Back to Journals » Infection and Drug Resistance » Volume 15
How Good are Bacteriophages as an Alternative Therapy to Mitigate Biofilms of Nosocomial Infections
Authors Singh A , Padmesh S , Dwivedi M, Kostova I
Received 10 November 2021
Accepted for publication 27 January 2022
Published 17 February 2022 Volume 2022:15 Pages 503—532
DOI https://doi.org/10.2147/IDR.S348700
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Suresh Antony
Aditi Singh,1 Sudhakar Padmesh,1 Manish Dwivedi,1 Irena Kostova2
1Amity Institute of Biotechnology, Amity University Uttar Pradesh, Lucknow Campus, Lucknow, 226028, India; 2Department of Chemistry, Faculty of Pharmacy, Medical University, Sofia, 1000, Bulgaria
Correspondence: Aditi Singh, Email [email protected]; [email protected]
Abstract: Bacteria survive on any surface through the generation of biofilms that provide a protective environment to grow as well as making them drug resistant. Extracellular polymeric matrix is a crucial component in biofilm formation. The presence of biofilms consisting of common opportunistic and nosocomial, drug-resistant pathogens has been reported on medical devices like catheters and prosthetics, leading to many complications. Several approaches are under investigation to combat drug-resistant bacteria. Deployment of bacteriophages is one of the promising approaches to invade biofilm that may expose bacteria to the conditions adverse for their growth. Penetration into these biofilms and their destruction by bacteriophages is brought about due to their small size and ability of their progeny to diffuse through the bacterial cell wall. The other mechanisms employed by phages to infect biofilms may include their relocation through water channels to embedded host cells, replication at local sites followed by infection to the neighboring cells and production of depolymerizing enzymes to decompose viscous biofilm matrix, etc. Various research groups are investigating intricacies involved in phage therapy to mitigate the bacterial infection and biofilm formation. Thus, bacteriophages represent a good control over different biofilms and further understanding of phage-biofilm interaction at molecular level may overcome the clinical challenges in phage therapy. The present review summarizes the comprehensive details on dynamic interaction of phages with bacterial biofilms and the role of phage-derived enzymes – endolysin and depolymerases in extenuating biofilms of clinical and medical concern. The methodology employed was an extensive literature search, using several keywords in important scientific databases, such as Scopus, Web of Science, PubMed, ScienceDirect, etc. The keywords were also used with Boolean operator “And”. More than 250 relevant and recent articles were selected and reviewed to discuss the evidence-based data on the application of phage therapy with recent updates, and related potential challenges.
Keywords: phage, extracellular polymeric substance, biofilm, antibiotic resistance, mitigating strategy, phage cocktail, phage lysins, depolymerases
Graphical Abstract:
Summary
Bacterial biofilms are serious causes of nosocomial and device-related infections. Phages are ubiquitous viruses which infect bacteria and are being extensively explored as alternatives to antibiotics. Phages employ many strategies to better penetrate hard-to-reach bacterial cells in biofilms and have been found to be effective against many resistant nosocomial bacterial strains.
Introduction
Bacteria have a unique protection layer called biofilm constructed by cell aggregation, that facilitate the bacterial immobilization on an extracellular polymeric matrix and form higher-order structures.1 This extracellular polymeric matrix is comprised of long chain sugars, DNA, and other biological macromolecules.2 With the help of such biofilms, bacteria have additional resistant strength against antimicrobial agents like biocides and antibiotics, etc.3 The bacterial cells within the biofilm are protected and tolerant to antibiotics, antiseptic, antimicrobials, and host immune responses.4 Although commensal flora also resides in the form of biofilms, especially in sites like the oral cavity, gastrointestinal tract, vagina; this heterogenous community is normally in a homeostatic relationship with the host. Any disturbance in that may lead to the formation of dysbiotic biofilms inhabited largely by opportunistic pathogens and ultimately lead to persistent infections.5 Such biofilms of pathogenic bacteria could be present on the surface of various medical instruments and materials too, in addition to the surface of the patient’s tissue.6
One of the most prevalent and extensively studied polymicrobial biofilms is oral biofilm. Imbalance and loss of the healthy oral microbial population may lead to various oral diseases. In a previous report, more than 700 bacterial species were observed to be located in the oral cavity7 where they continued to survive by constructing biofilm, however excessive synthesis of biofilms on teeth, tongue and other cavity areas may show deregulated immune responses, leading to further human oral diseases,8 for example, the dental plaque consisting of Porphyromonas gingivalis and Fusobacterium nucleatum is the crucial agent of gingivitis and periodontitis.9 Researchers could not classify many of the oral pathobionts like Porphyromonas gingivalis whether they are bystander microbiota or disease initiators.10 In addition, to cure oral diseases, alternative treatment processes are required that should target specifically microbes associated with disease in the biofilm without affecting the other beneficial microbiota.11
Orthopedic devices are also one of the most common surfaces colonized by bacteria. Joint replacement is a procedure where prosthetic joint infections due to bacterial biofilms on medical implants can be a frequent complication and may require surgical removal of the implant and prolonged antibiotic therapy.12 Some of the major pathogens isolated from medical devices and implants are given in Table 1. Similarly, the formation of biofilm on catheters and implants is another major cause of chronic, recurrent, or persistent infections. The urinary bladder usually does not have dangerous levels of bacteria, but when a foreign object like a catheter is put inside, the planktonic bacteria easily get attached to those surfaces forming biofilms. The urease-producing bacteria can form crystalline biofilms, leading to blockage of catheters, urine retention and a medical emergency.15 Major species involved in crystalline biofilms are Proteus mirabilis, P. vulgaris, Providencia rettgeri.16 While non-crystallized biofilms are formed by low urease producing bacterial species, namely Pseudomonas aeruginosa, Staphylococcus aureus, Klebsiella pneumoniae, Escherichia coli, Morganella morganii, Providencia stuartii.14
 |
Table 1 Most Common Pathogens Isolated from Biofilms Formed Over Medical Prostheses and Devices |
It is established that bacteria achieve a good level of resistance to various biocides and antibiotics due to the presence of biofilm. Investigations have suggested that more than 1000 times antibacterial compounds may be required for bacteria within biofilm compared to planktonic forms.17 Since mechanisms like horizontal gene transfer (HGT) and hypermutability are favoured within biofilms, these structures are now recognized as a reservoir of antibiotic genes.18 The collective recalcitrance toward antibiotics in a biofilm is majorly dependent on, the developmental stage of biofilm, its EPS composition, the biofilm architecture.19 These recalcitrant biofilms can also initiate an increase in multidrug resistant (MDR) bacteria. MDR infections are frequently associated with common human pathogens, known as ESKAPE (Enterococcus faecium, S. aureus, K. pneumoniae, Acinetobacter baumannii, P. aeruginosa and Enterobacter spp), which are the leading causes of nosocomial infections.20 WHO has declared antimicrobial resistance as one of the top 10 global public health threats facing humanity.21 In addition, the economic burden for providing treatment to six MDR ESKAPE pathogens is almost $4.6 billion annually.22 Thus, new therapies acting by totally different mechanisms are highly desired. In WHO’s global action plan on antimicrobial resistance, one of the five objectives is to develop new medicines, diagnostic tools, vaccines, and other interventions.23 In the present scenario, phage therapy represents a potential non-antibiotic approach worth exploring and current research has re-established phage therapy as an effective antibacterial treatment that works equally well against sensitive as well as resistant bacterial strains.24,25 Similarly, sufficient evidence is available confirming the potential of phages in biofilm removal under various conditions.26,27
The aim of the present review is, therefore, to focus on the role of phages as an alternative to antibiotics in the removal of bacterial biofilms, particularly biofilms of common drug resistant pathogens. It primarily summarizes the recent advancements in different phage-therapy strategies, starting from the whole phage to phage cocktail, phage-derived enzymes and engineered phages for mitigating biofilms. Lastly, the potential challenges and future potential of the approach are also highlighted.
Process of Biofilm Generation and Molecular Mechanisms During its Formation
Referring to various investigations, biofilm generation may include three major steps ie, attachment, maturation and dispersion (Figure 1). At the initial stage, attachment can happen in two ways, eg, reversible and irreversible attachment.28 Bacterial cells achieve better tolerance for physical or chemical shear forces due to irreversible attachment. To form a biofilm, motile bacterial cells employ flagella which are crucial for the initial attachment of the cell to the surface, however, O’Toole et al have demonstrated the development of biofilm by non-motile bacteria as well, eg S. aureus lacking flagella.29 During biofilm formation bacterial cells first interact with the surface through their flagella and type IV pili-mediated motilities, further this interaction leads to accumulation and aggregation of cells on the surface and eventually produce microcolonies. Maturation is done through the recruitment of more cells from the same species or other species to the biofilm from the bulk fluid and acquires the “mushroom” or “tower” shape appearance. At this stage, cells are arranged as per their metabolism and aero tolerance state. The bacterial cells within the biofilm secrete more proteins, DNA, polysaccharides for the biofilm scaffold during the maturation time. Figure 2 represents a bacterial biofilm composition, in which the bacterial cells in microcolonies are present as planktonic or embedded centrally in metabolically active or dormant forms. At the final stage, biofilm matures into a mushroom-like structure where bacterial cells achieve their nutrients through channels.29 Various factors have been observed for the dispersed condition of biofilm that may include expanded population, nutrients scarcity, high competition etc. Cell–cell disruptive factors are also present along with the matrix that causes the cell dispersal or detachment from mature biofilms and this may occur simply due to shear stresses. The other passive forces resulting in the breaking of a biofilm can be abrasion with environmental particles, grazing by eukaryotic organisms or sloughing due to frictional forces caused by the flow of ligands.30 During dispersion, the planktonic bacteria are released and spread to other areas, further beginning the formation of new biofilms.31
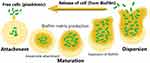 |
Figure 1 Graphical representation of various steps involved in generation of biofilm by bacterial cells. |
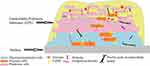 |
Figure 2 Schematic representation of a bacterial biofilm composition. A mature biofilm consists of bacterial cells (2–35%) in close proximity with the help of extracellular matrix, EPS containing exopolysaccharides (40–95%), proteins (1–60%), lipids (1–40%), enzymes, extracellular DNA and RNA (<1%).30 The bacterial cells in microcolonies are present as planktonic or embedded centrally in metabolically active or as dormant form (persister cells). |
The very first step of biofilm formation is attachment to the abiotic surface. This attachment is governed by some physicochemical properties of bacterial surface like hydrophobicity and its composition. There are several factors observed crucial for the attachment and proliferative stages of biofilm generation and pathogens have their own weapons and arsenal for formation of biofilms. Various studies have reported different molecular and genomic mechanisms employed by common pathogens for biofilm formation (Table 2), like uropathogenic E. coli (UPEC) have various adhesins and their receptors on host cells, of which chaperone-usher (CU) fimbrial adhesins are strongly associated with chronic colonization and urosepsis in UTI32 and Type 9 fimbriae in biofilm formation.36 Along with FimH adhesin on Type 1 pili, “curli” amyloid fibers and antigen 43 are also important mediators for attachment and progression of bacteria on abiotic surfaces.40 Investigators44 have found teichoic acids and the surface protein autolysin associated with abiotic surface attachment in the case of S. aureus, Enterococcus faecalis and Staphylococcus epidermidis. These cell wall anchored (CWA) surface proteins adhesion to a different extracellular matrix including collagen, fibrinogen, fibronectin promotes the formation of biofilms and are being studied in many antibiotic-resistant strains of S. aureus and S. epidermidis, common pathogens forming biofilms in indwelling medical devices.41 Similarly, researchers have identified the biofilm-associated genes in A. baumannii clinical isolates, which are highly conserved such as csuE and csuA, the proposed tip subunit of the Csu pili,51 Bap gene for biofilm associated proteins (Bap)52 and OmpA for outer membrane protein A (OmpA reported in 81–100% detection).55 In contrast to these gram-negative pathogens, the gram-positive ESKAPE member Enterococci though non-motile has also been found to be possessing a number of adhesins, viz. SegA, Acm (in E. faecium), Ace (in E. faecalis), Esp and Ebp, which contribute to biofilm formation and urinary tract infections.50 P. aeruginosa employs its Type IV pili and flagella and adhesins and lectins to adhere to epithelial cells in host lungs;58 whereas K. pneumoniae has hemagglutinins, Type 1 and Type 3 pili for the purpose.32 After attachment and during proliferation bacteria secretes extracellular matrix that may be composed of various compounds with different chemical properties including exopolysaccharides, proteins, eDNA, and other polymers.
