Back to Journals » Cancer Management and Research » Volume 10
Human leukocyte antigen gene polymorphisms are associated with systemic inflammation in hepatitis B virus-related hepatocellular carcinoma
Authors Wu XL, Li ZY, Bi XY, Zhao H, Zhao JJ, Zhou JG, Han Y, Huang Z, Zhang YF, Cai JQ
Received 7 March 2018
Accepted for publication 7 May 2018
Published 31 July 2018 Volume 2018:10 Pages 2315—2324
DOI https://doi.org/10.2147/CMAR.S167574
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Lu-Zhe Sun
Xiao-long Wu,1,* Zhi-yu Li,1,* Xin-yu Bi,1 Hong Zhao,1 Jian-jun Zhao,1 Jian-guo Zhou,1 Yue Han,2 Zhen Huang,1 Ye-fan Zhang,1 Jian-qiang Cai1
1Department of Hepatobiliary Surgery, 2Department of Interventional Therapies, National Cancer Center/Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, 100021, China
*These authors contributed equally to this work
Background: Systemic inflammation (SI) is associated with tumor progression and overall survival (OS) in patients with hepatocellular carcinoma (HCC). The presence of some single nucleotide polymorphisms (SNPs) in the human leukocyte antigen (HLA) region can influence the prognosis of patients with hepatitis B virus (HBV)-related HCC, although the mechanism remains unknown. This study aimed to analyze the correlations between HLA gene polymorphisms and SI.
Patients and methods: This study included 330 patients with HCC. The clinical parameters were reviewed, and five SNPs, namely rs2647073, rs3997872, rs3077, rs7453920, and rs7768538, were genotyped using the MassARRAY system.
Results: The rs3997872, rs7453920, and rs7768538 genotypes were found to be significantly associated with OS (P<0.05). The rs7453920 genotype was significantly associated with the neutrophil/lymphocyte ratio (NLR; P=0.001), which was used as an SI index with a threshold determined by receiver operating characteristic analysis. An elevated NLR was also an independent predictor of OS according to univariate and multivariate analyses (P<0.001).
Conclusion: Our data show that HLA gene polymorphisms are associated with SI in patients with HBV-related HCC, and the absence of minor allele A (rs7453920) promotes SI and shortens OS.
Keywords: human leukocyte antigen, single nucleotide polymorphism, systemic inflammation, hepatitis B virus, hepatocellular carcinoma
Introduction
Hepatocellular carcinoma (HCC) is among the most prevalent malignancies worldwide and is the fourth most common cancer and the third leading cause of cancer-related deaths in China.1 Development and prognosis of HCC depend not only on the intrinsic properties of tumor cells but also on several host-related factors. In the Chinese population, most HCCs develop as a complication of chronic liver inflammation and cirrhosis caused by hepatitis B virus (HBV) or hepatitis C virus (HCV) infection.2–4
Recent evidence suggests that systemic inflammation (SI) is associated with tumor progression and overall survival (OS) in patients with HCC.5–8 White blood cell counts and ratios, as well as C-reactive protein levels and the Glasgow prognostic score, are often used as indices of SI. The neutrophil/lymphocyte ratio (NLR) is the most commonly used index to predict prognoses following HCC treatment (which can include surgical resection, liver transplantation, transarterial chemoembolization [TACE], and sorafenib administration).6,8–12 The neutrophil count in the peripheral blood is associated with tumor-promoting inflammation, while lymphocyte counts reflect the antitumor immune function. An elevated NLR may indicate an imbalance between SI and the host immune response in favor of the former.13,14
Meanwhile, the host immune response plays a key role in liver carcinogenesis and progression. The human leukocyte antigen (HLA) region, which is located on chromosome 6p21.3, is important for the immune response to viral and cancer antigens. Recently, several genome-wide association studies (GWASs) have found that some loci in the HLA region are associated with susceptibility to chronic hepatitis B (CHB) or HBV-related HCC.15–20 Furthermore, our previous study produced evidence that some of these GWAS-identified susceptibility loci affect the prognosis of patients with HBV-related HCC.21
However, the mechanisms by which these susceptibility loci affect the survival of these patients are unknown. Based on the relationship between immunity, inflammation, and cancer, we hypothesized that HLA gene polymorphisms cause an imbalance between SI and immune response that results in poorer outcomes of patients with HBV-related HCC. Finally, this study aimed to analyze the correlation between HLA gene polymorphisms and SI.
