Back to Journals » International Journal of General Medicine » Volume 14
Identification and Construction of a Predictive Immune-Related lncRNA Signature Model for Melanoma
Received 26 September 2021
Accepted for publication 25 November 2021
Published 1 December 2021 Volume 2021:14 Pages 9227—9235
DOI https://doi.org/10.2147/IJGM.S340025
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Scott Fraser
Fang-Wei Li, Sheng-Kang Luo
Department of Plastic and Reconstructive Surgery, Guangdong Second Provincial General Hospital, Guangzhou City, Guangdong Province, 510317, People’s Republic of China
Correspondence: Sheng-Kang Luo
Department of Plastic and Reconstructive Surgery, Guangdong Second Provincial General Hospital, 466 Middle Xin Gang Road, Guangzhou City, Guangdong Province, 510317, People’s Republic of China
Tel +8615626045022
Email [email protected]
Objective: The occurrence and development mechanisms of melanoma are related to immunity and lncRNAs. Therefore, it is necessary to systematically explore immune-related lncRNA profiles to help improve the prognosis of melanoma.
Methods: We integrated immune-related lncRNAs and the basic clinical information of melanoma patients in the TCGA dataset. Immune-associated lncRNAs were selected by differential expression screening and enriched for analysis. After univariate and multivariate Cox regression analyses, a new prognostic indicator based on immune-associated lncRNAs was established.
Results: Overall, differentially expressed immune-related lncRNAs were significantly associated with clinical outcomes in patients with melanoma. A prognostic model was then established based on 14 immune-associated lncRNAs (LRRC8C-DT, AC021188.1, MALINC1, CCR5AS, EIF2AK3-DT, AC022306.2, AC242842.1, AL034376.1, AL662844.4, AC009065.3, AC099811.3, AC125807.2, SPINT1-AS1 and AC009495.2). Melanoma patients in the high-risk group had worse overall survival than those in the low-risk group. The AUC of the risk score was 0.786.
Conclusion: This study identified several clinically significant immune-related lncRNAs and established a relevant prognostic model, which provided a molecular analysis of immunity in melanoma and potential prognostic lncRNAs for melanoma.
Keywords: lncRNA, immune-related, model, overall survival, melanoma
Introduction
Cutaneous melanoma is a tumor characterized by the abnormal proliferation of epidermal melanocytes that are genetically altered by the interaction of genetic, physical, and environmental factors.1,2 Melanoma is the most invasive form of skin cancer. Risk factors for melanoma can be divided into internal and external factors and include light skin color, UV exposure, nevi, genetic predisposition, and a family history of melanoma.3–6 In 2019, there were an estimated 95,830 cases of skin melanoma and 7000 deaths from skin melanoma in the United States.7 Depending on the features of the tumor (location, stage, and genetic profile), the therapeutic options may include surgical resection, chemotherapy, radiotherapy, photodynamic therapy (PDT), immunotherapy, or targeted therapy.8 The development of targeted therapies (such as BRAF and MEK inhibitors) and immunotherapies (such as anti-PD-1 antibodies alone or in combination with anti-CTLA-4 antibodies) has revolutionized the systemic treatment of advanced melanoma, and these new drugs have become pretreatments recommended by many international guidelines for melanoma treatment.9 The crosstalk between tumor and immune cells significantly affects tumor invasion, clinical response and treatment outcome.10 The immune system can detect and destroy tumor cells through immunosurveillance or play a tumor-promoting role via suppressed immune activity and enhanced anti-inflammatory responses; the immune environment varies greatly from one individual to another, and thus, the prognosis and response to treatment also vary among individuals.11 Currently, no prognostic markers for melanoma have been widely recognized, and it is of great significance to study the immune molecular mechanism underlying melanoma and to discover new immune checkpoints.
Many molecules, especially long noncoding RNAs (lncRNAs), play an important role in the progression of melanoma. For example, studies showed that HOTAIR was overexpressed in metastatic tissues, which indicated that HOTAIR could promote the ability of melanoma cells to migrate and invade and that lncRNAs may be involved in the metastasis of melanoma.12,13 In in vitro studies, knocking out the UCA1 or MALAT-1 lncRNA could reduce the migration of melanoma cells, which indicates that the increased expression of the UCA1 and MALAT-1 lncRNAs might be related to melanoma metastasis.14 Li et al found that abnormally upregulated expression of a new lncRNA, BANCR, was involved in the proliferation of melanoma cells in vitro and in vivo.15 However, there are still few studies on the application of immune-related lncRNAs in melanoma.
