Back to Journals » International Journal of General Medicine » Volume 15
Identification and Validation of Hub Genes with Poor Prognosis in Hepatocellular Carcinoma by Integrated Bioinformatical Analysis
Authors Guo J , Li W, Cheng L , Gao X
Received 21 December 2021
Accepted for publication 1 April 2022
Published 11 April 2022 Volume 2022:15 Pages 3933—3941
DOI https://doi.org/10.2147/IJGM.S353708
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Scott Fraser
Jiang Guo,1 Wei Li,2 Long Cheng,1 Xuesong Gao3
1Department of Interventional Oncology, Beijing Ditan Hospital, Capital Medical University, Beijing, 100015, People’s Republic of China; 2Center of Liver Disease, Beijing Ditan Hospital, Capital Medical University, Beijing, 100015, People’s Republic of China; 3Department of General Medicine, Beijing Ditan Hospital, Capital Medical University, Beijing, 100015, People’s Republic of China
Correspondence: Xuesong Gao, Department of General Medicine, Beijing Ditan Hospital, Capital Medical University, Beijing, 100015, People’s Republic of China, Tel +86 13718689825, Fax +861084322146, Email [email protected]
Background: Hepatocellular carcinoma (HCC) is the reason for the world’s second largest cancer-related death. It is clinically valuable to study the molecular mechanisms of HCC occurrence and development for formulating more effective diagnosis and treatment strategies.
Methods: The five microarray data sets GSE45267, GSE101685, GSE84402, GSE62232 and GSE45267 were downloaded from Gene Expression Omnibus (GEO) database, including 165 HCC tissues and 73 normal tissues. Differential expressed genes (DEGs) between HCC tissues and normal tissues were determined by GEO2R. Gene ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG) and the protein–protein interaction network (PPI) network analysis were employed to identify DEGs and to evaluate the clinical significance in prognosis of HCC.
Results: A total of 152 genes differentially expressed in HCC tissues and normal tissues were identified. GO and KEGG functional enrichment analysis revealed that 39 up-regulated genes were mainly enriched in mitosis, cell cycle and oocyte meiosis, while those down-regulated genes (113) were concentrated in exogenous drug catabolism and the metabolism of cytochrome P450 on exogenous drugs. Totally, 19 hub genes were chosen by PPI network and module analysis and verified by The Cancer Genome Atlas (TCGA) database. Finally, 8 hub genes were selected, including CDK1, CYP2C8, CCNB1, AURKA, CYP2C9, BUB1B, MAD2L1 and TTK, which were associated with the overall survival rate of HCC patients.
Conclusion: This study presented eight target genes connected to the prognosis of HCC patients. Those mainly exists in cell cycle and drug catabolism, which may be latent targets for clinical treatment.
Keywords: hepatocellular carcinoma, bioinformatic analysis, differentially expressed genes, prognostic
Introduction
Hepatocellular carcinoma (HCC) is the sixth most familiar malignancy and cause of the world’s second largest cancer-related death.1 It is estimated that there are about 841,000 new cases of liver cancer worldwide in 2018, with 782,000 deaths, causing serious economic burden.2 After a great progress has been made in the prevention, diagnosis and treatment of HCC, a significant decrease occurred in the number of viral cases and this was compensated by the progressive expansion of non-viral cases. The improved overall survival was observed due to the wider use of semi-annual surveillance, expanding the proportion of tumors that qualified for curative treatments, and the improved outcome of loco-regional treatments.3 In recent years, more evidence shows that the occurrence and development of HCC is relevant to the abnormal expression of various oncogenes and the inactivation of tumor suppressor genes; these genes have also been increasingly used in the early diagnosis and treatment.4,5 Thus, it’s crucial to study the target molecules and molecular mechanisms of HCC occurrence and development for formulating more effective diagnosis and treatment strategies.