Patients and methods
Patients
A total of 360 Chinese patients with HBV-related HCC who were admitted to the Cancer Hospital, Chinese Academy of Medical Sciences, Beijing, China, between January 1999 and December 2012 and had not received any previous treatment were enrolled in this study. A total of 30 patients (8.3%) were excluded because of missing clinicopathological data; thus, 330 patients were ultimately included. Imaging studies including ultrasonography, computed tomography (CT), magnetic resonance imaging (MRI), and angiography were performed for diagnosis. HCC was diagnosed based on the European Association for the Study of the Liver-European Organization for Research and Treatment of Cancer (EASL-EORTC) clinical practice guidelines; biopsies were acquired if patients did not have typical imaging features.22 All the enrolled patients were serologically positive for both the hepatitis B surface antigen and core antibody; patients with HCV coinfection were excluded. The patient’s baseline clinical characteristics were collected, including white blood cell counts, neutrophil counts, lymphocyte counts, and levels of alanine aminotransferase, γ-glutamyl transpeptidase (GGT), lactate dehydrogenase (LDH), alkaline phosphatase (ALP), albumin, and alpha-fetoprotein (AFP). The type of treatment (surgical resection, TACE, radiofrequency ablation [RFA], or radiotherapy) was also recorded. The numbers and sizes of hepatic lesions were measured via CT or MRI. Liver function was evaluated using a liver enzyme test and graded according to the Child–Pugh classification. Liver cirrhosis was determined by imaging; the Barcelona Clinic Liver Cancer (BCLC) stage was also determined. This study was performed according to the Declaration of Helsinki and was approved by the institutional review board of the Chinese Academy of Medical Sciences Cancer Institute. All patients provided written informed consent.
Follow-up
Patients treated with radical resection, RFA, or radiotherapy underwent enhanced CT, ultrasonography, or MRI every 3–4 months to screen for recurrence; appropriate treatment was administered if recurrence was confirmed. For patients treated with TACE, tumor responses were evaluated regularly; if residual lesions were found, another round of TACE was performed after 1 month. TACE was performed until a patient could no longer tolerate the procedure or achieved a complete response. The patients were followed up every 3–4 months with regular laboratory tests and imaging. OS was defined as the interval between the date of initial treatment and that of the last follow-up visit or death. The final follow-up date was January 31, 2017; none of the patients were lost to follow-up.
Single nucleotide polymorphism (SNP) selection, DNA extraction, and genotyping
According to the previous GWAS findings on the susceptibility to chronic HBV infection or HBV-related HCC in Asian populations, five SNPs located in the HLA region (rs2647073 and rs3997872 at HLA-DRB1, rs3077 at HLA-DPA, and rs7453920 and rs7768538 at HLA-DQB2) were analyzed.15–20 All the five SNPs were identified with a minor allele frequency of >5% in the Chinese Han population based on the 1000 Genomes browser (http://www.1000genomes.org/). Genomic DNA was extracted from whole blood samples obtained from patients using a blood DNA kit (Tiangen, Beijing, China). The extraction procedure was performed following the manufacturer’s instructions, which included cell lysis using sodium dodecyl sulfate plus proteinase K, purification using phenol/chloroform, and precipitation using ethanolamine and ice-cold 100% ethanol. SNP genotyping was performed on the Sequenom MassARRAY system (Sequenom, San Diego, CA, USA) according to the manufacturer’s protocol. The genotyping primers for the five SNPs are summarized in Table S1, which were designed using the Sequenom Assay Design software v3.1. The resulting data management and analysis were performed using the Sequenom Typer software v4.0. All assays were performed in 384-well plates, and negative controls were included for quality control. Moreover, a second round of analysis was performed on 20% of the samples, which were selected randomly; all replicate results were found to be completely concordant. The genotyping success rates in the initial 360 patients were 98.9%, 99.2%, 99.4%, 98.6%, and 95.0% for rs2647073, rs3077, rs3997872, rs7453920, and rs7768538, respectively.
