Back to Journals » Cancer Management and Research » Volume 12
Identification of BRMS1L as Metastasis Suppressing Gene in Esophageal Squamous Cell Carcinoma
Authors Zhou R , Tang X, Li L, Zhang F, Sun J, Ju C, Zhou Y, Liu R, Liang Y, Lv B, Zhang Z, Hu H, Lv XB
Received 28 September 2019
Accepted for publication 4 January 2020
Published 23 January 2020 Volume 2020:12 Pages 531—539
DOI https://doi.org/10.2147/CMAR.S232632
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Chien-Feng Li
Ruihao Zhou, 1, 2,* Xiaofeng Tang, 1,* Liping Li, 3,* Feifei Zhang, 1 Jun Sun, 1 Cheng Ju, 1, 4 Yan Zhou, 5 Renfeng Liu, 1, 4 Yiping Liang, 1 Bin Lv, 1, 4 Zhiping Zhang, 1, 4 Haiyan Hu, 5 Xiao-Bin Lv 1
1Jiangxi Key Laboratory of Cancer Metastasis and Precision Treatment, The Third Affiliated Hospital of Nanchang University, Nanchang 30008, People’s Republic of China; 2Department of Pain Management, West China Hospital, Sichuan University, Chengdu, Sichuan 610041, People’s Republic of China; 3Department of Clinical Laboratory, The Third Affiliated Hospital of Nanchang University, Nanchang 330008, People’s Republic of China; 4Department of Orthopedics, The Third Affiliated Hospital of Nanchang University,Nanchang 330008, People’s Republic of China; 5Department of Oncology,Shanghai Jiao Tong University Affiliated Sixth People’s Hospital of Shanghai, Shanghai 200233, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Xiao-Bin Lv
Jiangxi Key Laboratory of Cancer Metastasis and Precision Treatment, The Third Affiliated Hospital of Nanchang University, North 128 Xiangshan Road, Nanchang 30008, Jiangxi, People’s Republic of China
Email [email protected]
Haiyan Hu
Department of Oncology, Shanghai Jiao Tong University Affiliated Sixth People’s Hospital, No. 600 Yishan Road, Xuhui District, Shanghai City 200233, People’s Republic of China
Email [email protected]
Introduction: Breast cancer metastasis suppressor 1 like (BRMS1-like)was first reported to be a component of the Sin3-HDAC complex, but the role in the progression of cancers was largely unknown. Our previous study reported that BRMS1L promoted the metastasis of breast cancer through facilitating the recruitment of HDAC complex to the promoter FZD10, and hence suppressing the transcription of FZD10.
Methods: In this study, we detected the expression level of BRMS1L in esophageal squamous cell carcinoma (ESCC). The effect of BRMS1L in TE-1D (knockdown) and ECA-109 (overexpression) cell lines was explored by transwell assays, wound healing assays, and cell adhesion assays. Quantitative real‑time PCR, Western blot analysis, and luciferase assays were used to detect the interaction of the CBP/P300-BRMS1L-ITGA7 axis.
Results: In the present study, we found that knockdown of BRMS1L promoted the migration, invasion, and epithelial–mesenchymal transition (EMT). Conversely, overexpression of BRMS1L inhibited the migration and invasion of ESCC. Mechanistically, BRMS1L exerted their metastasis-suppressing role via transcriptionally repress ITGA7 expression. Moreover, we revealed that CBP/p 300 regulated the expression of BRMS1L and might be responsible for the down-regulation of BRMS1L in ESCC.
Conclusion: Collectively, we identified the role of CBP/p300-BRMS1L-ITGA7 axis in the metastasis of ESCC.