This study identified immune-related lncRNAs in melanoma and established a model related to melanoma survival based on these immune-related lncRNAs. This study provides a reference for future molecular diagnosis and treatment.
Materials and Methods
Acquisition and Analysis of Melanoma Expression Data
We downloaded the RNA sequencing (RNA-seq) expression and clinical data of melanoma specimens from The Cancer Genome Atlas (TCGA) database (https://cancergenome.nih.gov/). Moreover, immune-related genes were retrieved from immunological gene sets on the official gene set enrichment analysis (GSEA) website. Immune-related lncRNAs with a P value less than 0.05 and a coefficient greater than 0.4 were identified through correlation analysis between immune-related genes and lncRNAs.
Cox Regression Analysis of the Immune-Associated lncRNA Signature
The combination of clinical survival data and immune-related lncRNA expression data was used to identify prognostic immune-associated lncRNAs by univariate Cox regression analysis. Multivariate regression analysis was also performed to calculate the risk score. The formula of the risk score was as follows: Risk score = coefficient1 * Expressiongene1 + … + coefficientN * ExpressiongeneN. Melanoma specimens were categorized into high- and low-risk groups based on the median risk score.
Independent Prognostic Analysis in Melanoma Patients
Clinical characteristics, including age, sex, stage, T stage, M stage and N stage, and the risk score were chosen as independent prognostic risk variables according to univariate and multivariate Cox regression analyses. In addition, principal component analysis (PCA) is a dimensionality reduction method often used in image processing. PCA was utilized to display all genes, immune-related lncRNAs, and high- and low-risk score-associated genes.
Statistical Analysis
R software 3.6.0 (ATandT Labs Research - Software Tools, RRID:SCR_002937, https://www.r-project.org/) was used for all the statistical analyses involving differential expression, Kaplan-Meier curves, Cox regression analysis, forest plots and PCA. A difference of P<0.05 was considered statistically significant.
Results
Differentially Expressed lncRNAs in Melanoma Patients
The clinical materials and lncRNA expression data of both normal specimens and melanoma specimens were collected from TCGA database. Additionally, the relationship between immune gene expression and lncRNA expression in melanoma patients was determined, and 6359 immune-associated lncRNAs with P < 0.05 and coefficient > 0.4 were extracted. There were 6351 positively regulated immune-related lncRNAs and 8 negatively regulated immune-related lncRNAs (see Supplementary Table 1). Interestingly, the former was much higher than the latter. There were a total of 180 females and 290 males included in the study, with a mean age at diagnosis of 58.22 years. A total of 248 patients survived, and 222 patients died.
Construction of a Model for Melanoma Patients
From the univariate Cox regression analysis, 48 prognostic immune-associated lncRNAs with P < 0.01 were identified (see Supplementary Table 2). Furthermore, 14 independent prognostic immune-related lncRNAs (Table 1) were chosen to construct a survival model based on the multivariate Cox regression analysis: risk score = 0.554 * AL662844.4 + 0.257 * AC009065.3 + (−0.295) * LRRC8C-DT + 0.308 * AC099811.3 + (−0.243) * AC021188.1 + (−0.767) * MALINC1 + 0.254 * AC125807.2 + 0.386 * SPINT1-AS1 + (−0.437) * CCR5AS + (−0.568) * EIF2AK3-DT + (−0.342) * AC022306.2 + 0.349 * AC009495.2 + (−0.334) * AC242842.1 + (−0.081) * AL034376.1. As suggested by the results, LRRC8C-DT, AC021188.1, MALINC1, CCR5AS, EIF2AK3-DT, AC022306.2, AC242842.1 and AL034376.1 had a positive impact on the overall survival of melanoma patients, whereas AL662844.4, AC009065.3, AC099811.3, AC125807.2, SPINT1-AS1 and AC009495.2 had a negative impact on the overall survival of melanoma patients. On the basis of the median risk score value, the melanoma samples were divided into high- and low-risk groups. The high-risk group had a lower overall survival than the low-risk group (Figure 1). Figures show the distribution of the risk score, overall survival status and heatmap of the expression of the fourteen immune-related lncRNAs in melanoma patients (Figure 2).