SI index
Several clinical parameters are used to reflect SI. In the current study, NLR was used to evaluate the correlation between SI, HLA gene SNPs, and the survival of patients with HBV-related HCC. The neutrophil and lymphocyte counts were obtained from hematologic blood tests within 1 week before the initial treatment. NLR was calculated by dividing the neutrophil counts by the lymphocyte counts and was considered as a continuous variable.
Statistical analyses
All statistical analyses were performed using SPSS 19.0 for Windows (SPSS Inc., Chicago, IL, USA). Categorical variables were expressed using frequencies and percentages, while continuous variables were expressed using mean with SD or median with range. Differences between the subgroups were analyzed using chi-squared test or Fisher’s exact test for categorical variables and Student’s t-test for continuous variables. OS and progression-free survival (PFS) rates between different groups were analyzed using the Kaplan–Meier method, and differences were compared using the log-rank test. The selected SNPs were analyzed using the abovementioned method to confirm the association of unfavorable genotypes with poor outcomes, and the Cox proportional hazards model was used to estimate the hazard ratios. The receiver operating characteristic (ROC) curve was measured to determine the NLR cutoff values to predict the presence or absence of the unfavorable genotype by determining the maximum sensitivity and specificity. Factors analyzed using Cox proportional hazard model for univariate analysis of OS with a P-value of <0.05 were subjected to stepwise multivariate analysis. P-values of <0.05 were considered statistically significant.
Results
Patients’ characteristics
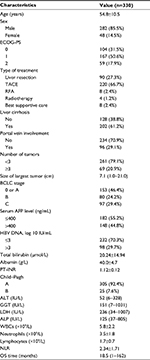
As summarized in Table 1, the patients in this study included 282 men (85.5%) and 48 women (14.5%) with a mean age of 54.8±10.5 years; 262 patients (79.4%) died after a median survival time of 18.5 months. None of the patients had an Eastern Cooperative Oncology Group performance status (ECOG-PS) score of >2. A total of 90 patients (27.3%) underwent liver resection, while 240 (72.7%) received TACE, RFA, radiotherapy, or best supportive care as initial treatment. Serum AFP levels were elevated (>400 ng/mL) in 148 patients (44.8%), while 305 patients (92.4%) had Child–Pugh class A disease. The mean NLR was 2.34±1.71.
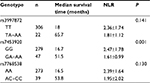
Association of selected SNPs with the NLR
In univariate analysis using the Cox proportional hazards model, rs3997872, rs7453920, and rs7768538 were significantly associated with OS (all P<0.05; Table 2). Patients with the TT genotype of rs3997872, GG genotype of rs7453920, and AA genotype of rs7768538 had poor OS; hence, these genotypes were considered negative prognostic predictors. Moreover, unfavorable genotypes were associated with poor PFS (Figure 1). However, only the rs7453920 genotype group was significantly associated with the NLR; the GG genotype had a mean NLR of 2.47±1.78, while the GA+AA genotype had a mean NLR of 1.61±0.99 (P=0.001; Table 3).
  | Table 3 Association of genotype of SNPs with NLR Note: Continuous variables are expressed as mean ± SD. Abbreviations: NLR, neutrophil/lymphocyte ratio; SNP, single nucleotide polymorphism. |
Optimal NLR cutoff value
The optimal NLR cutoff value was used to predict the absence or presence of the rs7453920 GG genotype and was determined using ROC curve analysis. The area under the curve (AUC) for the NLR was 0.708 (P<0.001), and the optimal cutoff value was 1.43 based on the Youden index (sensitivity, 75.3%; specificity, 60.0%; Figure 2A). ROC curve analysis to predict the median survival time revealed an AUC of 0.753 (P<0.001), with an NLR value of 2.19 (sensitivity, 61.2%; specificity, 80.6%) corresponding to the Youden index (Figure 2B). To differentiate between rs7453920 genotypes, patients were divided into two subgroups, such as NLR<1.43 (n=100; 30.3%) and NLR≥1.43 (n=230; 69.7%). Kaplan–Meier survival analysis showed that patients in the higher NLR subgroup had significantly poorer OS and PFS (P<0.001; Figure 3).