 |
Table 2 Mechanisms of Biofilm Formation Employed by Common Bacterial Pathogens |
Finally, biofilm enters the dispersed step which determines its further spread. However, besides the passive dispersal forces as described in the previous section, bacteria also have other signalling methods to become planktonic again. Some of the stressors can be limited availability of nutrients, accumulation of toxic by-products, changes in oxygen level etc.66 For example increased iron level in the extracellular environment for UPEC or high carbon and nitrogen levels for P. aeruginosa have been reported for biofilm dispersal. Alteration in gene expression in response to small molecules or autoinducers also promotes dispersal, like c-di-GMP upregulation favors sessility in E. coli and P. aeruginosa.50,67 The alginate lyase, an EPS hydrolysing enzyme produced by P. aeruginosa helps in bacterial detachment and dispersal. In an extensive review on biofilm dispersal, Guilhen et al have elaborated on multiple strategies shown by bacteria for dispersal as a necessary step which results in a planktonic state with enhanced colonization properties.68 In one such example, increased production of rhamnolipids aids in P. aeruginosa dispersal forming new microcolonies and a central void.67 In another proof-of-concept study, the in vitro evaluation of P. aeruginosa biofilm dispersal was studied for antimicrobial efficacy. The arabinose-induced biofilm dispersal in the engineered P. aeruginosa strain showed increased efficacy for imipenem and tobramycin.69
Biofilm as a Protective Shelter for Bacteria
Biofilms become significant because of characteristic properties and traits, expressed by microbial cells only when in a biofilm. These recalcitrant structures have conferred higher antimicrobial potential to the sessile cells in the biofilm than their planktonic counterparts. For example, cystic fibrosis patients are highly susceptible to chronic infections of P. aeruginosa and A. baumannii, which are frequently identified as biofilm-associated and multidrug-resistant thus highly difficult to treat.70 The basis of this biofilm-mediated bacterial antibiotic survival is largely dependent on the developmental stage, the composition of extracellular matrix or EPS and the architecture of biofilm.19 The EPS in a biofilm comprises a heterogenous mixture of water, polysaccharides, proteins, lipids and glycolipids, extracellular DNA and RNA and ions like Ca2+. However, the composition is dynamic and depends on bacterial strain as well as environmental conditions. The EPS protects bacteria by providing nutrients keeping them in close proximity for crosstalk and genetic material exchange and maintaining hydration to protect against desiccation.71
Many mechanisms, also referred to as “emergent properties” like long-term cell–cell interaction, gradients of pH, nutrients and oxygen, presence of enzymes, persister cells, stress responses and high level of biofilm heterogeneity,72 etc are identified. These mechanisms are extensively elaborated in some recent reviews.19,73,74 Biofilm recalcitrance mechanisms can be a combination of antimicrobial resistance and antimicrobial tolerance and this combination will depend on the type of bacteria, type of antimicrobial agent, age of biofilm, and growth conditions.73 The antimicrobial resistance mechanisms involve horizontal gene transfer (HGT), hypermutation, efflux pumps, quorum sensing,75 reduced outer membrane permeability and production of neutralizing enzymes. HGT has been seen at a higher rate in biofilms than in planktonic cells.76,77 Similarly, a 100 to 1000-fold rise in mutations is reported in a biofilm owing to limited diffusion of antimicrobials due to its architecture.78 Another effective mechanism is efflux pumps, which confer higher resistance to bacteria against antimicrobials. Efflux pumps are specialized membrane-bound proteins to expel or throw out different compounds from inside the bacterial cells, thereby reducing the cytoplasmic concentration of antimicrobial compounds below the effective level.79 The generation of biofilm starts from initiating and establishing communication by a bacterial cell with other neighboring cells by releasing specific small chemical molecules. This phenomenon which is governed by bacterial population density is known as “Quorum sensing (QS)”.75 Blocking of quorum sensing has shown an enhanced susceptibility of biofilm of S. aureus for various classes of antibiotics.80 These bacterial cells thus behave in a different way as a community rather than simply a cluster of independent cells.2
In addition, the tolerance mechanisms which enable bacteria to withstand higher concentrations of antimicrobials, include stress-response activation,74 hypoxia, initiation of quiescent state, decrease membrane potential, anaerobic metabolism and increased expression of efflux pump. Small colony variants (SCV) in a biofilm give rise to genetic diversity and are correlated with a higher level of persistent and recurrent infection.81 In a biofilm significant heterogeneity occurs and almost 60% of the cells may show phenotypic variations.82 SCVs are slow-growing and dormant cells and the presence of different cell states in the specific zone within a biofilm is dependent on various factors involving the gaseous, as well as nutrient stratifications. Nutrient gradients within a biofilm lead to the formation of dormant “persister cells”.72 These metabolically inactive cells present in the matrix have a major role to support the regrowth and resistance of the bacteria within the biofilm and could be the reason behind the drug resistance through biofilm that further led to the revival of the biofilm after treatment.83 Also, the gene expression of bacteria within a biofilm is coordinated by the production and detection of extracellular signal molecules, “anti-inducers”.84
Recently, Rasool et al have reported that plastic litter provides space for pathogenic bacteria to form biofilms, leading to the emergence of multidrug resistance.85 Sometimes these biofilms are composed of more than one bacterial species too. This multispecies biofilm is more resistant to external stress conditions like the presence of antibiotics or any other antimicrobial agents due to cooperative interactions among them. The existence of this special strength could be associated with an increase in biomass or modified constitution of EPS matrix,86,87 through which biofilms create a protective environment for resident organisms and is believed to be an important trait for millions of years.88 These bacterial reservoirs may lead to noncurable, recurrent or persistent infections at the site or spread as emboli.12
Usually, bacteria located on the surface of the biofilms are physiologically different from those located inside the more internal region of the biofilm matrix. The surface bacteria of biofilm are always renewed and help in the initializing primary phage infection.73,89,90 Whereas bacteria residing in the internal region of the biofilm have restricted entry as well as very limited access to nutrients and oxygen. It is noticeable that bacteria present in the inner matrix of biofilm adapt to environmental anoxia and nutrient limitation leading to lower metabolic rate and a slower rate of cell division.91 Although it gives them slower growth, it also provides less sensitivity against different antibiotics as well as the bacteriophage infection.73
Impact of Antibiotics in Generation of Antibiotic Resistant Strain and Biofilm
Toward the generation of antibiotic-resistant strains, β-lactamase enzyme that hydrolyses β-lactam antibiotics has been found to be crucial. Studies have been done on enzymes utilizing serine at the active site as well as on those needing divalent Zn ions for hydrolysis. β-Lactamase enzymes comprise more than 2000 unique sequences and some of them have substantial clinical implication such as class A penicillinases, the extended-spectrum β-lactamases (ESBLs), the AmpC cephalosporinases. Therefore, due to this adaptability of β-lactamase enzymes, a new strategy of antimicrobial therapy is urgently needed.92 In order to assist researchers, a Beta-Lactamase Database (BLDB) has been established that contains the latest structural and functional information on the β-lactamase superfamily of enzymes and has effective expression on antibiotic resistance. Naas et al has provided a detailed review on functional and structural characteristics of β-lactamase enzymes belonging to various families and subfamilies based on BLDB.93
The different type of antibiotic-resistant genes has been observed in some microbial pathogens belonging to Enterobacteriaceae family and its chromosome constitutes the extended-spectrum-ß-lactamases (ESBLs) and metallo-ß-lactamases (MBLs) along with the plasmids and transposons as mobile genetic elements. Further various characteristics of MBLs were explained by the Behzadi et al that demonstrated the opportunities in production of effective inhibitors against MBLs.64 In some other in vitro works, antibiotics with subminimal inhibitory concentration illustrated its agonist relation with bacterial biofilm formation that represents significant clinical prospects. The induction of biofilm formation can be achieved through the supply of a low dose of antibiotics. But still we need to resolve the intricacies involved in antibiotic-induced biofilm formation and its clinical use as antimicrobial therapy for infection in biomedical devices.94
Further, investigators have found out the substantial association among various pathogenic strains of K. pneumoniae linked with pervasiveness of antibiotic resistance and virulent genes.95 During this study of 9 months from Dec. 2018 to Aug. 2019, clinical samples were collected from various hospitals located in Tehran and Iran.
Bacteriophages and Their Dynamics in Biofilm
As one of the most abundant (~1031) living species on planet earth, bacteriophages—the viruses infecting bacteria, have acquired a lot of attention from researchers for decades.96 They are found both in aquatic and terrestrial ecosystems, thus influence the microbial communities in various ways. There is a huge heterogeneity in terms of their morphology and genomic organization. Different morphology like tailed, icosahedral, filamentous, or pleomorphic have been described; however, the majority (>90%) of identified phages are tailed.97 Similarly, phage with single- or double-stranded DNA and single- or double-stranded RNA are identified and documented with genome sizes from as less as 4 kb to much larger of 550 kb.97–99 Bacteriophages have significant importance because of their specificity to infect one bacterial species, though it is also reported that many phage species may infect one bacterial type, eg more than 25 phage species have been identified which infect E. coli.100
Having been discovered more than a century ago, phage research has come a long way in terms of understanding not only its biology, genetics or role in ecology and biodiversity but also the huge applications in various fields. Potential applications of bacteriophage in medical and clinical, agriculture, and food industry has been documented, with few acquiring necessary regulatory approvals and are commercially available. Similarly, the ability of phage to affect bacteria in planktonic and within biofilms makes them a potential candidate to be explored more. And when it comes to bacterial biofilms, phage also faces the same fate as other bactericidal modes, it becomes quite hard for them to penetrate biofilm due to its thick polymeric matrix. Usually, every biofilm differs in context, matrix, and physiological heterogeneity within the species and even differs from strain to strain. Before going into further detail, it is important to understand the mechanism of penetration, multiplication, and proliferation of bacteriophage within a biofilm.