Gene mutation, cell environment and other factors are bound up with the occurrence, development and metastasis of tumors.6 A unique immune response profile of the liver microenvironment where CD4+CD25+ Foxp3 regulatory T-cells (Tregs) also play a crucial role through their immunosuppressive role in HCC development and progression.7 During the slow development of HCC, a large number of genomes changed, and accumulated into the phenotypic changes of hepatocytes, resulting in cell intermediates and multiple monoclones, which eventually evolved into HCC.8,9 High-throughput technology and gene chip have become quicker methods to identify differential expressed genes (DEGs) and functional pathways in the occurrence and development of different diseases over recent years. Hence, bioinformatics has become an essential way to assess gene expression profile. Gene Expression Omnibus (GEO) and The Cancer Genome Atlas (TCGA) are the two largest public resource gene expression databases in the world. Molecular variations in HCC can be found through descriptive data generated by different microarray platforms.10–12 Pathway analysis shows that the changes of some important cellular signaling pathways are related to the main pathogenic mechanism.13 A few hub genes were classified as pivotal regulators of HCC metastasis through the protein-protein interaction (PPI) network.14 But owing to the heterogeneity, small sample size and different statistical methods of independent studies, the number of identified functional genes is not nearly enough for verifying HCC pathogenesis, and the most remarkable maladjusted genes in previous studies are not consistent.
Here, in this study, five microarray data sets GSE45267, GSE101685, GSE84402, GSE62232 and GSE45267 downloaded from NCBI-GEO included 165 HCC tissues and 73 normal tissues, and DEGs between them were identified by GEO2R. Gene ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG) and PPI network analysis were used to identify DEGs, and to evaluate the clinical significance of DEGs in the prognosis of HCC. Eight hub genes were identified as latent molecular targets for exploring new HCC intervention strategies. This work will further explore the development of HCC at molecular level.
Materials and Methods
Microarray Data
The gene expression profiles GSE45267, GSE101685, GSE84402, GSE62232 and GSE45267 were downloaded from GPL570 platform originate from the comprehensive database of gene expression of National Center for Biotechnology Information (NCBI) (https://www.ncbi.nlm.nih.gov/geo/). According to the labeling information on the platform, the probes were transformed to corresponding gene symbols. After normalizing the gene expression data, log2 transformation was performed in limma R package (http://www.bioconductor.org/).15
Recognition of DEGs
GEO2R (http://www.ncbi.nlm.nih.gov/geo/geo2r) was employed to define DEGs between HCC and normal tissues. GEO2R is a software for differential analysis of expression microarray based on GEO database, which allows users to compare two or more data sets in GEO series to select different genes expressed under different experimental conditions. FDR and Benjamini-Hochberg control the possibility of large “false positive”. Probe sets without corresponding gene symbols were filtered out, and the average value of log2FC was used when one gene corresponds to multiple probe sets. LogFC (fold change) >2 and P-value <0.01 were statistically remarkable.
Enrichment of DEGs by GO and KEGG Pathways
To better assess the selected DEGs, GO and KEGG pathway enrichment analysis were performed by David online tool (http://david.ncifcrf.gov) (Version 6.7); a P-value of <0.01 was statistically marked. David is a website for gene annotation, visualization and integrated discovery, which offers a comprehensive set of gene and protein annotation information to extract biological information. GO is the main bioinformatics tool for annotating genes and analyzing their biological functions.16 The GO mainly describes genes and their products from three aspects: molecular function (MF), biological process (BP) and cellular component (CC). KEGG is a database resource used for the advanced functions and utilities of biological systems from genomic and molecular information.17
Construction of PPI Network and Analysis of Selected Modules
To understand the relationship between DEGs, PPI network is established by using STRING online database (http://string-db.org) and Module analysis, and a confidence score of ≥0.7 is considered to be remarkable. STRING is a database for searching the interplay between known protein and predicting protein.18 Furthermore, PPI network is visualized by using Cytoscape (Version 3.4.0) (http://www.cytoscape.org/) (degree cut-off = 2, node score cut-off = 0.2, k-core = 2, and max depth = 100). By using the potential information of DEGs in GO and KEGG analysis module, genes with intersection of GO and KEGG ≥10 were selected as hub genes.