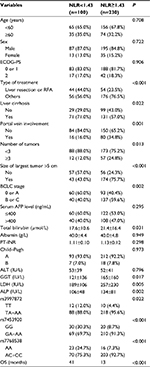
Association of clinical or genetic variables with the NLR subgroups
As summarized in Table 4, the type of treatment, liver cirrhosis, tumor number, tumor size, and BCLC stage in the two NLR subgroups were significantly different (all P<0.05). The levels of total bilirubin, GGT, LDH, and ALP were also significantly different between the NLR subgroups (all P<0.05). Interestingly, all three SNPs had significantly different genotype ratios between the two subgroups (all P<0.05).
Association of clinical variables with the rs7453920 genotype
Univariate analysis showed that the BCLC stage, portal vein involvement, Child–Pugh class, and NLR were significantly associated with the rs7453920 genotype. On multivariate analysis for the abovementioned variables, only NLR was independently associated with the rs7453920 genotype (hazard ratio, 4.21; 95% confidence interval [CI], 2.21–8.05; P<0.001; Table 5).
Prognostic factors for OS
To identify the predictive factors for OS, NLR and other clinical variables were subjected to univariate and multivariate analyses using the Cox proportional hazards model. The results showed that ECOG-PS, BCLC stage, portal vein involvement, tumor number, tumor size, AFP level, treatment type, and NLR were all significantly associated with OS. Multivariate analysis showed that ECOG-PS, BCLC stage, portal vein involvement, tumor number, tumor size, treatment type, and NLR were independent predictors of OS (all P<0.05; Table 6).
Discussion
In recent years, several GWASs have identified several loci associated with HBV infection and HBV-related HCC. Mbarek et al15 reported that some SNPs within the HLA-DQ loci were independently associated with susceptibility to CHB and persistent HBV infection in a Japanese population. Clifford et al20 also reported that 10 SNPs, all located in the gene region involved with immune response, were associated with HCC development. Among the identified SNPs, those located in the HLA region may affect immune response and the clinical outcome of patients with HBV-related HCC. The HLA region plays a key role in HBV clearance, and mutations in HLA genes may result in persistent HBV infection. Such infection, and the cirrhosis that may ensue, can exhaust the host immune system.23 At the same time, the activation of the immune system increases the production of pro-inflammatory cytokines and the upregulation of cell inflammatory markers.23–25 Furthermore, the development of liver tumors can affect the balance between SI and immune response, leading to poorer outcomes.4,26,27 The presence of unfavorable genotypes may indicate that the immune systems of these patients are compromised by HBV infection and HCC. Clinical inflammation indicators are commonly used to evaluate the effects of SI.5,12,14,28,29 The abovementioned evidence suggests a close correlation of HLA gene polymorphisms with inflammation indicators.
We found that rs3997872 at HLA-DRB1, as well as rs7453920 and rs7768538 at HLA-DQB2, influenced the clinical outcomes of patients with HBV-related HCC. On a related note, Matoba et al30 discovered that tumor HLA-DR expression was linked to early intrahepatic recurrence of HCC. Other studies showed that rs7453920 and rs7768538 are associated with persistent HBV infection and HCC development.31,32 In addition, Li et al33 discovered that rs7453920 was related to the prognosis of liver transplant recipients. The exact mechanism by which gene polymorphisms in HLA-DRB1 and HLA-DQB2 affect the clinical outcome of patients with HBV-related HCC is unclear. However, both HLA-DR and HLA-DQ molecules are encoded by major histocompatibility complex (MHC) class II genes, and SNPs might alter the MHC class II-mediated immune responses in a manner that is detrimental to the patients’ outcomes. MHC class II proteins expressed by antigen-presenting cells such as Kupffer and B cells are crucial for antigen presentation to CD4+ T cells,34 and Clifford et al20 reported that rs3997872 might be associated with altered MHC class II protein activity that impairs the T cell response.