Bacterial Resistance Mechanisms vs Phage Penetration into Biofilms
There are many factors that affect the penetration of phage into bacterial biofilm matrix such as the age of biofilm, density of biofilm, its molecular structure as well as the extracellular phage-inactivating enzymes secreted by different bacteria. In a detailed review on bacteria and phage interactions in biofilm, Hansen et al have elaborated on important mechanisms and factors for bacterial protection against phage attack while in a biofilm.101 Major factors which affect phage infection are phage diffusion within the biofilm,102 presence of spherical bacterial “microcolonies” resulting in poor adsorption of phages in such tightly packed bacterial cells,103 presence of EPS and cell debris forming “phage sinks” around high density areas,104,105 production of amyloid fibers “curli” enhancing matrix density and retaining phages at the outer periphery of biofilms.106 In addition, the metabolically inactive persister cells inhibit the growth of phage; however, this may delay the process but does not completely inhibit it. Pearl et al in their work have shown that coliphage lambda could infect persister cells and resume its lytic cycle once persisters become metabolically active again.107 Another factor affecting phage infection is coinfection, wherein many phage particles infect the same host due to clusters of host cells and limited mobility. This also decreases the population of progeny virions.108
Lastly, QS has also been found to have an important role in biofilm development and phage-resistance by regulating CRISPR-associated (Cas) gene expression, phage receptors and phage inactivating proteases.109–111 QS alters the population of phage receptors on particular cell surfaces. Its mode of action has been described in E. coli, where, in response to the N-acyl-1-homoserine lactone, a reduction in the number of λ receptors on the bacterial surface is observed, which directly reduces the rate of phage adsorption.110 This mechanism has also been observed in the phages of P. aeruginosa, when a quorum-sensing inhibitor, penicillinic acid was introduced along with the phage dilution, phage efficacy has been exponentially increased in the biofilm matrix.112 Some phages can make use of these autoinducers and can induce lysis, eg, a family of temperate phages of Bacillus subtilis uses a bacterial QS system to initiate lysis-lysogeny cycles.113 In a recent work by Shah et al, a novel QS-sensing anti-activator protein Aqs1 from Pseudomonas phage DMS3 was characterized to inhibit LasR, the main receptor of QS. The small protein also inactivated the pilus assembly protein PilB which results in inhibition of super infection by phages. This has highlighted the ability of small phage proteins to bind with multiple host proteins and disrupt key biological pathways.114
Conversely, old or aged biofilm matrix is less favourable for bacteriophage penetration, resulting in the limitation of phage therapy, especially during chronic bacterial infection. However, the least metabolically active bacteria inside biofilm matrix, still can produce new virion particles, thus resulting in the infection of freshly divided biofilm matrix. In such cases phage therapy is effective to some extent, depending upon the vulnerability of bacteria to phage virion particles. Usually, more aggressive, and extensive treatment is required in the eradication of biofilm. The density of biofilm is a common indicator of whether the phage dilution can penetrate through the biofilm or not. Since biofilm contains a good water channel inside its matrix, they provide a good mode of penetration through these channels.115
Phage Derived Enzymes as Potential Bactericidal Agents
Of all phage therapy methods, EPS-degrading enzymes against biofilms are gaining a lot of interest. They can be used as an alternative to conventional antibiotics. It is also estimated that the use of phage products is easier and safer. The two major categories of degrading enzymes, found useful in mitigating biofilms, are lysins and depolymerases. Both are described below in separate sections:
Bacteriophage Lysins
Although lytic phages are being reported widely for controlling biofilms; their penetration, propagation, and diffusion rate through EPS matrix are important characteristics that may limit a phage’s use. To overcome this, phage-derived endolysins have been studied for the purpose. Bacteriophage-encoded enzymes, endolysin, are a group of unrelated proteins with a molecular weight range of 15–40 kDa, produced by double-stranded DNA phage at the final stage of the lytic cycle. Lysins are reported to be fast acting bactericidal proteins that will instantly cause hydrolysis of bacterial cell walls, thus helping in the release of phage particles. Vincent Fischetti has identified the endolysin that was used to control several important gram-positive pathogens viz. S. pyogenes, S. aureus, S. pneumoniae, E. faecalis, E. faecium and Group B Streptococci.116 Structurally endolysins against gram-positive bacteria have a cell wall binding (CBD) domain to help in enzyme diffusion and an enzymatically active domain (EAD) towards the N-terminal of lysins. EAD has three distinct catalytic modes of action on the host cell wall (Figure 3) which are (i) glycosidases which cleave β-1,4 glycosidic bonds, (ii) amidases which cleave the link between N-acetylmuramoyl residues and L-alanine amino acid residues in certain cell walls, and (iii) endopeptidases cleaving the peptide bond of stem amino acids.119–121 In contrast to that, most endolysins of gram-negative bacteria have only EAD and no CBD; but have an individual catalytic domain with terminal charged residues for binding. During the lytic cycle, phages produce holin protein which creates holes in the cytoplasmic membrane of host bacteria and ensures the delivery of lysins to the peptidoglycan layer.122 Whereas, for gram-negative hosts, an additional protein, spanin is required for outer membrane degradation.121
 |
Figure 3 Representation of the natural and engineered endolysins. For natural endolysins, the site of action for three major enzyme classes on bacterial cell wall is illustrated on the upper right-side box; the amidases cleave amide bond between N-acetyl muramoyl residues and L-alanine; the glycosidases target the β-1,4-glycosidic bond between NAM (N-acetylmuramic acid) and NAG (N-acetylglucosamine), whereas endopeptidases act on peptide bridges.117 In case of modified and engineered lysins, different approaches have been tried.118 Some examples of next generation approaches are—chimeric lysins—engineered by shuffling, eg HY-133 a recombinant lysin of N-terminal domain of phage K and cell wall binding domain of lysostaphin used against Staphylococcus aureus biofilms in vascular graft infections; artilysins—is a fusion protein of lysin and outer membrane destabilising peptide thus targeting both gram-positive and gram-negative organisms, eg Art-175; virion associated lysins (VALs) or peptidoglycan hydrolases are tail-associated muralytic enzymes, not containing own cell wall binding domain thus are fused with that of other endolysins, EC300 is one such example to target Enterococcus faecalis; truncated lysins—are modified proteins where cell wall binding domain is removed to increase activity, single-domain truncated enzyme, CHAPk is one such example. |
Because lysins have a narrow antibacterial spectrum range, they have an advantage over broad-spectrum antibiotics and are suitable for drug-resistant pathogenic bacteria without affecting the host microbiome much, thus inhibiting dysbiosis.123 Lysins are much less likely to show resistance, therefore they are seen as a more suitable alternative for antibiotic-resistant pathogens. In some recent reports, the efficacy of endolysin in treatment of biofilms is reviewed.121,124,125 Table 3 represents the list of endolysins and their use in mitigation of gram-positive and gram-negative bacteria-induced biofilms. Lysins are also thermostable to a certain degree, high ion intolerant bodies and have shown synergistic effects as well with antibiotics. The lysins can exchange catalytic and binding domains efficiently with other lysins, have independent activity, or can fuse with other antimicrobial peptides to develop new lysins, which may have enhanced bactericidal properties. They are also found in phage tail proteins or virion-associated lysins, working as receptor recognition proteins to break the cell wall locally and allowing the transfer of phage genomic material into host bacterial cells.148 In a recent review on “endolysins in a clinical setting” by Murray et al118 the development and applications of next generation lysins are elaborated. Compared to natural endolysins, these engineered lysins – chimeric lysin, artilysin, virion-associated lysin, truncated lysin will have improved thermostability, better bioavailability, protease-resistance, improved target infections, etc. Along with natural endolysins, the proposed bioengineering modifications for these lysins are also broadly represented in Figure 3, which is adapted from Murray et al.118
 |
Table 3 Major Endolysins, Their Target Pathogens and Potential Applications |
Bacteriophage Depolymerases
Depolymerases are the enzymes that bind and digest the polysaccharide components of the host bacteria’s cell wall, thus helping the phage in transferring its genome into the host cell. Depolymerases are divided into two types, hydrolases and lyases. Triacylglycerol lipase is a third type of depolymerase, that acts on the carboxylic ester bonds of triacylglycerols. These enzymes can be further subdivided based on their target polysaccharides which is either capsular polysaccharides, lipopolysaccharides (LPS), O-polysaccharides or exopolysaccharides of biofilms. Such depolymerases are also reported which break proteins or lipids present on the cell wall.149 Modes of action of depolymerases are very diverse due to the co-evolution of phages with their host bacteria which result in horizontal gene transfer.150 The enzyme can be found in free rapidly diffusible soluble form or it can be found attached to the phage particle. The soluble form which is released during the lysis of the host,148 is seen as “halo” or a circular bull’s eye outside the plaque in soft agar gel and is characteristic of capsular hydrolytic activity.151 Many times, phages attacking gram-negative bacteria use tail spikes that specifically bind to a particular capsule site and release depolymerase enzyme which initiates sequential cleavage of polymer bonds.152,153
The major advantages of depolymerases are their narrow spectrum of activity without causing dysbiosis, can be used against multidrug resistant bacteria, or recombinantly modified to improve activity. Interestingly, such depolymerases have been recognized, which degrade EPS in biofilm thus helping in penetration and targeting resident bacteria. KPO1K1, a depolymerase synthesized by lytic phage can specifically target K. pneumoniae B5055, which is a frequent cause of nosocomial infection. The enzyme decreased the size of the microcolony with subsequent alteration of even old biofilm matrix.149 Depolymerases can be used as tail-spike proteins (TSP) along with whole virions where biofilms are present as a barrier in the effective delivery of drugs during clinical infections. However, if the phages can pose safety problems in terms of the transfer of genes for encoding toxins and antibiotic resistance, then depolymerase proteins may be employed as more genetically stable, faster, safer, and effective options.
Bacteriophage or Phage-derived Enzymes as Bacterial Biofilm Mitigating Agents—Advantages Offered
Phage studies have given sufficient evidence for the versatile and potential application of phage therapy in the field of medical, agriculture and food processing.154 Phage therapy is mainly focused on the lytic phages since they completely rupture the bacterial cells. Being highly host-specific, they recognize only one bacterial species or sometimes one particular strain only. As phages are ubiquitous in all-natural environments; it is possible to isolate them against any target bacteria. These “green” alternatives also have a much less environmental impact compared to standard antibiotics.155 In addition, being viruses for bacteria, phages are unlikely to infect mammalian cells. A number of studies have shown that they have minimum or low inherent toxicity, no major side effects, or localized reactions.156,157 Another advantage is that phages will have minimum impact on commensal bacterial flora in contrast to broad-spectrum antibiotics.158
The ability of bacteriophages to penetrate the host cell can be used to treat the bacterial infection such as in the treatment of multidrug resistant bacterial infection or infections due to the development of biofilm around medical devices where it is hard to treat the infection due to the thick biofilm matrix. Using lytic phages also prevents the horizontal gene transfer because these phages lack the integrases and other enzymes which are mainly responsible for the horizontal gene transfer.159 The specificity of phages to counter any particular bacteria is generally limited to a single strain which also limits the strain bio-spreading. This property of phages can be problematic in the process of treatment of acute infections, where a swift response is required. However, these issues can be countered by using multiple phages at the same time. These mixtures of phages are called phage cocktails. As the different phages in a cocktail target different receptors on host bacteria, studies have reported that such cocktails delay the development of phage-resistant mutants. Moreover, these additional phages also work as puncture devices that lead to the lysis of bacterial cells,160 though incompatible interactions between phages are also possible in certain conditions.
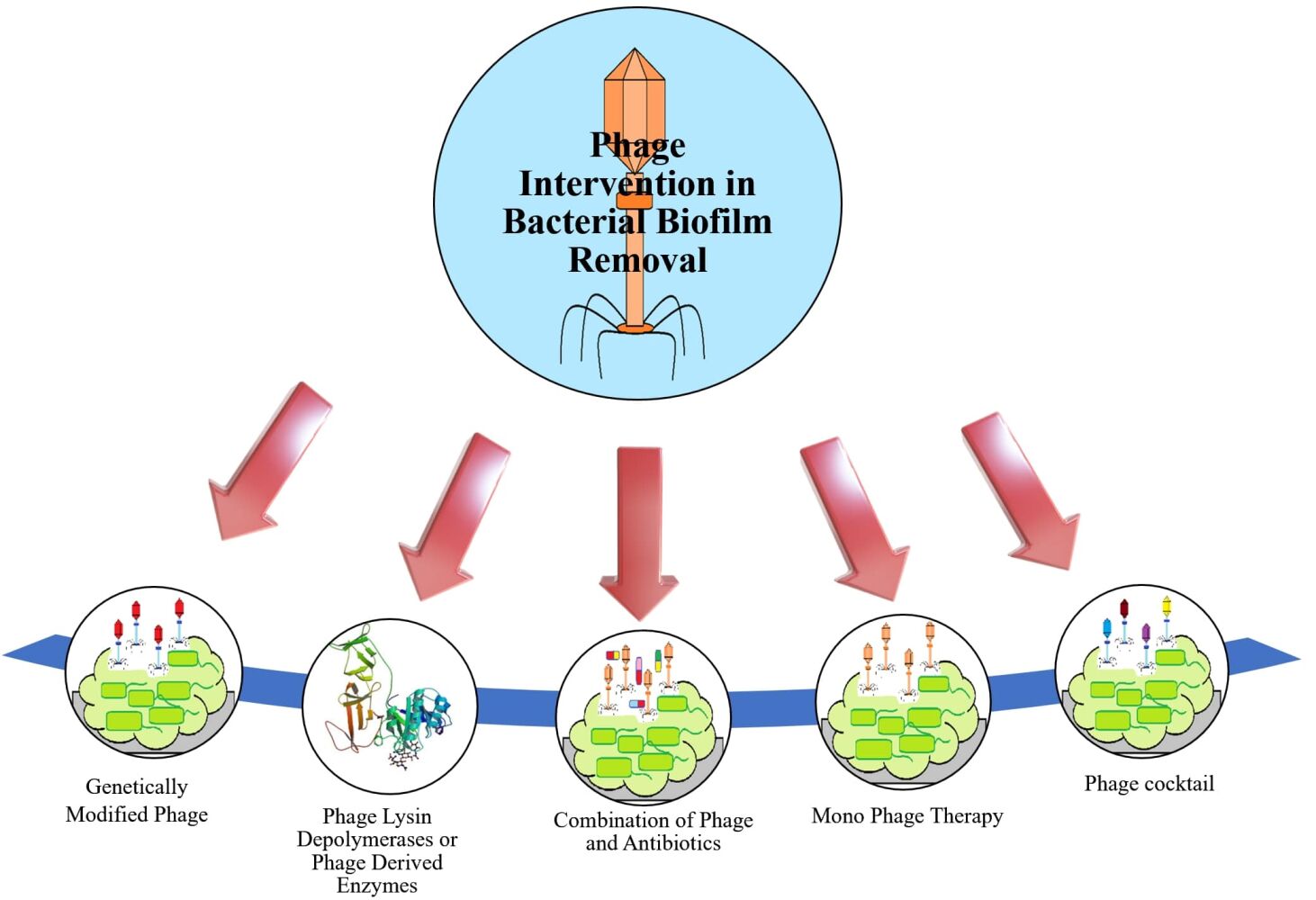
The applications and potential of phages or phage derived products for biofilm control have been reviewed in some recent works, in which multiple techniques of phage usage, like single phage dilution, multiphage cocktails, a phage derived proteins, a mixture of phages along with different antibiotics as well as genetically modified phages have been described as the mitigating strategy for biofilms.90,105,115,123,124,150,161,162 Most of the bacteria given in the WHO priority pathogens list,163 have been targeted by phage therapy in various ways (Figure 4). We present here some evidence-based data for all major approaches of phage therapy, namely, monophage, phage cocktail, a cocktail with antibiotics, phage-derived proteins, and genetically modified phages or enzymes, against common opportunistic and drug-resistant pathogens including ESKAPE in clinical settings.