 |
Table 1 Independent Prognostic Risk Immune-Related lncRNAs |
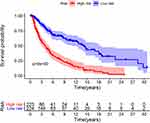 |
Figure 1 Kaplan-Meier overall survival curves of the risk stratification groups. The tick marks on the curve represent the censored subjects. The number of patients at risk is listed below the curve. |
Prognostic Signature of the Risk Score Combined with Clinical Variables
To assess whether the risk score predicts the prognosis of melanoma independently from other clinicopathological features, we performed a Cox regression analysis. The results of the univariate analyses showed a statistically significant relationship between age, stage, T-stage, N-stage, risk score and survival outcome (P < 0.001) (Figure 3A and Table 2). Multifactorial analyses showed that age, T stage, N stage, and risk score were independent prognostic factors for melanoma patients (P < 0.001) (Figure 3B and Table 2). To assess the sensitivity and specificity of the risk score in predicting the prognosis of melanoma patients, we performed receiver operating characteristic analysis. The area under the ROC curve of risk scores was 0.786, which was larger than the areas under the ROC curves of other clinical variables (Figure 4). Finally, the PCA results are presented in Figure 5. There were no significant differences between the high-risk and low-risk groups in terms of the expression of all genes (Figure 5A) or immune-related lncRNAs (Figure 5B). There was a significant difference between the two risk groups in the expression of the fourteen immune-related lncRNAs (Figure 5C) used in the prognostic model. In conclusion, these results indicate that the immune-related lncRNA signature identified above is an independent prognostic factor of melanoma.
 |
Table 2 Univariate and Multivariate Cox Proportional Hazards Regression Analysis of the 14-lncRNA Signature and Clinical Risk Factors in the Entire Dataset |
 |
Figure 3 Univariate and multivariate Cox regression analyses of clinical factors associated with overall survival. (A) Univariate Cox regression analyses. (B) Multivariate Cox regression analyses. |
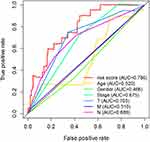 |
Figure 4 Receiver operating characteristic (ROC) curve. The ROC curve of the risk score had the largest AUC of 0.786 compared with the other clinical variables. |
 |
Figure 5 Principal component analysis. PCA of all genes (A), immune-related lncRNAs (B), and the two risk groups (C). |
Discussion
Cutaneous melanoma is the most common malignant skin carcinoma and has a high incidence. For patients with early-stage (stage I–IIIB) melanoma, surgery is the primary treatment, while immunotherapy has played an active role in treating unresectable or metastatic tumors.8 However, similar to the problems encountered in many tumor treatments, immunotherapy has various immunosuppressive mechanisms, the immune environment varies greatly from one individual to another, the prognosis and response to treatment vary among individuals, and only a small percentage of patients benefit from immunotherapy.11,16 Hence, in-depth exploration of the immunomolecular treatment of melanoma will provide new treatment and diagnosis options. For example, SNHG5 promoted melanoma development by inhibiting miR-26a-5p expression and inducing TRPC3 expression, suggesting the potential of SNHG5 as a novel target therapy for melanoma.17 In this study, we identified fourteen immune-related lncRNAs and used these lncRNAs to build a prognostic model. The AUC of the risk score was higher than that of other clinical variables.
Recent sequencing discoveries have demonstrated that there are various mechanisms underlying lncRNA expression in cells, ranging from gene expression to protein translation and stability.18 LncRNAs have been proven to participate in different processes of melanoma, such as progression and metastasis. Fourteen immune-related lncRNAs, LRRC8C-DT, AC021188.1, MALINC1, CCR5AS, EIF2AK3-DT, AC022306.2, AC242842.1, AL034376.1, AL662844.4, AC009065.3, AC099811.3, AC125807.2, SPINT1-AS1 and AC009495.2, were shown to have a great influence on melanoma. To investigate whether the immune-associated lncRNA signature can be an independent prognostic factor for cutaneous melanoma, we performed univariate and multivariate Cox analyses, which showed that the lncRNA signature is an independent prognostic factor for cutaneous melanoma independent of clinicopathological features. ROC curves and PCA proved the accuracy of the prognostic model.