Keywords: BRMS1L, ESCC, EMT, ITGA7, CBP/P300, cell invasion and migration
Introduction
Esophageal cancer is one of the most aggressive cancers in the world and the sixth leading cause of cancer-related deaths.1 Histologically, the most common subtype of this cancer, ESCC, has a distinct geographic distribution variation with Asia having the highest incidence area.1,2 Once diagnosed, most ESCC patients have progressed to a late stage or metastasis, and the overall 5-year survival rate of ESCC patients is lower than 10%. Although findings from molecular biology studies have increased our general understanding of the pathogenesis of ESCC, the mechanism of ESCC metastasis and ideal biomarkers for clinical prognosis have not yet been fullyillustrated.1,3 Therefore, identifying the mechanism metastasis of ESCC will be crucial to improve the survival rate of ESCC patients.
Epithelial–mesenchymal transition (EMT) was an important molecular mechanism that promotes the metastasis of cancers.4,5 When EMT occurred in tumor cells, the intercellular junction disappeared and the morphology of cells changed from round shape to spindle. In the mean time, epithelial cell markers were down-regulated, and mesenchymal cell markers were up-regulated. During EMT, the migratory and invasive ability of tumor cells was significantly enhanced, leading to tumor metastasis.6 BRMS1L was first reported as a component in the histone deacetylase (HDAC)complex.7 Gong et al8 found that BRMS1L could mediate the directed recruitment of HDAC complexes into the promoter of FZD10, resulting in decreased levels of histone acetylation such as H3K9 in the FZD10 promoter region and inhibition of FZD10 expression. In breast cancer cells and tissues with highly metastatic potential, BRMS1L expression was significantly down-regulated, resulting in increased expression of FZD10 and promotion of breast cancer cell metastasis.
Integrins were transmembrane cell surface receptors composed of 18 α subunits and 8 β subunits.9 Integrins directly bound to components of the extracellular matrix (ECM) and provided the traction needed for cell movement and invasion. Studies have shown that the expression of integrin abnormalities was a hallmark of tumorigenesis and development.10 Abnormal expression of integrin allowed tumor cells to acquire migratory and invasive ability, alter intracellular signal transduction, and survive in other micro environments without triggering internal apoptosis mechanisms.11 Moreover, the abnormal expression of integrin could also lead to the occurrence of drug resistance in tumor cells.12 Integrin a7 (ITGA7) was a receptor for the ECM protein laminin and formed a heterodimer with integrin β1. Studies have shown ITGA7 was abnormally expressed in invasive gliomas and acted as a key functional receptor for GSC via activating AKT.13 Besides, Li et al14 found that circITGA7 inhibited proliferation and metastasis of CRC cells by inhibiting the Ras signaling pathway and promoting transcription of ITGA7, suggesting that circITGA7 was a potential target for CRC. In-depth study of integrin would help us reveal the molecular mechanism of tumor metastasis and provide new ideas for the clinical treatment of tumors.
In this study, we explored the role of BRMS1L in the progression of ESCC and demonstrated the mechanism of BRMS1L affecting ESCC metastasis. We further elucidated that BRMS1L suppressed ESCC metastasis via suppression of the ITGA7 expression and was transcriptionally regulated by CBP/P300.
Materials and Methods
Cell Culture
The human ESCC cell lines (TE-1D, KYSE-180, KYSE-520, ECA-109) were purchased from Shanghai Institute of Biochemistry and Cell Biology (Shanghai, China). The use of 293T cells gifted from Dr. Kang was approved by the research ethics committee of The Third Hospital, Nanchang university. All the cells were cultured in DMEM medium containing 10% fetal bovine serum (FBS) in the incubator with 5% CO2 at 37°C.
Plasmid Constructs
The promoter sequences of BRMS1L and ITGA7 were cloned into the PGL3-Basic vector for luciferase experiments. The full-length sequences of BRMS1L, CBP, P300 in frame with an HA tag in the N-terminal were inserted into the pcDNA3.1 vector to obtain the overexpression plasmid. The siRNA sequences of BRMS1L, ITGA7, CBP, P300 purchased from GenePharma Co. Ltd. (Suzhou, China). The oligonucleotide sequences targeting the above listed genes were listed in Table 1.
 |
Table 1 Primer Sequences Used in This Article |
Transwell Assay
105cells with serum-free DMEM medium were added to the top chamber of each well and 500 uL of DMEM containing 10% FBS was added to the bottom chamber. To assess cell invasion, 105 cells per chamber were added to the upper chamber coated matrigel. After 24 hrs of incubation, the unfiltered cells were removed with a cotton swab. Then, the filtered cells in the bottom of the membrane of top chamber were fixed with 4% paraformaldehyde, stained with 0.1% crystal violet solution, and counted under a microscope.