Among the three SNPs associated with OS, only the rs7453920 genotype was significantly associated with the NLR, which was elevated in patients with the GG genotype. Mano et al35 reported that preoperative NLR is a predictor of survival after hepatectomy for patients with HCC; their threshold for high NLR was 2.81. Another study by Pinato et al8 set the threshold at 5, while Hu et al36 concluded that an NLR of ≥1.505 was an independent factor for adverse recurrence-free survival (RFS) and an NLR of ≥1.945 was a predictor of early recurrence. Recent studies have set the threshold in the 2.1–3 range, which was determined mainly from ROC curves for predicting median survival, OS, or RFS; however, the optimal value has yet to be determined.37–42 In our study, the NLR cutoff of 1.43 was derived from ROC analyses of the effects of rs7453920 and was found to have an AUC, sensitivity, and specificity of 0.708, 75.3%, and 60.0%, respectively. Our value, which ultimately aimed to reflect the effect of HLA gene polymorphisms on SI, was lower than those in previous studies and only had a sensitivity and specificity of 73.3% and 44.1%, respectively, according to ROC analysis of median survival. However, an NLR of ≥1.43 was still a negative predictor of OS in multivariate analysis. As the optimal NLR cutoff value for predicting median survival was 2.19 in our cohort, we posited that the SI and immune response might be imbalanced in patients with NLR values between 1.43 and 2.19, but not enough to affect their survival. Moreover, it is also possible that other factors such as tumor size, portal vein tumor thrombosis, or the severity of CHB can affect NLR.
The mechanism of how HLA gene polymorphisms affect the immune response and SI remains unknown. However, our study is the first to demonstrate that the rs7453920 genotype at HLA-DQB2 is associated with NLR and that an NLR threshold determined from effects of this genotype is an independent predictor of OS in patients with HBV-related HCC. Although it is unlikely that a single gene locus is a superior predictor of clinical outcomes compared to the BCLC staging system or other identified clinical prognostic factors, our findings highlight a novel aspect of SNP function in the post-GWAS era.
Our study had some limitations. First, only five SNPs in the HLA gene were analyzed, and only one was found to be of clinical significance. A large-scale study of additional SNPs involved in the immune response in other gene regions (such as interleukin-10 or STAT4) is required to validate our results. Second, some BCLC stage C patients were included in our study; most of them were treated with TACE. However, the EASL-EORTC clinical practice guidelines recommend targeted therapies for these patients. Unfortunately, sorafenib was only available for treating HCC in China after 2008, and none of our patients received it as an initial treatment; thus, we did not investigate the association between SI, immune response, and targeted therapies. Third, the study was performed only in a Chinese population, and all participants had HBV infection. Further studies are required to validate our findings in other populations. Moreover, similar studies can focus on gene loci associated with the susceptibility to HCC from other etiologies, such as HCV, alcohol, and nonalcoholic fatty liver disease.
Conclusion
rs7453920 G>A was associated with an elevated NLR, which in turn was an independent factor associated with shorter OS. Our results indicate that the absence of the minor allele A of rs7453920 promotes SI and adversely affects the OS of patients with HBV-related HCC.
Acknowledgment
This work was supported by the Capital Health Research and Development of Special (grant no. 2014-1-4022), the State Key Project on Infection Diseases of China (grant no. 2012ZX10002016 and no. 2018ZX10723204-005), and the Chinese Academy of Medical Sciences Innovation Fund for Medical Sciences (grant no. 2016-I2M-1-001 and no. 2017-I2M-4-002).
Disclosure
The authors report no conflicts of interest in this work.