Verification of Hub Genes
The expression of hub genes in HCC and their relationship with prognosis were obtained from TCGA database. The expression of hub genes in HCC was analyzed by online database Oncomine, and Kaplan-Meier curve was employed to assess the hub genes and the overall survival and disease free survival rate of HCC patients on the online platform of cBioPortal (http://www.cbioportal.org) and GEPIA2 (http://gepia2.cancer-pku.cn/#index).
Results
Recognition of DEGs in HCC
The gene chips selected this time are GSE45267, GSE101685, GSE84402 and GSE62232 of GPL570 platform. GSE45267 contains 46 HCC and 41 normal samples. GSE101685 consists of 24 HCC and 8 normal samples. GSE84402 includes 14 HCC and 14 normal samples. GSE62232 involves 81 HCC and 10 normal samples. With P<0.01 and [logFC]>2 as the screening criteria, we extracted 308, 459, 463 and 292 DEGs from GSE45267, GSE101685, GSE84402 and GSE62232 respectively, and found 152 overlapping DEGs (Figure 1). Finally, it was confirmed that 39 genes were up-regulated and 113 genes were down-regulated in HCC (Table 1).
 |
Table 1 All 152 Differentially Expressed Genes (DEGs) |
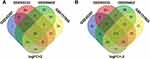 |
Figure 1 Venn diagrams of DEGs in the four datasets. (A) Up-regulated DEGs; (B) down-regulated DEGs. |
GO Functional Annotation of DEGs
To explore the biological functions of 152 DEGs, we used online database DAVID 6.7 to do GO analysis, and divided them into three functional groups: molecular function (MF), cellular component (CC) and biological process (BP). According to the survey, the up-regulated genes in HCC were concentrated in BP and CC. BP included mitosis, cell division, G2/M transition of mitotic cell cycle, protein ubiquitination involved in ubiquitin-dependent protein catabolism, complex-dependent catabolism, mitotic spindle tissue and the regulation of spindle microtubules attachment to centromere, etc. CC mainly includes mitotic spindle, cytoplasm, motile, condensed chromosome centromere, nucleus, spindle, spindle microtubules, spindle central region, centrosome, spindle pole and so on. In MF, the genes up-regulated in HCC mainly showed calmodulin-binding protein kinase, binding protein, microtubule-binding region, protein serine/threonine kinase activity, etc. (Table S1). The down-regulated genes in HCC are mainly concentrated in BP, including redox process, cyclooxygenase P450 pathway, exogenous drug catabolism process, cell response to cadmium ion, cell response to zinc ion, negative growth regulation, metabolic process of heterotypic biomass, etc. In CC, there are organelle membrane, extracellular region, blood particles, high density lipoprotein and endoplasmic reticulum membrane. The MF mainly includes oxygen binding, heme binding site, oxidoreductase activity, iron ion binding, monooxygenase activity and so on (Table S2).
KEGG Pathway Enrichment Analysis of DEGs
KEGG is a database resource used for the advanced functions and biological systems of large-scale molecular data sets generated from high-throughput experimental techniques. The up-regulated DEGs are accumulated on cell cycle, oocyte meiosis and p53 signaling pathway, while those down-regulated are rich in retinol metabolism, drug metabolism-cytochrome P450, chemical carcinogenesis, metabolism of cytochrome P450 to exogenous drugs, metabolic pathway, etc. (Table S3).
Construction of PPI Network and Analysis of Selected Modules
A PPI network of 152 DEGs (39 up-regulated and 113 down-regulated genes) was established by STRING. There are 7 up-regulated genes: RRM2, CCNB1, CDK1, AURKA, BUB1B, MAD2L1, TTK, and 25 down-regulated genes: CYP1A2, MT1G, MT1E, CNDP1, CYP26A1, CYP2A6, NAT2, AADAT, CYP2C8, ADH4, RDH16, AKR1D1, CYP8B1, MT1M, CYP2B6, CYP3A4, MT1F, KMO, ADH1C, CYP2C9, ADH1B, IDO2, MT1X, MT1H, TDO2 (Figure 2A). With Module analysis, the genes obtained above are intersected with the genes obtained from GO analysis and KEGG analysis (degree cut-off = 2, node score cut-off = 0.2, k-core = 2, and max depth = 100); 4 clusters of genes are obtained (Figure 2B–E, Table S4). At last, 19 hub genes are selected, that is, CCNB1, CDK1, AURKA, BUB1B, MAD2L1, TTK, CYP1A2, MT1E, CYP26A1, CYP2A6, NAT2, CYP2C8, MT1M, CYP2B6, CYP3A4, MT1F, CYP2C9, MT1X and MT1H.