Cell Adhesion Assay
Cells were seeded to 24-well plates coated with matrigel-coated wells and incubated for 1 hr. Then, the wells were washed with PBS to remove the non-adherent cells, and the adherent cells were fixed, stained, and counted under a microscope.
Wound Healing Assays
The acellular area created by a 200-µL pipette tip, and the cells were cultured for 24 hrs. The progression of migration was observed and photographed at 0, 12, and 24 hrs after scratching.
Quantitative Real- Time PCR (qRT- PCR)
Total RNA was extracted with TRIzol reagent (Invitrogen, USA) according to the manufacturer’s instructions. cDNA was synthesized with PrimeScript™ RT reagent Kit with gDNA Eraser (TaKaRa, Kusatsu, Japan). Real-timePCR was performed with CFX Connect Real-time PCR system (BioRad) using SYBR Premix Ex Taq kit (TaKaRa). The above listed primers are listed in Table 1.
Western Blot Analysis
The procedure was conducted as previously described.8 The cells were washed three times with PBS and lysed in RIPA buffer with protease inhibitor on ice for 30 mins. The total protein extracts were separated by 10% SDS-PAGE,transferred to a PVDF membrane (Millipore, Bedford, MA, USA) and incubated overnight at 4°C with the corresponding antibody. Peroxidase conjugated secondary antibodies (anti-mouseand anti-rabbit) were used at room temperature for 1 hr, and then developed using an ECL Western Blotting Substrate.
Luciferase Assay
The promoter sequence of ITGA7 and BRMS1L were constructed into PGL3-Basicplasmids. The promoters and overexpression plasmids or siRNAs were co-transfected into 293T cells using Lipofectamine 2000 (Invitrogen), and pRL-TK vector (Promega) served as an internal control. A dual luciferase assay (Promega, USA) was performed 24 h after transfection.
Statistical Analysis
All of statistical analyses were performed using SPSS 22.0 software and GraphPad Prism 8.0. The p-values were calculated using a one-way analysis of variance (ANOVA). The P values <0.05 were considered to be statistically significant.
Results
Knockdown of BRMS1L Promoted the Migration and Invasion of ESCC
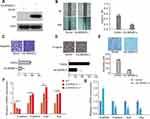
To investigate the role of BRMS1L in ESCC, we firstly examined the mRNA level of BRMS1L in ESCC cells (TE-1D, KYSE-180, KYSE-520, ECA-109) by qRT-PCR (Figure 1A). According to the differential expression level of BRMS1L in ESCC cells, we transfected BRMS1L siRNAs into TE-1D cells with higher endogenous BRMS1L level and the efficient knockdown of BRMS1L were validated by qRT-PCR(Figure 1B). Wound healing assay showed that silence of BRMS1L increased the healing ability in the scratched area of TE-1D cells (Figure 1C). Besides, silence of BRMS1L increased the migratory and invasive ability of TE-1D cells evaluated by transwell assay (Figure 1D and E). Finally, adhesion assay showed that knockdown of BRMS1L increased cell adhesion of ESCC cells (Figure 1F). Altogether, these results indicated that knockdown of BRMS1L improved the metastatic potential of ESCC cells.