 |
Figure 4 Bacteriophage therapies have shown promising results with WHO priority 1 and 2 pathogens. |
Evidence-based Data for Major Pathogens Using Monophage
Conventional phage therapy is using a single phage against the bacterial pathogen and multiple studies have reported successful results in ex vivo and in vivo settings. Some significant examples were oral and buccal biofilms of E. faecalis, F. nucleatum, Streptococcus mutans, or Aggregatibacter actinomycetemcomitans were treated with specific phages and survival of bacteria was studied. For E. faecalis, EFDG-1 phage brought a 5–7 log fold reduction in viable counts in an in vitro model;164 whereas the specific phage ΦAPMO1 brought complete inhibition of biofilm activity and 5 log10 reduction in viable counts for S. mutans.165 In another in vitro experiment, biofilms of A. actinomycetemcomitans, commonly associated with aggressive periodontitis and even infective endocarditis, were treated with AabΦO1, AabΦO1-1 which brought down viable count by 2.3 log10 without much affecting amount of biofilm.109 The synergistic effects of phage with antibiotics have been reported to cover a broader range of periodontopathogens.5
A recent work by Dakheel et al has reported two novel phages against MRSA biofilms. Two phages, UPMK-1 and UPMK-2 were isolated from lake and sewage water, using 25 biofilm-producing MRSA strains and both were assessed for biofilm degradation in in vitro and in situ settings. Though the biofilm was not completely removed even after 24 h treatment of phages in the microtiter plate assay method, there was an increase in phage titer reflecting the replication and propagation of phages within the biofilm.166 Another lytic, double-stranded DNA staphylophage, Stau2 was also identified and characterized, which was able to lyse 80% of clinical isolates (164/205) of S. aureus. Complete lysis within three hours of phage treatment was seen in most isolates and 100% protection was reported in experimental mice after lethal infection with S. aureus.152 Bacteriophages have shown good antimicrobial activity against biofilm-producing A. baumannii strains along with their therapeutic potential against drug-resistant A. baumannii in urinary tract infections.167
In a recent report by Monto et al, the efficacy of eight specific and novel phages was assessed for their biofilm disruption and susceptibility towards nine MDR E. coli O177 strains on artificially contaminated beef samples. A log reduction in viable E. coli cells was observed for seven days and during which two individual and three phage cocktails showed a reduction of cell counts below the detection limit (1.0 log10 CFU/g); whereas greater efficacy of phage cocktails than monophage was obtained in the destruction of preformed biofilms.168
Roach et al have used monophage therapy in the treatment of respiratory infection by P. aeruginosa (10 CFU). When this modeled monophage therapy, using PAK_P1 was used in the treatment of acute pneumonia by MDR P. aeruginosa in a mouse model, it provided the experimental basis to reveal that neutrophil-phage synergy is essential for resolution of pneumonia. This immunophage synergy led to the tolerance of these therapeutic phages by lung immune effector cells. It was also explained that phage resistance can emerge but does not result in therapeutic failure in healthy immunocompetent hosts. Treatment mainly fails when the host’s innate immune system fails to eradicate phage-resistant subpopulations.169
The phage TSK1 of Siphoviridae family has demonstrated a narrow host range against K. pneumoniae, including the MDR strains. TSK1 was able to remove 85–100% biomass in bacterial biofilms of different ages, whereas pretreatment by phage reduced >99% of K. pneumoniae biofilm within 24 h of incubation.170 In another study by Li et al, a recombinant of T4-like phage, WGqlae is designed by modifying the receptor specificity determinant region of gene 37 and has been found to lyse four additional E. coli hosts. WGqlae has demonstrated significant antimicrobial effects on E. coli DE192 and DE205B strains in planktonic tests, along with improved inhibitory effects on biofilm formation as well as clearance of mature biofilms.171 Similarly, Arumugam et al have identified 94 bacteriophage samples from sewage water against P. aeruginosa. These phages were able to infect 34 MDR P. aeruginosa strains too including hundred other pathogenic strains of P. aeruginosa with variable infectivity patterns, however, improved results were obtained when used in combinations.172 A novel lytic mycobacteriophage, PDRPxv is characterized by having a short latency period and a large burst size thus making it a promising therapeutic candidate against pathogenic Mycobacterium spp.173 The successful removal of P. aeruginosa biofilm by phage therapy in the mouse model of cystic fibrosis is another such example. In this study, an artificial sputum medium biofilm model and biofilm-associated murine model of chronic lung infection were developed to study the efficacy of a novel phage, PELP20 against P. aeruginosa LESB65 strain. After 24 h treatment of biofilm with phage, a 3-log reduction in P. aeruginosa CFU is reported. In addition, in the murine model studies, the novel phage showed complete clearance of bacteria after 48 h postinfection and also from the lungs of mice in established infection of six days.174
Evidence-based Data for Major Pathogens Using Phage Cocktail
Recent observations favor the in vivo use of phage cocktails against bacterial biofilms, in particular, with the multispecies biofilm. More than monophage therapy, the approach of using two or more phages ie, polyphage therapy is more useful because this gives additional advantages in terms of widening of host range, lesser chances of developing phage-resistant bacteria and targeting two or more pathogens. When more than one phage is used, each will have a different receptor for recognizing and attaching with bacteria. For instance, in a study by Alves et al, they have isolated six novel lytic phages against a wide range of P. aeruginosa clinical isolates. Three DL52, DL60, DL68 were identified to belong to Myoviridae, and the rest DL54, DL62 and DL68 of the Podoviridae family. This phage cocktail was assessed for biofilm-inhibiting activity in static and dynamic biofilm models. The six phages were able to eliminate 95% of biofilm biomass within four hours in the static model. Whereas in the dynamic biofilm model, the majority of the cells (>4 log) were eliminated after 48 h of phage cocktail treatment.175 In another in vivo study, a six phage cocktail was evaluated for its bactericidal activity in mice model with acute respiratory infection caused by P. aeruginosa. The cocktail of four novel phages (PYO2, DEV, E215, E217) and two previously identified phages (PAK_P1 and PAK_P4) was used against 40 clinical strains of P. aeruginosa, including isolates from Italian cystic fibrosis patients. The phage cocktail cured acute respiratory infection in mice and prevented bacteremia in Galleria mellonella larvae. This was found to be superior in terms of its ability to provide prophylaxis, target MDR or mucoid pseudomonal strains and was able to infect 97% of all P. aeruginosa strains tested.176 Another promising phage cocktail is EFDG1 and EFLK1, effective against planktonic and biofilm cultures of vancomycin-resistant Enterococci. The two phages together have shown improved activity profile against antibiotic resistant E. faecalis V583 strains and even phage-resistant mutant strain EFDG1r.164 However, an unknown and uncontrolled orientation of phages in a cocktail can be a potential drawback of this strategy.
A major pathogen in orthopedic and joint implant infections is Staphylococcus, specifically coagulase negative Staphylococcus or methicillin-resistant S. aureus (MRSA) in the majority of the cases. In one such study by Kaur et al, the combination of phage and linezolid was coated on naked wire and was surgically implanted in the intramedullary canal of mouse femur bone; along with hydroxypropyl methylcellulose (HPMC) coated and naked wire in other study groups. This was followed by inoculation of MRSA and results have shown that the combination of phage linezolid has the maximum reduction in adherence of MRSA and related inflammation. No emergence of the resistant mutant was also reported by the authors.177 In a phase I clinical trial, the safety profile and tolerability of phage cocktail was investigated in patients with recalcitrant chronic rhinosinusitis by S. aureus. Nine patients (median age 45) who received chronic rhinosinusitis due to S. aureus were divided into three cohorts and two intranasal irrigants of phage cocktail AB-SA01 at 3×108 PFU daily for seven days or 14 days or 3×109 PFU for 14 days. All groups completed the trial with no serious adverse effects and AB-SA01 was well-tolerated up to a concentration of 3×109 PFA for 14 days. No changes in body temperature and biochemical parameters were observed in all cohorts.178
Yilmaz et al have successfully demonstrated the reduction in P. aeruginosa and S. aureus biofilm formation by phage-treatment alone (2.3-fold) and improved results by an antibiotic-phage combination (8.6-fold). They have implanted a plastic intravenous catheter having established biofilm into rat tibial medullary canal and assessed all study group animals after two weeks for implant-associated osteomyelitis.179 In an MRSA osteomyelitis rabbit model, a cocktail mix of seven phages was tested by Kishor et al. These phages were isolated against clinical MRSA strains and were demonstrated to clear infection in active as well as chronic osteomyelitis within one week.180 Gibb and Hadjiargyrou, in an extensive review on phage therapy for bone and joint infections, have elaborated on its potential for orthopedic infections and the current status of in vitro, in vivo, and animal studies.181
For crystalline or non-crystalline biofilm-related urinary tract infections (UTIs), single phage or phage cocktails have been studied with promising results. For instance, when a phage cocktail containing three phages, 39APmC32, 65APm2833 and 72APm5211, were tested against 50 uropathogenic P. mirabilis strains related to catheter-associated urinary tract infections, a better biofilm destruction profile (2–3 strains more targeted compared to a single phage) was demonstrated, without inhibiting each other’s activity when used in gel or liquid form to wipe the surface of the catheter.182 As reported by Fu et al the effectiveness of a single phage against P. aeruginosa was found to be only for 24–48 h, after which the regrowth of biofilm and appearance of resistant strains was observed. However, when a five-phage cocktail was used in the same setting, a 99.9% reduction in biofilm formation and equally significant delay in the emergence of resistant strains was reported.19,183 Another important catheter-associated organism, P. mirabilis can also progress to complicated UTI infection due to the organism’s unique ability to form crystalline biofilms. In a detailed review on P. mirabilis biofilms associated with catheters, one of the potential documented approaches is phage therapy—either single, cocktail or with antibiotics. The phages against P. mirabilis biofilms were identified from the order Caudovirales, with the majority belonging to Siphoviridae, Podoviridae and Myoviridae lytic phages.184 In addition, a two-phage cocktail was used for P. mirabilis in a dynamic model simulating a catheter-associated biofilm. A significant reduction in the number of viable bacteria in 96 h after treatment with phage cocktail was demonstrated by electron microscopy.185 In another study, multidrug resistant P. mirabilis strains were 99.9% reduced by a mixture of five specific phages isolated from sewage water.186 A cocktail of six specific phages were used with a significant reduction in bacterial count in a mixed biofilm of P. aeruginosa and P. mirabilis.187
The phage cocktails studied for UPEC are reviewed in detail by Malik et al.188 In one such example, multidrug resistant E. coli was effectively removed (67% bacteriolytic activity) by T4 and KEP10 phage cocktails in an in vivo study using a mouse model.189 A three-phage cocktail showed 80% lytic activity when used against a subset of biofilm forming UPEC strains.190 In an in vivo study on burn wound infection in mice, a combination of five novel phages (Kpn1 – 5) against K. pneumoniae was used and bacterial load, wound contraction and histopathological analysis were done at different periods posttreatment. It was found that in the test group of phage cocktail, the bacterial load in skin tissue was reduced by 3 log CFU/mL on day three. The combination was also reported to give maximum protection against K. pneumoniae B5055 compared to any monophage group.191
Evidence-based Data for Major Pathogens Using Phages in Combination with Antibiotics
Antibiotics and phages work very differently on bacterial infection because phages are usually strain-specific, conversely antibiotics are multispectral drugs that treat a range of bacterial species. Still, phages have an advantage over antibiotics since they are strain-specific and can only target a single strain leaving the rest of the microflora untouched.192 In addition, the phage can emerge and mutate according to the stress and conditions given, which makes them better than the antibiotics because in the case of antibiotic use bacteria usually develop the resistance resulting in the restriction of antibiotic effect inside biofilms. In combating bacterial resistance, phage-antibiotic therapy has shown better results in multiple reports. It has also shown reduced chances of emergence of double resistance in bacterial pathogens, which means simultaneous resistance against antibiotics and phage is unlikely.193 Furthermore, the staggered application of antibiotics and phage cocktail have been found to be more effective in eradicating biofilms. In their latest peer-reviewed paper, Morrisette et al194 have discussed in detail the interactions between phage and antibiotics and the resultant effects on improved biofilm eradication, reduced bacterial growth, and alterations in bacterial resistance patterns.