Expression of Hub Genes in HCC and Its Relationship with Prognosis of Patients
We obtained the expression of hub genes in HCC and their relationship with prognosis from TCGA database. It was found that only 8 genes, CDK1, CYP2C8, CCNB1, AURKA, CYP2C9, BUB1B, MAD2L1 and TTK, were significantly different in HCC expression (P<0.05) (Figure 3), among which the up-regulated genes CDK1, CCNB1, AURKA, BUB1B, MAD2L1, TTK and down-regulated gene CYP2C8, CYP2C9 were interrelated to the overall survival (Figure 4) and disease-free survival (Figure S1) of patients.
Discussion
HCC, accounting for 90% of liver cancers, is one of the most familiar and deadly malignancies in those tumors.19 At present, the molecular mechanism of HCC is not completely clear. Precise diagnosis and prognosis evaluation are still enormous challenges in treatment. Hence, discovering new functional genes is helpful to assess the pathogenesis of HCC and enhance its diagnosis and prognosis. In order to identify new functional genes involved in HCC, we screened the differential genes tied to HCC prognosis through bioinformatics analysis.
Totally, 152 genes differentially expressed in HCC and normal tissues were determined. GO and KEGG enrichment analysis found that 39 up-regulated genes were concentrated on mitosis, cell cycle and oocyte meiosis, while those down-regulated (113) were rich in exogenous drug catabolism, drug metabolism-cytochrome P450, and the metabolism of cytochrome P450 on exogenous drugs. Cell cycle and mitosis crucial in tumor occurrence and development, and are also one of the main targets of tumor drug therapy.20–22 In the treatment of anti-tumor drugs, the gradual emergence of drug resistance is a crucial reason leading to poor prognosis of patients. Thus, at first, we selected 19 hub genes by PPI network and Module analysis; then, we verified them by TCGA database, and finally discovered 8 hub genes. Those genes, CDK1, CYP2C8, CCNB1, AURKA, CYP2C9, BUB1B, MAD2L1 and TTK, are relevant to the overall survival rate of HCC patients. These genes may be promising biomarkers for HCC prognosis.
Most of these genes participate in the process of cell cycle and cell mitosis, such as CDK1, CCNB1, BUB1B, AURKA, MAD2L1 and TTK. CDK1 can form a complex with cyclin B1 (CCNB1) and cyclin B2 (CCNB2), regulate G2/M phase of mammalian cell cycle, and exert important effects in mitosis.23 Recent studies have suggested that CDK1 suppresses the proliferation, migration and invasion of HCC cells.24,25 AURKA, also described as Aurora kinase A, is a vital serine/threonine kinase responsible for regulating cell mitosis, which has a major part to play in centrosome replication and separation, spindle assembly, maturation, chromosome arrangement, spindle assembly checkpoint and cytoplasm division.26 The increased expression of AURKA may bring about chromosome instability, transformation and centrosome amplification in mammalian cells, and promote the carcinogenesis of c-myc.27,28 AURKA is over-expressed in many tumors, especially in liver cancer.29,30 BUB1B is an integral part to spindle assembly checkpoint, including MAD1, MAD2, MAD3/Bub1b, Mps1, Bub1 and Bub3.31 In one study, Qiu et al32 verified the carcinogenicity of BUB1B in HCC, including promoting the progression of G0/G1 cell cycle, inhibiting the apoptosis of HCC cells and causing poor prognosis. MAD2L1 is effective in spindle mitosis, and the imbalance of MAD2L1 brings about chromosome instability and chromosome aneuploidy.33 At the moment, many evidences demonstrated that MAD2L1 can be employed as a biomarker of poor prognosis of lung cancer,34,35 and MAD2L1 gene variation can result in the decrease of shuttle checkpoint function and increase the risk of lung cancer.36 The function of MAD2L1 in HCC needs to be verified by more clinical samples. In a basic experiment, miR-200c-5p suppressed HCC proliferation and metastasis by inhibiting MAD2L1.37 The human monopolar spindle 1 (hMps1/TTK) gene (NM_003318) locates on chromosome 6q13-q21 and encodes a bis-serine/threonine and tyrosine protein kinase.38 TTK is necessary for mitotic checkpoint and wrong chromosome attachment.39 Elevated TTK level will result in centrosome amplification, over-activation of spindle assembly checkpoint and chromosome instability, thus promoting tumor occurrence.40 According to Liang et al, TTK gene is a potential therapeutic target for anti-sorafenib resistance in HCC patients.41 CYP2C8 and CYP2C9 are members of CYP450 gene family, both of which are down-regulated in HCC tissues; the low level of CYP2C8 has been verified to be related to poor overall survival rate and disease-free survival rate.42 Overexpression of CYP2C8 suppressed the proliferation, clonality, migration, invasion and cell cycle of HCC cells via PI3K/Akt/p27 kip1 Axis. Members of CYP2C subfamily participate in the metabolism of many endogenous and exogenous substances, including drug metabolism. It may be the reason that low expression of CYP2C8 and CYP2C9 were associated with poor survival rate of HCC in part.43
All these theories are in conformity with our results. These genes may play a central role in the occurrence, development and drug resistance of HCC. There are some limitations. One of the main problems is that we did not collect clinical samples for verification. What’s more, we only analyzed the overall survival rate of patients, but did not evaluate the disease-free survival rate, clinical characteristics and other indicators. Others, after the functional enrichment analysis of DEGs, we ignored that in signaling pathway, so the results could be further explored.
Conclusion
To summarize, we identified 152 DEGs with differential expression in HCC through bioinformatics analysis, and verified them with TCGA database, and identified 8 hub genes associated with the prognosis of patients. These hub genes are accumulated on cell cycle and drug catabolism, which may be latent targets for HCC, and supply further strategies for clinical diagnosis, accurate treatment and prognosis analysis.
Ethics Statement
This study was approved by the Institutional Review Board of Beijing Ditan Hospital (2019-044-001).
Funding
There is no funding to report.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Xie S, Jiang X, Zhang J, et al. Identification of significant gene and pathways involved in HBV related hepatocellular carcinoma by bioinformatics analysis. Peer J. 2019;7:e7408. doi:10.7717/peerj.7408
2. Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.21492
3. Bucci L, Garuti F, Lenzi B, et al. The evolutionary scenario of hepatocellular carcinoma in Italy: an update. Liver Int. 2017;37(2):259–270. doi:10.1111/liv.13204
4. Xing T, Yan T, Zhou Q. Identification of key candidate genes and pathways in hepatocellular carcinoma by integrated bioinformatical analysis. Exp Ther Med. 2018;15(6):4932–4942. doi:10.3892/etm.2018.6075
5. Zhou L, Du Y, Kong L, Zhang X, Chen Q. Identification of molecular target genes and key pathways in hepatocellular carcinoma by bioinformatics analysis. Onco Targets Ther. 2018;11:1861–1869. doi:10.2147/OTT.S156737
6. Sanyal AJ, Yoon SK, Lencioni R. The etiology of hepatocellular carcinoma and consequences for treatment. Oncologist. 2010;15(Suppl 4):14–22. doi:10.1634/theoncologist.2010-S4-14
7. Granito A, Muratori L, Lalanne C, et al. Hepatocellular carcinoma in viral and autoimmune liver diseases: role of CD4+ CD25+ Foxp3+ regulatory T cells in the immune microenvironment. World J Gastroenterol. 2021;27(22):2994–3009. doi:10.3748/wjg.v27.i22.2994
8. Wang XW, Hussain SP, Huo TI, et al. Molecular pathogenesis of human hepatocellular carcinoma. Toxicology. 2002;181–182:43–47. doi:10.1016/S0300-483X(02)00253-6
9. Rinninella E, Zocco MA, De Gaetano A, et al. From small nodule to overt HCC: a multistep process of carcinogenesis as seen during surveillance. Eur Rev Med Pharmacol Sci. 2012;16(9):1292–1294.