BRMS1L Regulated the Migration and Invasion of ESCC via EMT Process
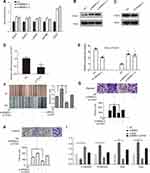
To further confirm the functional role of BRMS1L in ESCC, we performed a gain-of-function assay by transfecting HA-tagged BRMS1L into ECA-109 cells (Figure 2A). Contrary to knockdown of BRMS1L, overexpression of BRMS1L reduced the motility of ESCC cells evaluated by wound healing assay (Figure 2B). Besides, transwell assay results showed that overexpression of BRMS1L repressed the migration and invasion of ECA-109 cells (Figure 2C and D). Finally, adhesion assay showed that transfection of BRMS1L reduced the adhesive ability of ECA-109 cells (Figure 2E). EMT was an important molecular mechanism that correlated with the improvement of the metastatic capacity of tumorcells.4,5 We hence examined whether modulation of BRMS1L influenced the EMT of ESCC cells. Indeed, silence of BRMS1L increased N-cadherin, Snail and Slug mRNA level and decreased E-cadherin mRNA level (Figure 2F) in TE-1D cells. On the contrary, overexpression of BRMS1L obviously upregulated E-cadherin and downregulated N-cadherin, Snail and Slug (Figure 2G) in ECA-109 cells. These results indicated that BRMS1L regulated the EMT of ESCC cells.
BRMS1L Affected the Migration and Invasion of ESCC via Transcriptionally Suppressing ITGA7 Expression
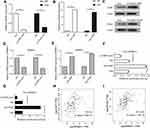
To explore the potential mechanisms by which BRMS1L repressed the migration, invasion, and EMT of ESCC cells, we analyzed the previous mRNA array results for differentially expressed genes related to the metastasis of cancercells.8 We selected SULF2, FOXC2, CDH23, KIF26B, and ITGA7 for examination using qRT-PCR.Among them, ITGA7 was the most prominently downregulated gene upon BRMS1L knockdown (Figure 3A). ITGA7was a marker of tumorigenesis and development, and upregulated ITGA7 increased the migratory and invasive ability of cancer cells. We hence focused on ITGA7 for further study. Western blot results showed that silence of BRMS1L increased the ITGA7 protein level (Figure 3B). On the contrary, overexpression of BRMS1L suppressed ITGA7 expression in protein level (Figure 3C). To further determine whether BRMS1L directly suppressed the transcription of ITGA7, we performed a luciferase reporter assay. A DNA fragment between −2000 and +200 relative to ITGA7 transcription start site (TSS) was amplified and inserted into a pGL3-Basicplasmid to obtain ITGA7 promoter reporter vector. Knockdown or overexpression of BRMS1L enhanced or reduced the luciferase activity of pGL3-ITGA7,respectively (Figure 3D), indicating that BRMS1L directly repressed the transcriptional activity of ITGA7 promoter. Since ITGA7 was a direct transcriptional target of BRMS1L, we explored whether ITGA7 mediated BRMS1L-exerted suppression of migration and invasion on ESCC cells. The rescue experiments showed that silence of ITGA7 (Figure 3E) could reverse the si-BRMS1Lexerted migratory and invasive promoting ability of ESCC cells (Figure 3F–H). Consistently, silence of ITGA7 also rescued BRMS1L silence exerted mRNA alterations of EMT marker (Figure 3I).
The Expression of BRMS1L Was Regulated by Transcription of CBP/P300
To explore the potential mechanisms by which BRMS1L was downregulated in ESCC cancer, we performed the bioinformatics analysis of the ENCODE project (https://www.encodeproject.org/), which showed that the promoter region of BRMS1L was enriched of many histone modification signals, such as H3K4Me3, and H3K27Ac signals. Furthermore, high-through ChIP-seqdata (UCSC) showed that p300 was enriched in the BRMS1L promoter. Therefore, we hypothesized that CBP/P300 might activate BRMS1L transcription as co-activators in ESCC.
To confirm our hypothesis, we knockdowned CBP and P300 expression using siRNAs-pool containing three different siRNAs (Figure 4A). The qRT-PCR results showed that both silencing CBP and P300 reduced BRMS1L expression (Figure 4B). Conversely, overexpression of CBP and P300 (Figure 4C) increased BRMS1L expression (Figure 4D and E). Then, we employed luciferase assay to investigate whether CBP and P300 could activate the transcriptional activity of BRMS1L promoter. Consistent with the qRT-PCR results above, knockdown or overexpression of CBP/P300 significantly decreased or increased the luciferase activity of pGL3-BRMS1L,respectively (Figure 4F and G). Most importantly, based on the public database (http://gepia.cancer-pku.cn/), CBP and p300 expression levels were positively correlated with BRMS1L in ESCC samples (Figure 4H and I). The above results suggested that CBP/P300 regulated the transcription of BRMS1L and might be responsible for the downregulation of BRMS1L in ESCC cells.