The phenomenon of stimulation of host bacterial cell’s production of some virulent phage, after treatment with a sub-lethal concentration of antibiotics, is given the term phage-antibiotic synergy (PAS). It is shown that the production of ϕMFP phage is increased by more than seven-fold in uropathogenic E. coli strain when it is treated with a subinhibitory concentration of cefotaxime. Similar stimulation was seen in T4 like phages with β-lactam and quinolone antibiotics.195 The idea is to increase the combined bactericidal effect.
A successful study was conducted where the Sb-1 S. aureus phage was given in combination with antibiotics to effectively eradicate the biofilm matrix. In the study, the biofilm of methicillin resistant S. aureus (MRSA) ATCC 43300 was treated with Sb-1 alone or in combination with five antibiotics, namely fosfomycin, rifampin, vancomycin, daptomycin or ciprofloxacin. Pretreatment with Sb-1 followed by subinhibitory concentration of antibiotics resulted in the eradication of MRSA biofilm in a dose-dependent manner. The phage at a concentration of 107 PFU/mL had a direct killing effect on persister cells (5×105 CFU/mL).196 Combination therapy was also found effective when T4 phage was given in the combination with tobramycin or cefotaxime and resulted in a 99% reduction in antibiotic resistant cells and 39% reduction of phage resistant cells in E. coli biofilms.197 In another study, a 12-phage cocktail PP1131 in combination with the different antibiotics was studied for its effectiveness in the treatment of experimental endocarditis due to P. aeruginosa. A single dose phage destroyed 7 log PFU within six hours in vitro and the phage resistant mutants which grew after 24 h were effectively removed by phage combined with ciprofloxacin at 2.5× MIC. Whereas, the combination of phage cocktail (≥3 log10 CFU/mL) with ciprofloxacin led to successful treatment of 64% of experimental rats, demonstrating that phage-antibiotic cocktail was highly effective in the treatment of fibrin clots compared to the monophage-therapy with antibiotic or even with the phage cocktail alone.198 A similar test was conducted for the eradication of the P. aeruginosa biofilm using PB-1 bacteriophage and tobramycin combination which gave 60% and 99% reduction in antibiotic and phage-resistant cells respectively.197 In another study, the efficacy of Pseudomonas specific phages, NP1 and NP2, isolated from sewage water was tested against P. aeruginosa PA14 and its resistant mutants, PA14Cip-R and PA14Gen-R, which were resistant against ciprofloxacin and gentamicin respectively. The biofilms of P. aeruginosa PA14 were treated in vitro either with phage alone or a combination of two phages and five antibiotics; in which phage NP1 and NP2 in combination with different antibiotics were found to be more effective compared to the monophage treatment or treatment via antibiotic alone. It was also observed that this combined treatment is specifically effective in killing of Pseudomonas biofilms grown on cultured epithelial cell layers.199
Evidence-based Data for Major Pathogens Using Phage-derived Endolysin Enzyme
Reports of the efficacy of lysins in elimination of biofilms of gram-positive organisms have started appearing since 2007 when Φ11 endolysin was successfully used for reduction of S. aureus biofilm.200 In a detailed review on endolysins as a therapeutic alternative for drug-resistant pathogens, Gondil et al have presented several examples where endolysins have been reported to be effective against fatal infections caused by S. aureus, S. pneumoniae, S. pyogenes, Mycobacterium spp., E. coli, K. pneumoniae, P. aeruginosa, A. baumannii, etc.201 Lysins have been found to be effective for catheter-associated biofilm removal. S. aureus lysin CF-301 was found to have a strong antibiofilm effect on human synovial fluid which frequently forms a resistant biofilm during joint infections. The lysin also removed biofilms within one hour from other surfaces such as surgical mesh, catheters and polystyrene. Apart from S. aureus, it was effective against coagulase negative S. aureus, Streptococcus pyogenes, Streptococcus agalactiae as well as effectively killing persister cells of S. aureus. Endolysins showed better activity against suspended S. aureus biofilm compared to intact or scraped biofilms.185 The activity of CF-301 was much improved by using it with lysostaphin, a cell wall hydrolase.202 Streptococcus uberis, a common cause of bovine mastitis, was effectively killed in vitro by two endolysins PlySs2 and PlySs9 isolated from Streptococcus suis serotype −2 and −9 prophage respectively. Of the two, PlySs9 was reported more potent than PlySs2, with lower MIC and one log higher killing.203
Though the outer membrane in gram-negative bacteria resists the action of endolysins from outside, many novel lysins have been studied and reported with specific and improved killing actions. P. aeruginosa, cause of nosocomial infections by this gram-negative MDR strains affecting the urinary tract, surgical wound as well as blood infections due to catheters and implants to have been a cause of high morbidity and mortality. PlyPa03 and PlyPa91 are two promising lysins that have shown good bactericidal properties against many clinical and biofilm/embedded strains of P. aeruginosa. PlyP91 has shown efficacy in P. aeruginosa skin infection and PlyPa03 has been reported to be effective in P. aeruginosa pneumonia, on mouse model in vivo studies.136 Lys PA26 is a novel endolysin that is used in P. aeruginosa biofilm disruption without the need to add outer membrane permeabilizers. This has also been found to be effective against A. baumannii, K. pneumoniae, and E. coli up to a temperature range of 50°C.129
Some of them have been studied in vivo too in animal models. Lood et al have for the first time shown the in vivo efficacy of gram-negative lysin Ply307, against multidrug resistant A. baumannii. Twenty-one distinct lysins were identified from prophages induced from 13 A. baumannii strains. Ply307 brought >5 log unit decrease in A. baumannii isolates, also significantly reducing planktonic and biofilm organisms in vitro as well as in vivo. The efficacy of PlyF307 was tested against A. baumannii biofilms formed on PVC catheters. The Scanning Electron Microscopy has shown that much of the EPS of biofilm on these catheters were destroyed.127 LysAB2 lysin isolated from A. baumannii ΦAB2 showed broad bactericidal activity against MDR A. baumannii, E. coli as well as S. aureus. The antimicrobial peptides from the C-terminal of LysAB2 were also found to be bactericidal for A. baumannii both in vitro and in vivo. Some other A. baumannii specific lysins are lysABP-01, PlyAB1, Ply6A3, ABgp46 which have shown promising results in vitro against several MDR A. baumannii strains.125 The phage lysin ABgp46 was additionally active against P. aeruginosa and Salmonella enterica; whereas Ply6A3 lysin showed a 70% bactericidal action against MDR A. baumannii and protected the experimental mouse model from a lethal A. baumannii septic challenge.131
Other gram-negative pathogens which have been targeted by lysins or their recombinant improved versions are E. coli, K. pneumoniae, Burkholderia cenocepacia, and Salmonella enterica. Lysin KP27 against drug resistant K. pneumoniae, EndoT5, and Lysep3 lysins coliphages against E. coli are few such examples that have been shown to be active even in the presence of different outer membrane factors. The recombinant version of Lysep3 fused with D8 domain has shown improved activity.204 Lastly, “amurins” which are a group of phage-encoded antimicrobial peptides, have shown remarkable bactericidal activity against gram-negative pathogens as well as for the removal of biofilm embedded bacteria. In a proof-of-concept study, the peptide App2-MI amurin was tested Stenotrophomonas maltophilia biofilms on hemodialysis catheters. The amurin was found to be effective at a concentration of 1 µg/mL in eradicating the biofilm.125 Table 3 gives a list of endolysins identified from different phages and targeted against common gram-positive and gram-negative pathogenic bacteria.
Evidence-based Data for Major Pathogens Using Phage-derived Depolymerase Enzyme
Among some of the significant examples for this enzyme is depolymerase Dpo7, derived from vB_SepiS-phiPLA7 phage, which has shown the reduction of Staphylococcus spp. biofilm biomass by 53–85% in 67% of the bacterial strains tested in a dose-dependent; but time-independent response.205 Another test based on the biofilm disruptive activity of depolymerase is Dpo42, derived from phage vB_EcoM_ECOO78. Its antibiofilm activity was tested against multiple bacteria like E. coli where it showed dose-dependent biofilm prevention activity.129 Depolymerase Dpo1, isolated from the phage Petty, was found to be effective in removing biofilms formed by Acinetobacter strains. The enzyme was able to reduce EPS viscosity, thus reducing the virulence of tested strains; however, it was not very effective in destroying Acinetobacter biofilms as only 20% reduction in that was observed.206 IME180 is another phage carrying a functionally active depolymerase gene, against P. aeruginosa biofilm. The purified enzyme inhibited bacterial biofilm formation and reduced preformed biofilm biomass.207 In a study by Olsen et al, lysin LysK and depolymerase DA7 have been tested in combination against S. aureus biofilms in static as well as in dynamic models, where they showed synergistic behavior, dropping the number of viable cells in biofilm.208 Another depolymerase enzyme Dep42, specific for K. pneumoniae is identified in bacteriophage SH-KP152226 of Podoviridae family. Dep42 was demonstrated to cause lysis of capsular K. pneumoniae. In biofilms, the depolymerase caused degradation of EPS causing the removal of attached cells. This may prove useful in combining it with other antimicrobial agents to effectively remove biofilms.131
The constitution of biofilm, whether single or multispecies also affects the depolymerase effectiveness in removing biofilms. As mixed bacterial communities are present mostly in naturally occurring biofilms than a single species, the use of a single depolymerase may have limited effect. The different types of EPS in such mixed communities may limit the penetration of antibiofilm agents through biofilms or may even trap phage in biofilm matrix or reduce their multiplication due to the presence of metabolically inactive cells or reduced availability of receptors or inhibited depolymerase activity.209 In addition, there can be huge heterogeneity in the polysaccharide content of EPS layers of bacterial biofilms; thus, a depolymerase that is effective for breaking cell wall polysaccharides may not be effective for degrading EPS layers. This may limit the range of polysaccharides that can be targeted by a specific depolymerase, even the cell surface polysaccharide of closely related bacterial hosts may not be recognized.210 Thus, a combination therapy using two or more agents has also been studied with promising results. In this approach, a phage derived enzyme depolymerase is mixed with different active antimicrobial agents like antibiotics, phages, lysins, detergents, chemicals or natural compounds to improve the efficacy. For example, Dpo42 mixed with polymyxin and KPO1K2 depolymerase mixed with gentamycin have given much improved results in reducing K. pneumoniae bacterial biofilms.131 Similarly, a combination of depolymerase with honey and EC3a phage with depolymerase activity against E.coli biofilm153 have more efficient antibiofilm activity than phage or its enzyme alone. Another strategy is to combine alginate lyase with antibiotics for targeting P. aeruginosa in biofilms. The mucoid strains of P. aeruginosa frequently produce alginate which resists antibiotic penetration thus presents a problem in managing drug-resistant and opportunistic P. aeruginosa. The effective killing of P. aeruginosa in biofilms by combining antibiotics with alginate lyase as an adjuvant has been reported. This effect of alginate lyase in dispersing through biofilm was independent of antibiotic action.211 Many more such examples can be obtained in the review on bacteriophage depolymerases by Topka-Bielecka et al.30
Evidence-based Data for Major Pathogens Using Genetically Modified Phage or Phage-derived Enzymes
Phages can be genetically modified to increase their host range and also to increase their survival in the biofilm matrix, eg a modified T7 E. coli phage was designed to express intracellularly a hydrolase, which is released during infection to the extracellular matrix, enhancing the biofilm degradation. This test showed 99% eradication of E. coli biofilm.212 Bacteriophage K1F-GFP is another engineered phage, using CRISPR/Cas selection method, is targeted specifically against E. coli K1, a common nosocomial pathogen for urinary tract infections. The engineered fluorescent phage, K1F-GFP, and E. coli EV36-RFP enter cells via phagocytosis, phage efficiently kills intracellular E. coli from T24 human urinary bladder epithelial cells, and finally, both are degraded by phagocytosis and xenophagy.213 In a study done by Tinoco et al, a lysogenic _Ef11 E. faecalis phage was genetically modified to eliminate all the genes related to lysogeny, eliminating transduction problem and achieving a noteworthy drop in the population of E. faecalis biofilms, wherein vancomycin-resistant as well as sensitive strains were used.159
One of the most important characters of genetically modified phages is their ability to infect multiple hosts. For example, a T7 coliphage was modified by the insertion of a sequence for 1080. It is a short peptide with a broad spectrum antibiofilm effect, making it a broad spectrum T7 phage.214 Temperate phages may be of such interest for delivering programmable DNA nucleases associated with CRISPR to reverse antibiotic resistance. This system can selectively annihilate plasmids that confer antibiotic resistance.215
In addition, lysins have also been engineered to modify their target specificity. As an example, chimeric lysin Cs12, obtained by fusion of the catalytic domain of Cp1-7 lysozyme to the CW-7 repeats of the LySMP lysin from a Staphylococcus spp. have increased the target specificity. It was designed to remove Staphylococcus spp. biofilm in vitro and validated in vivo with a zebrafish infection model.216 Another chimeric lysin P128 showed bactericidal activity against S. aureus and 99% reduction effects on S. epidermidis and Staphylococcus haemolyticus biofilms.217 The removal of biofilms in endocarditis and catheter associated infections is reported from an animal model study by this novel recombinant chimeric ectolysin, P128 which has potential antistaphylococcal activity.218 In the study, BALB/c mice were injected with a single dose of P128. The ectolysin exerted a quick bactericidal effect and inhibited the fatal MRSA and VRSA invasion in the test group.219 P128 ectolysin has also shown potent antibiofilm activity on planktonic and biofilm embedded cells. The effect was seen for both resistant and sensitive isolates of S. aureus and also in synergy with standard-of-care antibiotics.220 P128 has also completed a Phase II trial for safety and efficacy in healthy volunteers and patients (https://clinicaltrials.gov/ct2/show/NCT01746654).
