding the limits of large datasets. J Cancer Educ. 2012;27(4):664–669. | ||
Pollack R. Doctors, hospitals rethinking electronic medical records mandated by 2009 law; 2014. Available from: http://www.washington examiner.com/doctors-hospitals-rethinking-electronic-medical-records-mandated-by-2009-law/article/2554622. Accessed August 26, 2015. | ||
Hill RG Jr, Sears LM, Melanson SW. 4000 clicks: a productivity analysis of electronic medical records in a community hospital ED. Am J Emerg Med. 2013;31(11):1591–1594. | ||
Bowman S. Impact of electronic health record systems on information integrity: quality and safety implications. Perspect Health Inf Manag. 2013;10:1c. | ||
O’Donnell HC, Kaushal R, Barron Y, Callahan MA, Adelman RD, Siegler EL. Physicians’ attitudes towards copy and pasting in electronic note writing. J Gen Intern Med. 2009;24(1):63–68. | ||
Wrenn JO, Stein DM, Bakken S, Stetson PD. Quantifying clinical narrative redundancy in an electronic health record. J Am Med Inform Assoc. 2010;17(1):49–53. | ||
Armstrong EP, Malone DC, Krishnan S, Wessler MJ. Costs and utilization of hemophilia A and B patients with and without inhibitors. J Med Econ. 2014;17(11):798–802. | ||
Baker RI, Laurenson L, Winter M, Pritchard AM. The impact of information technology on haemophilia care. Haemophilia. 2004;10 Suppl 4:41–46. | ||
Cusack CM, Hripcsak G, Bloomrosen M, et al. The future state of clinical data capture and documentation: a report from AMIA’s 2011 Policy Meeting. J Am Med Inform Assoc. 2013;20(1):134–140. |
 © 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.