Discussion
The global incidence and mortality of ESCC are 8th and 6th, respectively.1,15 With the development of diagnosis and treatment, the five-year survival rate of patients with ESCC has been significantly improved. But the mortality rate of patients with metastasis was still very high, mainly because the mechanism of cancer metastasis was not completely demonstrated yet. Therefore, elucidating the mechanism of metastasis was important to reduce cancer-associated deaths of ESCCpatients.16,17
Accumulating evidence has shown that many metastasis-related genes were capable of participating in regulating many processes of cancer metastasis, including migration, invasion, lymph node metastasis, and distant organcolonization.18 For example, BRMS1 was shown to be closely related to metastasis suppression, which was a core component of the Sin3/HDAC complex. Interestingly, BRMS1 and BRMS1L were both important components of the Sin3A complex. Furthermore, BRMS1L shared homology with the yeast and mammalian Sds3 proteins as BRMS1, raising the possibility that they might serve similar functions. Downregulation of BRMS1 was closely related to metastasis of breast cancer,19 lung cancer,20 melanoma,20 and glioma.21 Compared with BRMS1, the function of BRMS1L in cancer development was little known. Our previous study8 revealed that BRMS1L inhibited the migratory and invasive ability of breast cancer cell via EMT process. Besides, BRMS1L mediated the directed accumulation of HDAC complexes into the promoter of FZD10, resulting in decreased levels of histone acetylation such as H3K9 in the FZD10 promoter region and inhibition of FZD10 expression. Some recent studies have found that BRMS1L was a novel target of p53 and might be a promising prognostic marker for breast cancer and glioma.22,23 And in epithelial ovarian cancer (EOC), BRMS1L regulated migration and invasion of EOC cells via Wnt/β-catenin signalpathway.24 In this study, we found that BRMS1L repressed ESCC metastasis via suppressing of the ITGA7 expression and BRMS1L itself was transcriptionally regulated by CBP/P300.
Different members of the integrin family played different roles in the tumor and performed different functions. Haas et al found that silence of ITGA7 inhibited growth and survival of glioblastoma stem cells (GSC) and was a potential therapeutic avenue and functional marker for glioblastoma.13 Ming et al found that ITGA7 was a functional surface marker in the cancer stem cells of ESCC. Functional studies have shown that ITGA7+and ITGA7 overexpressing cells exhibited enhanced stem cell characteristics, including increased expression of stemness-associated genes and induction of EMT characteristics, significantly promoting cell migration, invasion, and anti-chemotherapy capabilities.25 In this study, we demonstrated that BRMS1L suppressed ESCC metastasis via suppressing of the ITGA7 expression. Consistent with previously mentioned, our results showed that silence of BRMS1L reversed the increased invasive, and migratory ability of ESCC cells mediated by ITGA7 silencing.
CBP and p300 have a high sequence similarity and are often referred to as CBP/p300.26 As an important macromolecular protein in lysine acetyltransferases (KATs), p300 was involved in the transcriptional activation of multiple genes. Previous reports showed that p300 was capable of acetylating H3K14, H3K18, H4K12 and regulating the transcription of a series of genes.27 Liang et al revealed that CBP/P300 was capable of binging to linc00460 promoter by transcriptional co-activator through histone acetylation.28 Besides, researchers found that the expression of lncRNA HULC was upregulated by CREB via binding to the core promoter ofHULC,29 whereas CBP/P300 could interact with CREB directly. In our study, we revealed that CBP/P300 bound to BRMS1L promoter and transcriptional activated BRMS1L. CBP/p300 could also transcriptionally regulate many key tumor-associated proteins, such as NF-κB, β-catenin, myc, and p53.30,31 Thus, we might think that the above tumor-associatedproteins alteration lead to abnormal gene expression such as BRMS1L through CBP/P300 function, resulting in a differential expression of BRMS1L in ESCC with different metastatic potentials.