In another work by Singh et al, Plt187AN-KSH3b, a chimeric phage endolysin derived from Ply187 was evaluated for its therapeutic potential against ocular infections, specifically S. aureus endophthalmitis in a mouse model. They have reported strong antimicrobial activity of the chimeric endolysin against methicillin-sensitive S. aureus and MRSA, both without any resistance development.221 A novel chimeric lysin, ClyV, is constructed against S. agalactiae by fusing the EAD domain of PlyGBS lysin with the CBD domain of PlyV12 lysin. The engineered lysin has shown better bactericidal activity than the parental enzyme and a single intraperitoneal dose of 0.1 mg of ClyV was able to provide 100% protection against S. agalactiae infection in experimental mouse models, thus demonstrating the potential role of this chimeric lysin against antibiotic resistant beta hemolytic streptococcal infections.222 The engineered LysH5 lysin was studied for its potential to kill S. aureus persister cells with 100% efficacy at a minimum concentration of 0.15 µM.205
A recombinant version of CF-301, Exebacase a potent antistaphylococcal lysin with many improved features of lower resistance and synergy with conventional antibiotics, is the first direct lytic agent (DLA) clearing phase 2 clinical trial. In this clinical study done on 121 patients with S. aureus bloodstream infection or endocarditis, a single dose of exebacase given with standard antibiotics has caused a 43% point higher clinical response rate in the MRSA subgroup. This is the first proof of concept study for the therapeutic potential of exebacase for MRSA caused bloodstream infections and endocarditis.223 Watson et al have reported synergistic activity of exebacase with 12 broad range antibiotics.224
Many examples of recombinantly designed lysins with improved activity against gram-negative bacteria are also being reported. For example, Art-175 lysin is a fusion protein that contains lysin fused to an outer membrane destabilizing peptide and is active against both gram-negative and gram-positive bacteria.135 It has also shown the independent activity against the biofilms of multidrug resistant P. aeruginosa and caused the osmotic lysis in bacterial metabolism. This provides evidence that even at low metabolic rates the lysins can disrupt persistent bacteria within the biofilm.225 Lysocins are bioengineered lysin-bacteriocin fusion molecule which can translocate through the outer membrane of P. aeruginosa. This proof-of-concept study by Haselpoth et al has shown efficient peptidoglycan cleavage, the log-fold killing of P. aeruginosa, effective biofilm disruption, and no toxicity, thus outperforming standard-of-care antibiotics.226
Challenges in Phage Therapy and Future of Phage Research
Bacteriophages have been studied for a long time to treat infections in humans, however, bacteria also have different strategies to prevent infection of phages, namely, blocking surface receptors, alter mechanism, secrete extracellular polymeric capsule, quorum sensing, secretion of phage inactivating enzymes, presence of EPS matrix, etc. Thus, phage-bacteria interactions in a biofilm must be understood completely to get successful outcomes. Factors such as the metabolic activity of bacteria in biofilm, the density of biofilm, old or aged biofilm are some important aspects that govern whether bacteriophage can penetrate the biofilm or not and thus must be considered to achieve greater efficacy. In a recent work by Simmons et al, the coexistence of phage resistant and phage susceptible bacteria and dynamics of phage resistance in a biofilm is demonstrated, using resistant and susceptible E. coli strains. In these in vitro and simulation studies on the spatial structure of a biofilm, they have reported that at initial stages when phage is introduced, and phage-resistant bacteria are rare, large interaction of phages with susceptible cells leads to killing of host cells. Space thus created is occupied by resistant cell clusters; whereas when resistant bacteria are common at the time of addition of phage, they make a barrier for the phages to reach a susceptible host, and diffusion is impeded. That is how the susceptible cells are shielded by resistant cells. This barrier effect of resistant cells is an important factor to be considered for phage therapy efficacy and for it to be successful, the biofilm architecture must be disrupted for exposing target bacteria to phages.102
Moreover, in a multispecies biofilm matrix, the comparative health depends upon the amplification of phage within the biofilm matrix as well as on the availability of nutrients and oxygen to different bacterial species. The study by Harcombe and Bull suggested that multiple species composition of biofilm can influence the proliferation, penetration of phage inside biofilm matrix thus affecting the phage therapy.227 Usually, lab studies are focused on the single species biofilm matrices where it is hard to determine the success of bacteriophage infection inside the biofilm matrix. This leads to a constant debate on the usefulness of phages in targeting the bacterial population. Even though phages have been reported to be safe and no side effects have been documented, still it is hypothesized that phages should be administered locally at the site of infection and not systemically.228 There are almost no studies that have compared local vs systemic administration of phages or quantification of anti-phage antibiotics in biofilm infections. Therefore, for their effective clinical applications, extensive in vivo studies should be done for assessing the response of plasma proteins and components of the host immune system.30 Figure 5 reproduced with permission from Hassan et al depicts the friend and foe roles of bacteriophages, in which as bacteriophriend they have potential in phage therapy and biofilm removal, whereas bacteriophage as foe can be a vehicle for transfer of antibiotic resistance and other virulent factors.99
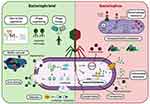 |
Figure 5 Bacteriophage—Friend vs Foe. The impact of bacteriophages with respect to antimicrobial resistance are summarized, depicting the beneficial applications on the left and the potential risks on right. Notes: Reproduced from Hassan AY, Lin JT, Ricker N, Anany H. The age of phage: Friend or foe in the new dawn of therapeutic and biocontrol applications? Pharmaceuticals. 2021;14(3):1–36. doi:10.3390/ph14030199.99 Copyright: © 2021 by the Her Majesty the Queen in Right of Canada as represented by the Minister of Agriculture and Agri-Food Canada; Licensee MDPI, Basel, Switzerland. Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/). |
Although phages are highly versatile and stable in storage229 and have been tried in all forms, such as liquids, creams or impregnated solids via different administration routes like oral, intramuscular, intravenous, topical, or superficial;157 therapeutic limitations of bioavailability, rapid clearance or non-targeted delivery have been reported. To overcome that, encapsulation of phage or endolysins into a suitable drug delivery system is an alternate approach that is being extensively explored.230 Lastly, though several clinical trials have been documented until now for chronic wound infections231 or infectious complications of surgical wounds, burn infections and infected ulcers;232 none of them were able to support completely the in vitro and in vivo studies. Górski et al have summarized some of the recent clinical trials and critically analyzed the reasons for the failure of these studies which were mainly due to insufficient phage coverage or overgrowth of other bacteria. According to them, the major focus points, which must be considered before initiating a clinical trial, should be quality (taking a well-characterized phage), titer of phage preparation (>106/mL of phage titer is recommended) and patient’s antibody response during phage therapy.233
Conclusion
With the constant rise of incidences of hospital-acquired and community-acquired infections by common pathogens; the need for alternate therapies is increasing day by day. Phage therapy is gaining attention in clinical medicine as we are getting more and more insight into phage mechanisms. For example, in a recent report, Yan et al have demonstrated incorporation of A. baumannii specific phage into a hydrogel, which has shown a 90% reduction in microbial survival after four hours of the treatment in an ex vivo wound infection model using pig skin.234 A Staphylococcal bacteriophage, (monophage Sb-1) has been used to treat foot ulcers in patients with osteomyelitis, as well as PYO bacteriophage, a complex preparation designed to treat wound infection is also available commercially in the market.235 Another potential approach is phage-loaded biocompatible nanofibers for wound dressings.236 Similarly, some phage-based compounds to treat biofilms, have also emerged in the market and they have shown favorable results. In a recent work, two commercially available formulations of phage namely monophage Sb-1 and PYO (a polyphage) were evaluated for their eradication potential on in vitro biofilms of MRSA and also in a G. mellonella model of MRSA systemic infection for in vivo experiments. The co-incubation with the highest phage titers (107 PFU/mL) was effective in the eradication of MRSA biofilm within seven days, along with preventing MRSA infection and increasing survival rate in G. mellonella larvae.237
Due to their specificity and narrow spectrum activity, bacteriophages provide an attractive alternative to overcome the increasing antimicrobial resistance,238 but not many promising results are coming out for their role in mitigating biofilms of medically important microbes. This may be due to the fact that most of the studies for bacteriophage or their derived proteins are done in vitro which have helped us to understand their role and ultimately their application in static environments, but a better understanding of phage dynamics, phage proteins, phage biology, and ecology is needed. Combining laboratory experimental data with newer approaches like metagenomics (especially “viromics”), empirical studies, bioengineering, mathematical models239 and computational studies have been helpful. However, it is important that we must have a robust clinical trial design and understanding of mechanisms involving co-evolution of bacteria and phage.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
The author Dr. Aditi Singh is thankful to Science and Engineering Research Board (SERB), a statutory body of the Department of Science and Technology (DST), Govt. of India for the financial support of TARE research grant (TAR/2019/000140). The author Dr. Manish Dwivedi acknowleges the financial support of DST-INSPIRE Faculty Award, Govt. of India (2017).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Rode DKH, Singh PK, Drescher K. Multicellular and unicellular responses of microbial biofilms to stress. Biol Chem. 2020;401(12). doi:10.1515/hsz-2020-0213
2. Flemming H-C. Biofilms. In: Encyclopedia of Life Sciences. John Wiley & Sons, Ltd; 2008. doi10.1002/9780470015902.a0000342.pub2
3. Hutchings M, Truman A, Wilkinson B. Antibiotics: past, present and future. Curr Opin Microbiol. 2019;51. doi:10.1016/j.mib.2019.10.008
4. Banerjee A, Batabyal K, Singh AD, et al. Multi-drug resistant, biofilm-producing high-risk clonal lineage of Klebsiella in companion and household animals. Lett Appl Microbiol. 2020;71(6). doi:10.1111/lam.13381
5. Szafrański SP, Winkel A, Stiesch M. The use of bacteriophages to biocontrol oral biofilms. J Biotechnol. 2017;250:29–44. doi:10.1016/j.jbiotec.2017.01.002
6. Abebe GM. The Role of bacterial biofilm in antibiotic resistance and food contamination. Int J Microbiol. 2020;2020. doi:10.1155/2020/1705814
7. Human Microbiome Project Consortium T. Structure, function and diversity of the healthy human microbiome The Human Microbiome Project Consortium*; 2012. doi:10.1038/nature11234
8. Kriebel K, Hieke C, Müller-Hilke B, Nakata M, Kreikemeyer B. Oral biofilms from symbiotic to pathogenic interactions and associated disease - Connection of periodontitis and rheumatic arthritis by peptidylarginine deiminase. Front Microbiol. 2018;9(JAN). doi:10.3389/fmicb.2018.00053
9. Caton JG, Armitage G, Berglundh T, et al. A new classification scheme for periodontal and peri-implant diseases and conditions – introduction and key changes from the 1999 classification. J Clin Periodontol. 2018;45. doi:10.1111/jcpe.12935
10. How KY, Song KP, Chan KG. Porphyromonas gingivalis: an overview of periodontopathic pathogen below the gum line. Front Microbiol. 2016;7(FEB):53. doi:10.3389/fmicb.2016.00053
11. Chatzigiannidou I, Teughels W, Van de Wiele T, Boon N. Oral biofilms exposure to chlorhexidine results in altered microbial composition and metabolic profile. Npj Biofilms Microbiomes. 2020;6(1). doi:10.1038/s41522-020-0124-3
12. van Belleghem JD, Manasherob R, Miȩdzybrodzki R, et al. The rationale for using bacteriophage to treat and prevent periprosthetic joint infections. Front Microbiol. 2020;11. doi:10.3389/fmicb.2020.591021
13. Chatterjee S, Maiti PK, Dey R, Kundu AK, Dey RK. Biofilms on indwelling urologic devices: microbes and antimicrobial management prospect. Ann Med Health Sci Res. 2014;4(1):100–104. doi:10.4103/2141-9248.126612
14. Cortese YJ, Wagner VE, Tierney M, Devine D, Fogarty A. Review of catheter-associated urinary tract infections and in vitro urinary tract models. J Healthc Eng. 2018;2018:1–16. doi:10.1155/2018/2986742
15. Feneley RCL, Hopley IB, Wells PNT. Urinary catheters: history, current status, adverse events and research agenda. J Med Eng Technol. 2015;39(8):459–470. doi:10.3109/03091902.2015.1085600
16. Jordan RP, Malic S, Waters MG, Stickler DJ, Williams DW. Development of an antimicrobial urinary catheter to inhibit urinary catheter encrustation. Microbiol Discov. 2015;3(1):1. doi:10.7243/2052-6180-3-1
17. Ceri H, Olson ME, Stremick C, Read RR, Morck D, Buret A. The Calgary biofilm device: new technology for rapid determination of antibiotic susceptibilities of bacterial biofilms. J Clin Microbiol. 1999;37(6):1771–1776. doi:10.1128/jcm.37.6.1771-1776.1999
18. Olsen I. Biofilm-specific antibiotic tolerance and resistance. Eur J Clin Microbiol Infect Dis. 2015;34(5):877–886. doi:10.1007/s10096-015-2323-z
19. Uruén C, Chopo-Escuin G, Tommassen J, Mainar-Jaime RC, Arenas J. Biofilms as promoters of bacterial antibiotic resistance and tolerance. Antibiot. 2020;10(1):1–36. doi:10.3390/antibiotics1001000321
20. Santajit S, Indrawattana N. Mechanisms of antimicrobial resistance in ESKAPE pathogens. Biomed Res Int. 2016;2016. doi:10.1155/2016/2475067