References
Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. | ||
Diakos CI, Charles KA, McMillan DC, Clarke SJ. Cancer-related inflammation and treatment effectiveness. Lancet Oncol. 2014;15(11):e493–e503. | ||
Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2011;140(6):883–899. | ||
Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. | ||
Aino H, Sumie S, Niizeki T, et al. The systemic inflammatory response as a prognostic factor for advanced hepatocellular carcinoma with extrahepatic metastasis. Mol Clin Oncol. 2016;5(1):83–88. | ||
Chan SL, Chan AWH, Chan AKC, et al. Systematic evaluation of circulating inflammatory markers for hepatocellular carcinoma. Liver Int. 2017;37(2):280–289. | ||
Howell J, Pinato DJ, Ramaswami R, et al. Integration of the cancer-related inflammatory response as a stratifying biomarker of survival in hepatocellular carcinoma treated with sorafenib. Oncotarget. 2017;8(22):36161–36170. | ||
Pinato DJ, Stebbing J, Ishizuka M, et al. A novel and validated prognostic index in hepatocellular carcinoma: the inflammation based index (IBI). J Hepatol. 2012;57(5):1013–1020. | ||
McDonald B, Spicer J, Giannais B, Fallavollita L, Brodt P, Ferri LE. Systemic inflammation increases cancer cell adhesion to hepatic sinusoids by neutrophil mediated mechanisms. Int J Cancer. 2009;125(6):1298–1305. | ||
Tajiri K, Baba H, Kawai K, et al. Neutrophil-to-lymphocyte ratio predicts recurrence after radiofrequency ablation in hepatitis B virus infection. J Gastroenterol Hepatol. 2016;31(7):1291–1299. | ||
Pinato DJ, Sharma R. An inflammation-based prognostic index predicts survival advantage after transarterial chemoembolization in hepatocellular carcinoma. Transl Res. 2012;160(2):146–152. | ||
Xu L, Yu S, Zhuang L, et al. Systemic inflammation response index (SIRI) predicts prognosis in hepatocellular carcinoma patients. Oncotarget. 2017;8(21):34954–34960. | ||
Qi X, Li J, Deng H, Li H, Su C, Guo X. Neutrophil-to-lymphocyte ratio for the prognostic assessment of hepatocellular carcinoma: a systematic review and meta-analysis of observational studies. Oncotarget. 2016;7(29):45283–45301. | ||
Xue TC, Jia QA, Ge NL, Chen Y, Zhang BH, Ye SL. Imbalance in systemic inflammation and immune response following transarterial chemoembolization potentially increases metastatic risk in huge hepatocellular carcinoma. Tumour Biol. 2015;36(11):8797–8803. | ||
Mbarek H, Ochi H, Urabe Y, et al. A genome-wide association study of chronic hepatitis B identified novel risk locus in a Japanese population. Hum Mol Genet. 2011;20(19):3884–3892. | ||
Hu Z, Liu Y, Zhai X, et al. New loci associated with chronic hepatitis B virus infection in Han Chinese. Nat Genet. 2013;45(12):1499–1503. | ||
Kim YJ, Kim HY, Lee J-H, et al. A genome-wide association study identified new variants associated with the risk of chronic hepatitis B. Hum Mol Genet. 2013;22(20):4233–4238. | ||
Nishida N, Sawai H, Matsuura K, et al. Genome-wide association study confirming association of HLA-DP with protection against chronic hepatitis B and viral clearance in Japanese and Korean. PLoS One. 2012;7(6):1–8. | ||
Li S, Qian J, Yang Y, et al. GWAS identifies novel susceptibility loci on 6p21.32 and 21q21.3 for hepatocellular carcinoma in chronic hepatitis B virus carriers. PLoS Genet. 2012;8(7):e1002791. | ||
Clifford RJ, Zhang J, Meerzaman DM, et al. Genetic variations at loci involved in the immune response are risk factors for hepatocellular carcinoma. Hepatology. 2010;52(6):2034–2043. | ||
Li C, Bi X, Huang Y, et al. Variants identified by hepatocellular carcinoma and chronic hepatitis B virus infection susceptibility GWAS associated with survival in HBV-related hepatocellular carcinoma. PLoS One. 2014;9(7):e101586. | ||
European Association For The Study Of The Liver; European Organisation For Research And Treatment Of Cancer. EASL-EORTC clinical practice guidelines: management of hepatocellular carcinoma. J Hepatol. 2012;56(4):908–943. | ||
Albillos A, Lario M, Álvarez-Mon M. Cirrhosis-associated immune dysfunction: distinctive features and clinical relevance. J Hepatol. 2014;61(6):1385–1396. | ||