In summary, we demonstrated the mechanism of BRMS1L affecting ESCC metastasis. We further elucidated that BRMS1L suppressed ESCC metastasis via suppressing of the ITGA7 expression and was regulated by the transcription of CBP/P300.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Chen J, Kwong DL, Cao T, et al. Esophageal squamous cell carcinoma (ESCC): advance in genomics and molecular genetics. Dis Esophagus. 2015;28(1):84–89. doi:10.1111/dote.2015.28.issue-1
2. Kano M, Seki N, Kikkawa N, et al. miR-145, miR-133a and miR-133b:tumor-suppressive miRNAs target FSCN1 in esophageal squamous cell carcinoma. Int J Cancer. 2010;127(12):2804–2814. doi:10.1002/ijc.v127:12
3. Sugihara H, Ishimoto T, Miyake K, et al. Noncoding RNA expression aberration is associated with cancer progression and is a potential biomarker in esophageal squamous cell carcinoma. Int J Mol Sci. 2015;16(11):27824–27834. doi:10.3390/ijms161126060
4. De Craene B, Berx G. Regulatory networks defining EMT during cancer initiation and progression. Nat Rev Cancer. 2013;13(2):97–110. doi:10.1038/nrc3447
5. Lu W, Kang Y. Epithelial-mesenchymal plasticity in cancer progression and metastasis. Dev Cell. 2019;49(3):361–374. doi:10.1016/j.devcel.2019.04.010
6. Mitra A, Mishra L, Li S. EMT, CTCs and CSCs in tumor relapse and drug-resistance. Oncotarget. 2015;6(13):10697–10711. doi:10.18632/oncotarget.v6i13
7. Nikolaev AY, Papanikolaou NA, Li M, et al. Identification of a novelBRMS1-homologue protein p40 as a component of the mSin3A/p33(ING1b)/HDAC1 deacetylase complex. Biochem Biophys Res Commun. 2004;323(4):1216–1222.
8. Gong C, Qu S, Lv XB, et al. BRMS1L suppresses breast cancer metastasis by inducing epigenetic silence of FZD10. Nat Commun. 2014;5:5406. doi:10.1038/ncomms6406
9. Manninen A, Varjosalo M. A proteomics view onintegrin-mediatedadhesions. Proteomics. 2017;17(3–4). doi:10.1002/pmic.201600022
10. Desgrosellier JS, Cheresh DA. Integrins in cancer: biological implications and therapeutic opportunities. Nat Rev Cancer. 2010;10(1):9–22. doi:10.1038/nrc2748
11. Winograd-Katz SE, Fassler R, Geiger B, et al. The integrin adhesome: from genes and proteins to human disease. Nat Rev Mol Cell Biol. 2014;15(4):273–288. doi:10.1038/nrm3769
12. Eke I, Dickreuter E, Cordes N. Enhanced radiosensitivity of head and neck squamous cell carcinoma cells by beta1 integrin inhibition. Radiother Oncol. 2012;104(2):235–242. doi:10.1016/j.radonc.2012.05.009
13. Haas TL, Sciuto MR, Brunetto L, et al. Integrin alpha7 is a functional marker and potential therapeutic target inglioblastoma. Cell Stem Cell. 2017;21(1):35–50.e9. doi:10.1016/j.stem.2017.04.009
14. Li X, Wang J, Zhang C, et al. Circular RNA circITGA7 inhibits colorectal cancer growth and metastasis by modulating the Ras pathway and upregulating transcription of its host gene ITGA7. J Pathol. 2018;246(2):166–179. doi:10.1002/path.2018.246.issue-2
15. Sakai NS, Samia-Aly E, Barbera M, et al. A review of the current understanding and clinical utility of miRNAs in esophageal cancer. Semin Cancer Biol. 2013;23(6, Part B):512–521. doi:10.1016/j.semcancer.2013.08.005