21. Antimicrobial resistance. Available from: https://www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance.
22. Nelson RE, Hatfield KM, Wolford H, et al. National estimates of healthcare costs associated with multidrug-resistant bacterial infections among hospitalized patients in the United States. Clin Infect Dis. 2021;72(Supplement_1):S17–S26. doi:10.1093/cid/ciaa1581
23. Global action plan on antimicrobial resistance. Available from: https://www.who.int/publications/i/item/9789241509763.
24. Manohar P, Tamhankar AJ, Lundborg CS, Nachimuthu R. Therapeutic characterization and efficacy of bacteriophage cocktails infecting Escherichia coli, Klebsiella pneumoniae, and Enterobacter species. Front Microbiol. 2019;10(MAR):574. doi:10.3389/fmicb.2019.00574
25. Moghadam MT, Amirmozafari N, Shariati A, et al. How phages overcome the challenges of drug resistant bacteria in clinical infections. Infect Drug Resist. 2020;13:45–61. doi:10.2147/IDR.S234353
26. Gutiérrez D, Fernández L, Rodríguez A, García P. Are phage lytic proteins the secret weapon to kill Staphylococcus aureus? mBio. 2018;9(1):e01923–e02017. doi:10.1128/mBio.01923-17
27. Sharahi JY, Azimi T, Shariati A, Safari H, Tehrani MK, Hashemi A. Advanced strategies for combating bacterial biofilms. J Cell Physiol. 2019;234(9):14689–14708. doi:10.1002/jcp.28225
28. Renner LD, Weibel DB. Physicochemical regulation of biofilm formation. MRS Bull. 2011;36(5). doi:10.1557/mrs.2011.65
29. O’Toole G, Kaplan HB, Kolter R. Biofilm formation as microbial development. Annu Rev Microbiol. 2000;54. doi:10.1146/annurev.micro.54.1.49
30. Topka-Bielecka G, Dydecka A, Necel A, et al. Bacteriophage-derived depolymerases against bacterial biofilm. Antibiotics. 2021;10:175–196. doi:10.3390/antibiotics10020175
31. Rabin N, Zheng Y, Opoku-Temeng C, Du Y, Bonsu E, Sintim HO. Biofilm formation mechanisms and targets for developing antibiofilm agents. Future Med Chem. 2015;7(4). doi:10.4155/fmc.15.6
32. Behzadi P. Classical chaperone-usher (CU) adhesive fimbriome: uropathogenic Escherichia coli (UPEC) and urinary tract infections (UTIs). Folia Microbiol (Praha). 2020;65(1):45–65. doi:10.1007/s12223-019-00719-x
33. Behzadi P. Uropathogenic Escherichia coli and fimbrial adhesins virulome. Urin Tract Infect. 2017. doi:10.5772/INTECHOPEN.71374
34. Werneburg GT, Thanassi DG. Pili assembled by the chaperone/usher pathway in Escherichia coli and salmonella. EcoSal Plus. 2018;8(1). doi:10.1128/ecosalplus.ESP-0007-2017
35. Baby S, Kumar Karnaker V. Adhesins of uropathogenic Escherichia coli (UPEC). IP Int J Med Microbiol Trop Dis. 2021;2(1):10–18.
36. Subashchandrabose S, Mobley HLT. Virulence and fitness determinants of uropathogenic Escherichia coli. Microbiol Spectr. 2015;3(4). doi:10.1128/MICROBIOLSPEC.UTI-0015-2012
37. Ribić R, Meštrović T, Neuberg M, Kozina G. Effective anti-adhesives of uropathogenic Escherichia coli. Acta Pharm. 2018;68(1):1–18. doi:10.2478/ACPH-2018-0004
38. Wurpel DJ, Totsika M, Allsopp LP, Webb RI, Moriel DG, Schembri MA. Comparative proteomics of uropathogenic Escherichia coli during growth in human urine identify UCA-like (UCL) fimbriae as an adherence factor involved in biofilm formation and binding to uroepithelial cells. J Proteomics. 2016;131:177–189. doi:10.1016/J.JPROT.2015.11.001
39. Hozzari A, Behzadi P, Kerishchi Khiabani P, Sholeh M, Sabokroo N. Clinical cases, drug resistance, and virulence genes profiling in Uropathogenic Escherichia coli. J Appl Genet. 2020;61(2):265–273. doi:10.1007/s13353-020-00542-y
40. Jahandeh N, Ranjbar R, Behzadi P, Behzadi E. Uropathogenic Escherichia coli virulence genes: invaluable approaches for designing DNA microarray probes. Cent Eur J Urol. 2015;68(4):452–458. doi:10.5173/ceju.2015.625
41. Foster TJ. Surface proteins of Staphylococcus epidermidis. Front Microbiol. 2020;11. doi:10.3389/fmicb.2020.01829
42. Ferreira A, Gray M, Wiedmann M, Boor KJ. Comparative genomic analysis of the sigB operon in Listeria monocytogenes and in other Gram-positive bacteria. Curr Microbiol. 2004;48(1):39–46. doi:10.1007/S00284-003-4020-X
43. Vorkapic D, Pressler K, Schild S. Multifaceted roles of extracellular DNA in bacterial physiology. Curr Genet. 2016;62(1):71–79. doi:10.1007/S00294-015-0514-X
44. Arciola CR, Campoccia D, Montanaro L. Implant infections: adhesion, biofilm formation and immune evasion. Nat Rev Microbiol. 2018;16(7):397–409. doi:10.1038/S41579-018-0019-Y
45. Boles BR, Horswill AR. agr-mediated dispersal of staphylococcus aureus biofilms. PLOS Pathog. 2008;4(4):e1000052. doi:10.1371/JOURNAL.PPAT.1000052
46. Xue T, Ni J, Shang F, Chen X, Zhang M. Autoinducer-2 increases biofilm formation via an ica- and bhp-dependent manner in Staphylococcus epidermidis RP62A. Microbes Infect. 2015;17(5):345–352. doi:10.1016/J.MICINF.2015.01.003
47. Patel JD, Colton E, Ebert M, Anderson JM. Gene expression during S. epidermidis biofilm formation on biomaterials. J Biomed Mater Res Part A. 2012;100A(11):2863–2869. doi:10.1002/JBM.A.34221
48. Lister JL, Horswill AR. Staphylococcus aureus biofilms: recent developments in biofilm dispersal. Front Cell Infect Microbiol. 2014;4(DEC). doi:10.3389/FCIMB.2014.00178
49. Wang R, Khan BA, Cheung GYC, et al. Staphylococcus epidermidis surfactant peptides promote biofilm maturation and dissemination of biofilm-associated infection in mice. J Clin Invest. 2011;121(1):238. doi:10.1172/JCI42520
50. Kostakioti M, Hadjifrangiskou M, Hultgren SJ. Bacterial biofilms: development, dispersal, and therapeutic strategies in the Dawn of the postantibiotic era. Cold Spring Harb Perspect Med. 2013;3(4). doi:10.1101/cshperspect.a010306
51. Sarshar M, Behzadi P, Scribano D, Palamara AT, Ambrosi C. Acinetobacter baumannii: an ancient commensal with weapons of a pathogen. Pathogens. 2021;10(4). doi:10.3390/pathogens10040387
52. Geisinger E, Huo W, Hernandez-Bird J, Isberg RR. Acinetobacter baumannii: envelope determinants that control drug resistance, virulence, and surface variability. Annu Rev Microbiol. 2019;73:481–506. doi:10.1146/annurev-micro-020518-115714
53. Tomaras AP, Flagler MJ, Dorsey CW, Gaddy JA, Actis LA. Characterization of a two-component regulatory system from Acinetobacter baumannii that controls biofilm formation and cellular morphology. Microbiology. 2008;154(11):3398–3409. doi:10.1099/MIC.0.2008/019471-0
54. Eze EC, Chenia HY, El Zowalaty ME. Acinetobacter baumannii biofilms: effects of physicochemical factors, virulence, antibiotic resistance determinants, gene regulation, and future antimicrobial treatments. Infect Drug Resist. 2018;11:2277–2299. doi:10.2147/IDR.S169894
55. Colquhoun JM, Rather PN. Insights into mechanisms of biofilm formation in Acinetobacter baumannii and implications for uropathogenesis. Front Cell Infect Microbiol. 2020;10. doi:10.3389/fcimb.2020.00253
56. Harding CM, Pulido MR, Di Venanzio G, et al. Pathogenic Acinetobacter species have a functional type I secretion system and contact-dependent inhibition systems. J Biol Chem. 2017;292(22):9075–9087. doi:10.1074/JBC.M117.781575
57. Scavone P, Iribarnegaray V, Caetano AL, Schlapp G, Härtel S, Zunino P. Fimbriae have distinguishable roles in Proteus mirabilis biofilm formation. Pathog Dis. 2016;74(5):1–9. doi:10.1093/femspd/ftw033
58. Behzadi P, Baráth Z, Gajdács M. It’s not easy being green: a narrative review on the microbiology, virulence and therapeutic prospects of multidrug-resistant Pseudomonas aeruginosa. Antibiotics. 2021;10(1):1–29. doi:10.3390/antibiotics10010042
59. Thuenauer R, Landi A, Trefzer A, et al. The pseudomonas aeruginosa lectin LecB causes integrin internalization and inhibits epithelial wound healing. MBio. 2020;11(2). doi:10.1128/MBIO.03260-19
60. Vanderwoude J, Fleming D, Azimi S, Trivedi U, Rumbaugh KP, Diggle SP. The evolution of virulence in Pseudomonas aeruginosa during chronic wound infection. Proc Biol Sci. 2020;287(1937):20202272. doi:10.1098/RSPB.2020.2272
61. Trunk K, Benkert B, Quäck N, et al. Anaerobic adaptation in Pseudomonas aeruginosa: definition of the Anr and Dnr regulons. Environ Microbiol. 2010;12(6):1719–1733. doi:10.1111/J.1462-2920.2010.02252.X
62. Li M, Long Y, Liu Y, et al. HigB of Pseudomonas aeruginosa enhances killing of phagocytes by up-regulating the Type III secretion system in Ciprofloxacin induced persister cells. Front Cell Infect Microbiol. 2016;6(OCT). doi:10.3389/FCIMB.2016.00125
63. Alonso VP, Queiroz MM, Gualberto ML, Nascimento MS. Klebsiella pneumonia carbapenemase (KPC), methicillin-resistant Staphylococcus aureus (MRSA), and vancomycin-resistant Enterococcus spp. (VRE) in the food production chain and biofilm formation on abiotic surfaces. Curr Opin Food Sci. 2019;26:79–86. doi:10.1016/j.cofs.2019.04.002
64. Behzadi P, García-Perdomo HA, Karpiński TM, Issakhanian L. Metallo-ß-lactamases: a review. Mol Biol Rep. 2020;47(8):6281–6294. doi:10.1007/s11033-020-05651-9
65. Zheng JX, Lin ZW, Chen C, et al. Biofilm formation in Klebsiella pneumoniae bacteremia strains was found to be associated with CC23 and the presence of wcaG. Front Cell Infect Microbiol. 2018;8(FEB):21–32. doi:10.3389/fcimb.2018.00021
66. Rowe MC, Withers HL, Swift S. Uropathogenic Escherichia coli forms biofilm aggregates under iron restriction that disperse upon the supply of iron. FEMS Microbiol Lett. 2010;307(1). doi:10.1111/j.1574-6968.2010.01968.x
67. Wood TL, Gong T, Zhu L, et al. Rhamnolipids from Pseudomonas aeruginosa disperse the biofilms of sulfate-reducing bacteria. Npj Biofilms Microbiomes. 2018;4:22. doi:10.1038/s41522-018-0066-1
68. Guilhen C, Forestier C, Balestrino D. Biofilm dispersal: multiple elaborate strategies for dissemination of bacteria with unique properties. Mol Microbiol. 2017;105(2). doi:10.1111/mmi.13698