Dirchwolf M, Ruf AE. Role of systemic inflammation in cirrhosis: from pathogenesis to prognosis. World J Hepatol. 2015;7(16):1974–1981. | ||
Dirchwolf M, Podhorzer A, Marino M, et al. Immune dysfunction in cirrhosis: distinct cytokines phenotypes according to cirrhosis severity. Cytokine. 2016;77:14–25. | ||
Gardini AC, Scarpi E, Faloppi L, et al. Immune inflammation indicators and implication for immune modulation strategies in advanced hepatocellular carcinoma patients receiving sorafenib. Oncotarget. 2016;7(41):67142–67149. | ||
Hernandez-Gea V, Toffanin S, Friedman SL, Llovet JM. Role of the microenvironment in the pathogenesis and treatment of hepatocellular carcinoma. Gastroenterology. 2013;144(3):512–527. | ||
Turner N, Wong HL, Templeton A, et al. Analysis of local chronic inflammatory cell infiltrate combined with systemic inflammation improves prognostication in stage II colon cancer independent of standard clinicopathologic criteria. Int J Cancer. 2016;138(3):671–678. | ||
Chan SL, Chan AWH, Chan AKC, et al. Systematic evaluation of circulating inflammatory markers for hepatocellular carcinoma. Liver Int. 2017;37(2):280–289. | ||
Matoba K, Iizuka N, Gondo T, et al. Tumour HLA-DR expression linked to early intrahepatic recurrence of hepatocellular carcinoma. Int J Cancer. 2005;115(2):231–240. | ||
Mathew S, Abdel-Hafiz H, Raza A, Fatima K, Qadri I. Host nucleotide polymorphism in hepatitis B virus-associated hepatocellular carcinoma. World J Hepatol. 2016;8(10):485–498. | ||
Ochi Y, Hashimoto S, Kawabe N, et al. HLA-DQ gene polymorphisms are associated with hepatocellular carcinoma and hepatitis B surface antigen in chronic hepatitis B virus infection. Hepatol Res. 2017;47:755–766. | ||
Li Y, Huang Q, Tang J, et al. Correlation of HLA-DP/DQ polymorphisms with transplant etiologies and prognosis in liver transplant recipients. Medicine (Baltimore). 2017;96(25):e7205. | ||
Rudolph MG, Stanfield RL, Wilson IA. How TCRs bind MHCs, peptides, and coreceptors. Annu Rev Immunol. 2006;24:419–466. | ||
Mano Y, Shirabe K, Yamashita Y-I, et al. Preoperative neutrophil-to-lymphocyte ratio is a predictor of survival after hepatectomy for hepatocellular carcinoma: a retrospective analysis. Ann Surg. 2013;258(2):301–305. | ||
Hu X, Mao W, Park Y, Xu W, Kim B, Wang H. Blood neutrophil-to-lymphocyte ratio predicts tumour recurrence in patients with hepatocellular carcinoma within Milan criteria after hepatectomy. Yonsei Med J. 2016;57(5):1115–1123. | ||
Yang T, Zhu J, Zhao L, et al. Lymphocyte to monocyte ratio and neutrophil to lymphocyte ratio are superior inflammation-based predictors of recurrence in patients with hepatocellular carcinoma after hepatic resection. J Surg Oncol. 2017;115(6):718–728. | ||
Li S-H, Wang Q-X, Yang Z-Y, et al. Prognostic value of the neutrophil-to-lymphocyte ratio for hepatocellular carcinoma patients with portal/hepatic vein tumour thrombosis. World J Gastroenterol. 2017;23(17):3122. | ||
Taussig MD, Irene Koran ME, Mouli SK, et al. Neutrophil to lymphocyte ratio predicts disease progression following intra-arterial therapy of hepatocellular carcinoma. HPB. 2017;19(5):458–464. | ||
Hung H-C, Lee J-C, Cheng C-H, et al. Impact of neutrophil to lymphocyte ratio on survival for hepatocellular carcinoma after curative resection. J Hepatobiliary Pancreat Sci. 2017;24:559–569. | ||
Arai K, Fukumoto T, Kido M, et al. Preoperative neutrophil-to-lymphocyte ratio as a predictor of survival after reductive surgery plus percutaneous isolated hepatic perfusion for hepatocellular carcinoma: a retrospective analysis. Surg Today. 2017;47(3):385–392. | ||
Son SH, Park EY, Park HH, Kay CS, Jang HS. Pre-radiotherapy neutrophil-to-lymphocyte ratio as an independent prognostic factor in patients with locally advanced hepatocellular carcinoma treated with radiotherapy. Oncotarget. 2017;8(10):16964–16971. |
Supplementary material
  | Table S1 Primers for genotyping of candidate SNPs Abbreviation: SNP, small nucleotide polymorphism. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.