For melanoma, which is a tumor with a poor prognosis, the establishment of an effective prognostic model and the exploration of its mechanism of action can improve the outcome of treatment. The expression levels of LRRC8C−DT were positively correlated with the abundances of M0 and M2 macrophages but negatively correlated with the abundance of CD8+ T cells.19 M1 macrophages exhibit proinflammatory activity and play an antitumor role, while M2 macrophages exhibit anti-inflammatory activity and promote tumor growth. It has been shown that the antitumor effects of immune checkpoint inhibitors can be enhanced by repolarizing M2 macrophages into M1 macrophages, and CD8 T cells located at the melanoma tumor invasive margin can predict the clinical response to PD-1 blockade therapy and are positively correlated with the response to pembrolizumab.20,21 AC021188.1 has been found to be associated with prognosis and was included in 5-disease prognostic signature lncRNAs in head and neck squamous cell carcinoma (HNSCC).22,23 Bida et al revealed the effect of the lncRNA MA-linc1 on cell cycle progression and cancer growth and found that its high expression was correlated with lower survival in breast cancer and lung cancer patients, and this might be related to the inhibition of MA-linc1-enhanced anti-mitotic drug paclitaxel-induced apoptotic cell death.24 CCR5 expression sustains myeloid-derived suppressor cell (MDSC) suppression activities, intratumoral Treg infiltration, and melanoma tumor growth and is highly attractive as a means to quench or eliminate unconstrained tumor cell growth.25,26 A study also showed that the expression of CCR5AS could affect HIV infection and disease progression. For example, inhibiting CCR5AS expression decreased the infection of CD4+ T cells in vitro.27,28 AC242842.1 is a pyroptosis-related and immune-related lncRNA signature that plays a key role in melanoma and can also be used for predicting prognosis.5,29 AL662844.4 is an autophagy-related lncRNA that plays a key role in bladder cancer.30 AC125807.2 is a potential prognostic biomarker in lung adenocarcinoma.31,32 LncRNAs such as AC009495.2 can distinguish acute myeloid leukemia types and significantly change the behavior of acute myeloid leukemia cells.33,34 Additionally, SPINT1-AS1 plays a role in esophageal squamous cell carcinoma,35 breast cancer,36 renal clear cell carcinoma and other tumors.37
The advantages of this study include that it was a TCGA-based study that included a large sample size and adjusted for the patient’s clinical and demographic characteristics. However, there was a limitation in this study: we did not provide information on experimental data for deep verification, and we will improve this aspect of the study in the future.
Conclusion
This study identified several clinically significant immune-related lncRNAs based on data from cutaneous melanoma patients in TCGA database and established a relevant prognostic model. This signature provides new potential biomarkers for prognosis and immunotherapy selection in cutaneous melanoma.
Abbreviations
lncRNAs, long noncoding RNAs; RNA-seq, RNA sequencing; TCGA, The Cancer Genome Atlas; GSEA, gene set enrichment analysis; AUC, area under the curve; PCA, principal component analysis; ROC, receiver operating characteristic.
Data Sharing Statement
The datasets supporting the conclusions of this article are available in TCGA database. (https://cancergenome.nih.gov/).
Ethics Approval and Informed Consent
This study was exempt from ethical review and approval from the Institutional Review Board of Guangdong Second Provincial General Hospital.
Acknowledgments
We acknowledge TCGA database for providing their platforms and contributors for uploading their meaningful datasets.
Author Contributions
FWL and SKL conceived and designed the study. FWL analyzed the data and drew the figures. All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Funding
There is no funding to report.