10. Brazma A, Parkinson H, Sarkans U, et al. ArrayExpress–a public repository for microarray gene expression data at the EBI. Nucleic Acids Res. 2003;31(1):68–71. doi:10.1093/nar/gkg091
11. Schulze K, Imbeaud S, Letouze E, et al. Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nat Genet. 2015;47(5):505–511. doi:10.1038/ng.3252
12. Tung EK, Mak CK, Fatima S, et al. Clinicopathological and prognostic significance of serum and tissue Dickkopf-1 levels in human hepatocellular carcinoma. Liver Int. 2011;31(10):1494–1504. doi:10.1111/j.1478-3231.2011.02597.x
13. Whittaker S, Marais R, Zhu AX. The role of signaling pathways in the development and treatment of hepatocellular carcinoma. Oncogene. 2010;29(36):4989–5005. doi:10.1038/onc.2010.236
14. Wang T, Yang N, Liang C, et al. Detecting protein-protein interaction based on protein fragment complementation assay. Curr Protein Pept Sci. 2020;21(6):598–610. doi:10.2174/1389203721666200213102829
15. Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi:10.1093/nar/gkv007
16. Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The gene ontology consortium. Nat Genet. 2000;25(1):25–29. doi:10.1038/75556
17. Kanehisa M. The KEGG database. Novartis Found Symp. 2002;247:
18. Szklarczyk D, Franceschini A, Wyder S, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43(Database issue):D447–452. doi:10.1093/nar/gku1003
19. Llovet JM, Kelley RK, Villanueva A, et al. Hepatocellular carcinoma. Nat Rev Dis Primers. 2021;7(1):6. doi:10.1038/s41572-020-00240-3
20. Saffar H, Okhovat H, Arbabsoleymani S, et al. The utility of phosphohistone H3 in inter observer variability of mitotic count in meningioma, is there any benefit? Asian Pac J Cancer Prev. 2021;22(7):2049–2052. doi:10.31557/APJCP.2021.22.7.2049
21. Wang Z, Yang B, Zhang M, et al. lncRNA epigenetic landscape analysis identifies EPIC1 as an oncogenic lncRNA that interacts with MYC and promotes cell-cycle progression in cancer. Cancer Cell. 2018;33(4):706–720 e709. doi:10.1016/j.ccell.2018.03.006
22. Zhang H, Christensen CL, Dries R, et al. CDK7 inhibition potentiates genome instability triggering anti-tumor immunity in small cell lung cancer. Cancer Cell. 2020;37(1):37–54 e39. doi:10.1016/j.ccell.2019.11.003
23. Zou Y, Ruan S, Jin L, et al. CDK1, CCNB1, and CCNB2 are prognostic biomarkers and correlated with immune infiltration in hepatocellular carcinoma. Med Sci Monit. 2020;26(e925289). doi:10.12659/MSM.925289
24. Li L, Huang K, Zhao H, Chen B, Ye Q, Yue J. CDK1-PLK1/SGOL2/ANLN pathway mediating abnormal cell division in cell cycle may be a critical process in hepatocellular carcinoma. Cell Cycle. 2020;19(10):1236–1252. doi:10.1080/15384101.2020.1749471
25. Jin J, Xu H, Li W, Xu X, Liu H, Wei F. LINC00346 acts as a competing endogenous RNA regulating development of hepatocellular carcinoma via modulating CDK1/CCNB1 axis. Front Bioeng Biotechnol. 2020;8:54. doi:10.3389/fbioe.2020.00054
26. Vader G, Lens SM. The Aurora kinase family in cell division and cancer. Biochim Biophys Acta. 2008;1786(1):60–72. doi:10.1016/j.bbcan.2008.07.003
27. Zhou H, Kuang J, Zhong L, et al. Tumour amplified kinase STK15/BTAK induces centrosome amplification, aneuploidy and transformation. Nat Genet. 1998;20(2):189–193. doi:10.1038/2496