16. Steeg PS, Theodorescu D. Metastasis: a therapeutic target for cancer. Nat Clin Pract Oncol. 2008;5(4):206–219. doi:10.1038/ncponc1066
17. Fidler IJ, Kripke ML. The challenge of targeting metastasis. Cancer Metastasis Rev. 2015;34(4):635–641. doi:10.1007/s10555-015-9586-9
18. Smith SC, Theodorescu D. Learning therapeutic lessons from metastasis suppressor proteins. Nat Rev Cancer. 2009;9(4):253–264. doi:10.1038/nrc2594
19. Zhang Z, Yamashita H, Toyama T, et al. Reduced expression of the breast cancer metastasis suppressor 1 mRNA is correlated with poor progress in breast cancer. Clin Cancer Res. 2006;12(21):6410–6414. doi:10.1158/1078-0432.CCR-06-1347
20. Smith PW, Liu Y, Siefert SA, et al. Breast cancer metastasis suppressor 1 (BRMS1) suppresses metastasis and correlates with improved patient survival innon-small cell lung cancer. Cancer Lett. 2009;276(2):196–203. doi:10.1016/j.canlet.2008.11.024
21. Mei P, Bai J, Shi M, et al. BRMS1 suppresses glioma progression by regulating invasion, migration and adhesion of glioma cells. PLoS One. 2014;9(5):e98544. doi:10.1371/journal.pone.0098544
22. Koyama R, Tamura M, Nakagaki T, et al. Identification and characterization of a metastatic suppressor BRMS1L as a target gene of p53. Cancer Sci. 2017;108(12):2413–2421. doi:10.1111/cas.2017.108.issue-12
23. Lv J, Yang H, Wang X, et al. Decreased BRMS1L expression is correlated with glioma grade and predicts poor survival in glioblastoma via an invasive phenotype. Cancer Biomark. 2018;22(2):311–316. doi:10.3233/CBM-171019
24. Cao P, Zhao S, Sun Z, et al. BRMS1L suppresses ovarian cancer metastasis via inhibition of the beta-catenin-wnt pathway. Exp Cell Res. 2018;371(1):214–221. doi:10.1016/j.yexcr.2018.08.013
25. Ming XY, Fu L, Zhang LY, et al. Integrin α7 is a functional cancer stem cell surface marker in oesophageal squamous cell carcinoma. Nat Commun. 2016;7:13568. doi:10.1038/ncomms13568
26. Arany Z, Sellers WR, Livingston DM, et al. E1A-associatedp300 and CREB-associated CBP belong to a conserved family ofcoactivators. Cell. 1994;77(6):799–800. doi:10.1016/0092-8674(94)90127-9
27. Kaypee S, Sudarshan D, Shanmugam MK, et al. Aberrant lysine acetylation in tumorigenesis: implications in the development oftherapeutics. Pharmacol Ther. 2016;162:98–119. doi:10.1016/j.pharmthera.2016.01.011
28. Liang Y, Wu Y, Chen X, et al. A novel long noncoding RNA linc00460up-regulated by CBP/P300 promotes carcinogenesis in esophageal squamous cell carcinoma. Biosci Rep. 2017;37(5):BSR20171019. doi:10.1042/BSR20171019
29. Wang J, Liu X, Wu H, et al. CREB up-regulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer. Nucleic Acids Res. 2010;38(16):5366–5383. doi:10.1093/nar/gkq285
30. Di Martile M, Del Bufalo D, Trisciuoglio D. The multifaceted role of lysine acetylation in cancer: prognostic biomarker and therapeutictarget. Oncotarget. 2016;7(34):55789–55810. doi:10.18632/oncotarget.v7i34
31. Chen LF, Greene WC. Regulation of distinct biological activities of the NF-kappaB transcription factor complex by acetylation. J Mol Med (Berl). 2003;81(9):549–557. doi:10.1007/s00109-003-0469-0
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.