Disclosure
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Slominski A, Wortsman J, Carlson AJ, Matsuoka LY, Balch CM, Mihm MC. Malignant melanoma. Arch Pathol Lab Med. 2001;125(10):1295–1306. doi:10.5858/2001-125-1295-MM
2. Bastian BC. The molecular pathology of melanoma: an integrated taxonomy of melanocytic neoplasia. Annu Rev Pathol. 2014;9:239–271. doi:10.1146/annurev-pathol-012513-104658
3. Pastor-Tomás N, Martínez-Franco A, Bañuls J, et al. Risk factors for the development of a second melanoma in patients with cutaneous melanoma. J Eur Acad Dermatol Venereol. 2020;34(10):2295–2302. doi:10.1111/jdv.16341
4. Raimondi S, Suppa M, Gandini S. Melanoma epidemiology and sun exposure. Acta Derm Venereol. 2020;100(11):adv00136. doi:10.2340/00015555-3491
5. Xie J, Li H, Chen L, et al. A novel pyroptosis-related lncRNA signature for predicting the prognosis of skin cutaneous melanoma. Int J Gen Med. 2021;14:6517–6527. doi:10.2147/IJGM.S335396
6. Müller C, Wendt J, Rauscher S, et al. Risk factors of subsequent primary melanomas in Austria. JAMA Dermatol. 2019;155(2):188–195. doi:10.1001/jamadermatol.2018.4645
7. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7–34. doi:10.3322/caac.21551
8. Davis LE, Shalin SC, Tackett AJ. Current state of melanoma diagnosis and treatment. Cancer Biol Ther. 2019;20(11):1366–1379. doi:10.1080/15384047.2019.1640032
9. Kandolf Sekulovic L, Guo J, Agarwala S, et al. Access to innovative medicines for metastatic melanoma worldwide: Melanoma World Society and European Association of Dermato-oncology survey in 34 countries. Eur J Cancer. 2018;104:201–209. doi:10.1016/j.ejca.2018.09.013
10. Margolin K. Introduction to the role of the immune system in melanoma. Hematol Oncol Clin North Am. 2014;28(3):537–558. doi:10.1016/j.hoc.2014.02.005
11. Attrill GH, Ferguson PM, Palendira U, Long GV, Wilmott JS, Scolyer RA. The tumour immune landscape and its implications in cutaneous melanoma. Pigment Cell Melanoma Res. 2021;34(3):529–549. doi:10.1111/pcmr.12926
12. Tang L, Zhang W, Su B, Yu B. Long noncoding RNA HOTAIR is associated with motility, invasion, and metastatic potential of metastatic melanoma. Biomed Res Int. 2013;2013:251098. doi:10.1155/2013/251098
13. Cantile M, Scognamiglio G, Marra L, et al. HOTAIR role in melanoma progression and its identification in the blood of patients with advanced disease. J Cell Physiol. 2017;232(12):3422–3432. doi:10.1002/jcp.25789
14. Tian Y, Zhang X, Hao Y, Fang Z, He Y. Potential roles of abnormally expressed long noncoding RNA UCA1 and Malat-1 in metastasis of melanoma. Melanoma Res. 2014;24(4):335–341. doi:10.1097/CMR.0000000000000080
15. Li R, Zhang L, Jia L, et al. Long non-coding RNA BANCR promotes proliferation in malignant melanoma by regulating MAPK pathway activation. PLoS One. 2014;9(6):e100893. doi:10.1371/journal.pone.0100893
16. Curran MA, Montalvo W, Yagita H, Allison JP. PD-1 and CTLA-4 combination blockade expands infiltrating T cells and reduces regulatory T and myeloid cells within B16 melanoma tumors. Proc Natl Acad Sci U S A. 2010;107(9):4275–4280. doi:10.1073/pnas.0915174107
17. Gao J, Zeng K, Liu Y, Gao L, Liu L. LncRNA SNHG5 promotes growth and invasion in melanoma by regulating the miR-26a-5p/TRPC3 pathway. Onco Targets Ther. 2019;12:169–179. doi:10.2147/OTT.S184078
18. Schmitz SU, Grote P, Herrmann BG. Mechanisms of long noncoding RNA function in development and disease. Cell Mol Life Sci. 2016;73(13):2491–2509. doi:10.1007/s00018-016-2174-5
19. Qiu Y, Li H, Xie J, Qiao X, Wu J. Identification of ABCC5 among ATP-binding cassette transporter family as a new biomarker for hepatocellular carcinoma based on bioinformatics analysis. Int J Gen Med. 2021;14:7235–7246. doi:10.2147/IJGM.S333904