28. Lu L, Han H, Tian Y, et al. Aurora kinase A mediates c-Myc’s oncogenic effects in hepatocellular carcinoma. Mol Carcinog. 2015;54(11):1467–1479. doi:10.1002/mc.22223
29. Chou CH, Chou YE, Chuang CY, Yang SF, Lin CW. Combined effect of genetic polymorphisms of AURKA and environmental factors on oral cancer development in Taiwan. PLoS One. 2017;12(2):e0171583. doi:10.1371/journal.pone.0171583
30. Simon EP, Freije CA, Farber BA, et al. Transcriptomic characterization of fibrolamellar hepatocellular carcinoma. Proc Natl Acad Sci USA. 2015;112(44):E5916–5925. doi:10.1073/pnas.1424894112
31. Fu X, Chen G, Cai ZD, et al. Overexpression of BUB1B contributes to progression of prostate cancer and predicts poor outcome in patients with prostate cancer. Onco Targets Ther. 2016;9:2211–2220. doi:10.2147/OTT.S101994
32. Qiu J, Zhang S, Wang P, et al. BUB1B promotes hepatocellular carcinoma progression via activation of the mTORC1 signaling pathway. Cancer Med. 2020;9(21):8159–8172. doi:10.1002/cam4.3411
33. Li Y, Benezra R. Identification of a human mitotic checkpoint gene: hsMAD2. Science. 1996;274(5285):246–248. doi:10.1126/science.274.5285.246
34. Shi YX, Zhu T, Zou T, et al. Prognostic and predictive values of CDK1 and MAD2L1 in lung adenocarcinoma. Oncotarget. 2016;7(51):85235–85243. doi:10.18632/oncotarget.13252
35. Wei R, Wang Z, Zhang Y, et al. Bioinformatic analysis revealing mitotic spindle assembly regulated NDC80 and MAD2L1 as prognostic biomarkers in non-small cell lung cancer development. BMC Med Genomics. 2020;13(1):112. doi:10.1186/s12920-020-00762-5
36. Guo Y, Zhang X, Yang M, et al. Functional evaluation of missense variations in the human MAD1L1 and MAD2L1 genes and their impact on susceptibility to lung cancer. J Med Genet. 2010;47(9):616–622. doi:10.1136/jmg.2009.074252
37. Li Y, Bai W, Zhang J. MiR-200c-5p suppresses proliferation and metastasis of human hepatocellular carcinoma (HCC) via suppressing MAD2L1. Biomed Pharmacother. 2017;92:1038–1044. doi:10.1016/j.biopha.2017.05.092
38. Liu X, Liao W, Yuan Q, Ou Y, Huang J. TTK activates Akt and promotes proliferation and migration of hepatocellular carcinoma cells. Oncotarget. 2015;6(33):34309–34320. doi:10.18632/oncotarget.5295
39. Liu X, Winey M. The MPS1 family of protein kinases. Annu Rev Biochem. 2012;81:561–585. doi:10.1146/annurev-biochem-061611-090435
40. Zhang L, Shi R, He C, et al. Oncogenic B-Raf(V600E) abrogates the AKT/B-Raf/Mps1 interaction in melanoma cells. Cancer Lett. 2013;337(1):125–132. doi:10.1016/j.canlet.2013.05.029
41. Liang XD, Dai YC, Li ZY, et al. Expression and function analysis of mitotic checkpoint genes identifies TTK as a potential therapeutic target for human hepatocellular carcinoma. PLoS One. 2014;9(6):e97739. doi:10.1371/journal.pone.0097739
42. Ren X, Ji Y, Jiang X, Qi X. Downregulation of CYP2A6 and CYP2C8 in tumor tissues is linked to worse overall survival and recurrence-free survival from hepatocellular carcinoma. Biomed Res Int. 2018;2018:5859415. doi:10.1155/2018/5859415
43. Zhou X, Li TM, Luo JZ, et al. CYP2C8 suppress proliferation, migration, invasion and sorafenib resistance of hepatocellular carcinoma via PI3K/Akt/p27(kip1) axis. J Hepatocell Carcinoma. 2021;8:1323–1338. doi:10.2147/JHC.S335425
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.