20. Wang H, Yang L, Wang D, Zhang Q, Zhang L. Pro-tumor activities of macrophages in the progression of melanoma. Hum Vaccin Immunother. 2017;13(7):1556–1562. doi:10.1080/21645515.2017.1312043
21. Choo YW, Kang M, Kim HY, et al. M1 macrophage-derived nanovesicles potentiate the anticancer efficacy of immune checkpoint inhibitors. ACS Nano. 2018;12(9):8977–8993. doi:10.1021/acsnano.8b02446
22. Wang Q, Yang W, Peng W, Qian X, Zhang M, Wang T. Integrative analysis of DNA methylation data and transcriptome data identified a DNA methylation-dysregulated four-LncRNA signature for predicting prognosis in head and neck squamous cell carcinoma. Front Cell Dev Biol. 2021;9:666349. doi:10.3389/fcell.2021.666349
23. Liu G, Zheng J, Zhuang L, et al. A prognostic 5-lncRNA expression signature for head and neck squamous cell carcinoma. Sci Rep. 2018;8(1):15250. doi:10.1038/s41598-018-33642-1
24. Bida O, Gidoni M, Ideses D, Efroni S, Ginsberg D. A novel mitosis-associated lncRNA, MA-linc1, is required for cell cycle progression and sensitizes cancer cells to Paclitaxel. Oncotarget. 2015;6(29):27880–27890. doi:10.18632/oncotarget.4944
25. Jacquelot N, Duong CPM, Belz GT, Zitvogel L. Targeting chemokines and chemokine receptors in melanoma and other cancers. Front Immunol. 2018;9:2480. doi:10.3389/fimmu.2018.02480
26. Weber R, Riester Z, Hüser L, et al. IL-6 regulates CCR5 expression and immunosuppressive capacity of MDSC in murine melanoma. J Immunother Cancer. 2020;8(2):e000949. doi:10.1136/jitc-2020-000949
27. Kulkarni S, Lied A, Kulkarni V, et al. CCR5AS lncRNA variation differentially regulates CCR5, influencing HIV disease outcome. Nat Immunol. 2019;20(7):824–834. doi:10.1038/s41590-019-0406-1
28. Zou S. Variation of CCR5AS lncRNA enhances HIV-1 infection through regulation of CCR5 expression. Viral Immunol. 2020;33(8):536–538. doi:10.1089/vim.2020.0034
29. Xiao B, Liu L, Li A, et al. Identification and validation of immune-related lncRNA prognostic signatures for melanoma. Immun Inflamm Dis. 2021;9(3):1044–1054. doi:10.1002/iid3.468
30. Wan J, Guo C, Fang H, Xu Z, Hu Y, Luo Y. Autophagy-related long non-coding RNA is a prognostic indicator for bladder cancer. Front Oncol. 2021;11:647236. doi:10.3389/fonc.2021.647236
31. Hou J, Yao C. Potential prognostic biomarkers of lung adenocarcinoma based on bioinformatic analysis. Biomed Res Int. 2021;2021:8859996. doi:10.1155/2021/8859996
32. Chen L, Ren Z, Cai Y. Construction and analysis of survival-associated competing endogenous RNA network in lung adenocarcinoma. Biomed Res Int. 2021;2021:4093426. doi:10.1155/2021/4093426
33. Zimta AA, Tomuleasa C, Sahnoune I, Calin GA, Berindan-Neagoe I. Long non-coding RNAs in myeloid malignancies. Front Oncol. 2019;9:1048. doi:10.3389/fonc.2019.01048
34. Affinito O, Pane K, Smaldone G, et al. lncRNAs-mRNAs co-expression network underlying childhood B-cell acute lymphoblastic leukaemia: a pilot study. Cancers. 2020;12(9):2489. doi:10.3390/cancers12092489
35. Shen FF, Pan Y, Yang HJ, et al. Decreased expression of SPINT1-AS1 and SPINT1 mRNA might be independent unfavorable prognostic indicators in esophageal squamous cell carcinoma. Onco Targets Ther. 2019;12:4755–4763. doi:10.2147/OTT.S206448
36. Zhou T, Lin K, Nie J, et al. LncRNA SPINT1-AS1 promotes breast cancer proliferation and metastasis by sponging let-7 a/b/i-5p. Pathol Res Pract. 2021;217:153268. doi:10.1016/j.prp.2020.153268
37. Qi-Dong X, Yang X, Lu JL, et al. Development and validation of a nine-redox-related long noncoding RNA signature in renal clear cell carcinoma. Oxid Med Cell Longev. 2020;2020:6634247. doi:10.1155/2020/6634247
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.