69. Roizman D, Vidaillac C, Givskov M, Yang L. In vitro evaluation of biofilm dispersal as a therapeutic strategy to restore antimicrobial efficacy. Antimicrob Agents Chemother. 2017;61(10). doi:10.1128/AAC.01088-17
70. Olivares E, Badel-Berchoux S, Provot C, Prévost G, Bernardi T, Jehl F. Clinical impact of antibiotics for the treatment of Pseudomonas aeruginosa biofilm infections. Front Microbiol. 2020;10:2894. doi:10.3389/fmicb.2019.02894
71. Flemming HC, Wingender J. The biofilm matrix. Nat Rev Microbiol. 2010;8(9):623–633. doi:10.1038/nrmicro2415
72. Flemming HC, Wingender J, Szewzyk U, Steinberg P, Rice SA. Biofilms: an emergent form of bacterial life. Nat Rev Microbiol. 2016;14(9). doi:10.1038/nrmicro.2016.94
73. Hall CW, Mah TF. Molecular mechanisms of biofilm-based antibiotic resistance and tolerance in pathogenic bacteria. FEMS Microbiol Rev. 2017;41(3). doi:10.1093/femsre/fux010
74. Hathroubi S, Mekni MA, Domenico P, Nguyen D. Biofilms: microbial shelters against antibiotics. Microb Drug Resist. 2017;23(2):147–156. doi:10.1089/mdr.2016.0087
75. Pena RT, Blasco L, Ambroa A, et al. Relationship between quorum sensing and secretion systems. Front Microbiol. 2019;10:1100. doi:10.3389/fmicb.2019.01100
76. Stalder T, Top E. Plasmid transfer in biofilms: a perspective on limitations and opportunities. Npj Biofilms Microbiomes. 2016;2:16022. doi:10.1038/npjbiofilms.2016.22
77. Brito IL. Examining horizontal gene transfer in microbial communities. Nat Rev Microbiol. 2021;19(7):442–453. doi:10.1038/s41579-021-00534-7
78. Mehta HH, Prater AG, Beabout K, et al. The essential role of hypermutation in rapid adaptation to antibiotic stress. Antimicrob Agents Chemother. 2019;63(7). doi:10.1128/AAC.00744-19
79. van Acker H, Coenye T. The role of efflux and physiological adaptation in biofilm tolerance and resistance. J Biol Chem. 2016;291(24):12565–12572. doi:10.1074/jbc.R115.707257
80. Brackman G, Breyne K, De Rycke R, et al. The quorum sensing inhibitor hamamelitannin increases antibiotic susceptibility of staphylococcus aureus biofilms by affecting peptidoglycan biosynthesis and eDNA release. Sci Rep. 2016;6:20321. doi:10.1038/srep20321
81. Tashiro Y, Eida H, Ishii S, Futamata H, Okabe S. Generation of small colony variants in biofilms by Escherichia coli harboring a conjugative F plasmid. Microbes Environ. 2017;32(1):40–46. doi:10.1264/jsme2.ME16121
82. Mirani ZA, Aziz M, Khan SI. Small colony variants have a major role in stability and persistence of Staphylococcus aureus biofilms. J Antibiot (Tokyo). 2015;68(2):98–105. doi:10.1038/ja.2014.115
83. Fauvart M, de Groote VN, Michiels J. Role of persister cells in chronic infections: clinical relevance and perspectives on anti-persister therapies. J Med Microbiol. 2011;60(6). doi:10.1099/jmm.0.030932-0
84. Ng WL, Bassler BL. Bacterial quorum-sensing network architectures. Annu Rev Genet. 2009;43:197–222. doi:10.1146/annurev-genet-102108-134304
85. Rasool FN, Saavedra MA, Pamba S, et al. Isolation and characterization of human pathogenic multidrug resistant bacteria associated with plastic litter collected in Zanzibar. J Hazard Mater. 2021;405. doi:10.1016/j.jhazmat.2020.124591
86. Govaert M, Smet C, Walsh JL, Van Impe JFM. Dual-species model biofilm consisting of Listeria monocytogenes and Salmonella typhimurium: development and inactivation with cold atmospheric plasma (CAP). Front Microbiol. 2019;10. doi:10.3389/fmicb.2019.02524
87. Rao Y, Shang W, Yang Y, Zhou R, Rao X. Fighting mixed-species microbial biofilms with cold atmospheric plasma. Front Microbiol. 2020;11. doi:10.3389/fmicb.2020.01000
88. Hall-Stoodley L, Costerton JW, Stoodley P. Bacterial biofilms: from the natural environment to infectious diseases. Nat Rev Microbiol. 2004;2(2). doi:10.1038/nrmicro821
89. Abedon ST. Bacteriophage exploitation of bacterial biofilms: phage preference for less mature targets? FEMS Microbiol Lett. 2016;363:246. doi:10.1093/femsle/fnv246
90. Abedon ST. Phage “delay” towards enhancing bacterial escape from biofilms: a more comprehensive way of viewing resistance to bacteriophages. AIMS Microbiol. 2017;3(2):186–226. doi:10.3934/microbiol.2017.2.186
91. Vestby LK, Grønseth T, Simm R, Nesse LL. Bacterial biofilm and its role in the pathogenesis of disease. Antibiotics. 2020;9(2). doi:10.3390/antibiotics9020059
92. Bonomo RA. β lactamases: a focus on current challenges. Cold Spring Harb Perspect Med. 2017;7:1–15.
93. Naas T, Oueslati S, Bonnin RA, et al. Beta-lactamase database (BLDB)–structure and function. J Enzyme Inhib Med Chem. 2017;32(1):917–919. doi:10.1080/14756366.2017.1344235
94. Kaplan JB. Antibiotic-induced biofilm formation. Int J Artif Organs. 2011;34(9):737–751. doi:10.5301/IJAO.5000027
95. Ahmadi M, Ranjbar R, Behzadi P, Mohammadian T. Virulence factors, antibiotic resistance patterns, and molecular types of clinical isolates of Klebsiella Pneumoniae. Expert Rev Anti Infect Ther. 2021. doi:10.1080/14787210.2022.1990040
96. Mushegian AR. Are there 1031virus particles on earth, or more, or fewer? J Bacteriol. 2020;202(9). doi:10.1128/JB.00052-20
97. Ackermann HW. 5500 Phages examined in the electron microscope. Arch Virol. 2007;152(2):227–243. doi:10.1007/s00705-006-0849-1
98. Devoto AE, Santini JM, Olm MR, et al. Megaphages infect Prevotella and variants are widespread in gut microbiomes. Nat Microbiol. 2019;4(4). doi:10.1038/s41564-018-0338-9
99. Hassan AY, Lin JT, Ricker N, Anany H. The age of phage: friend or foe in the new Dawn of therapeutic and biocontrol applications? Pharmaceuticals. 2021;14(3):1–36. doi:10.3390/ph14030199
100. Clokie MR, Millard AD, Letarov AV, Heaphy S. Phages in nature. Bacteriophage. 2011;1(1):31. doi:10.4161/BACT.1.1.14942
101. Hansen MF, Svenningsen LS, Røder HL, Middelboe M, Burmølle M. Big impact of the tiny: bacteriophage–bacteria interactions in biofilms. Trends Microbiol. 2019;27(9):739–752. doi:10.1016/j.tim.2019.04.006
102. Simmons EL, Bond MC, Koskella B, Drescher K, Bucci V, Nadell CD. Biofilm structure promotes coexistence of phage-resistant and phage-susceptible bacteria. mSystems. 2020;5(3):e00877–e00919. doi:10.1128/mSystems.00877-19
103. Eriksen RS, Svenningsen SL, Sneppen K, Mitarai N. A growing microcolony can survive and support persistent propagation of virulent phages. Proc Natl Acad Sci U S A. 2017;115(2). doi:10.1073/pnas.1708954115
104. Bull JJ, Christensen KA, Scott C, Jack BR, Crandall CJ, Krone SM. Phage-bacterial dynamics with spatial structure: self organization around phage sinks can promote increased cell densities. Antibiotics. 2018;7(1):8. doi:10.3390/antibiotics7010008
105. Tian F, Li J, Nazir A, Tong Y. Bacteriophage – a promising alternative measure for bacterial biofilm control. Infect Drug Resist. 2021;14:205–217. doi:10.2147/IDR.S290093
106. Perov S, Lidor O, Salinas N, et al. Structural insights into curli CsgA cross-β fibril architecture inspire repurposing of anti-amyloid compounds as anti-biofilm agents. PLoS Pathog. 2019;15(8):e1007978. doi:10.1371/journal.ppat.1007978
107. Pearl S, Gabay C, Kishony R, Oppenheim A, Balaban NQ. Nongenetic individuality in the host-phage interaction. PLoS Biol. 2008;6(5):e120. doi:10.1371/journal.pbio.0060120
108. Taylor BP, Penington CJ, Weitz JS. Emergence of increased frequency and severity of multiple infections by viruses due to spatial clustering of hosts. Phys Biol. 2016;13(6). doi:10.1088/1478-3975/13/6/066014
109. Castillo-Ruiz M, Vinés ED, Montt C, et al. Isolation of a novel Aggregatibacter actinomycetemcomitans serotype b bacteriophage capable of lysing bacteria within a biofilm. Appl Environ Microbiol. 2011;77(9):3157–3159. doi:10.1128/AEM.02115-10
110. Høyland-Kroghsbo NM, Paczkowski J, Mukherjee S, et al. Quorum sensing controls the Pseudomonas aeruginosa CRISPR-Cas adaptive immune system. Proc Natl Acad Sci U S A. 2017;114(1):131–135. doi:10.1073/pnas.1617415113
111. Moreau P, Diggle SP, Friman V-P. Bacterial cell-to-cell signaling promotes the evolution of resistance to parasitic bacteriophages. Ecol Evol. 2017;7(6):1936–1941. doi:10.1002/ece3.2818
112. Qin X, Sun Q, Yang B, Pan X, He Y, Yang H. Quorum sensing influences phage infection efficiency via affecting cell population and physiological state. J Basic Microbiol. 2017;57(2):162–170. doi:10.1002/jobm.201600510
113. Erez Z, Steinberger-Levy I, Shamir M, et al. Communication between viruses guides lysis–lysogeny decisions. Nature. 2017;541(7638):488–493. doi:10.1038/nature21049
114. Shah M, Taylor VL, Bona D, et al. A phage-encoded anti-activator inhibits quorum sensing in Pseudomonas aeruginosa. Mol Cell. 2021;81(3):571–583.e6. doi:10.1016/j.molcel.2020.12.011
115. González S, Fernández L, Gutiérrez D, Campelo AB, Rodríguez A, García P. Analysis of different parameters affecting diffusion, propagation and survival of Staphylophages in bacterial biofilms. Front Microbiol. 2018;9. doi:10.3389/fmicb.2018.02348
116. Fischetti VA. Development of phage lysins as novel therapeutics: a historical perspective. Viruses. 2018;10(6):310. doi:10.3390/v10060310
117. Roach DR, Donovan DM. Antimicrobial bacteriophage-derived proteins and therapeutic applications. Bacteriophage. 2015;5(3):e1062590. doi:10.1080/21597081.2015.1062590
118. Murray E, Draper LA, Ross RP, Hill C. The advantages and challenges of using endolysins in a clinical setting. Viruses. 2021;13(4):680. doi:10.3390/v13040680
119. Rodríguez-Rubio L, Gerstmans H, Thorpe S, Mesnage S, Lavigne R, Briers Y. DUF3380 domain from a Salmonella phage endolysin shows potent N-acetylmuramidase activity. Appl Environ Microbiol. 2016;82(16):4975–4981. doi:10.1128/AEM.00446-16
120. Matamp N, Bhat SG. Phage endolysins as potential antimicrobials against multidrug resistant Vibrio alginolyticus and Vibrio parahaemolyticus: current status of research and challenges ahead. Microorganisms. 2019;7(3):84. doi:10.3390/microorganisms7030084
121. Abdelrahman F, Easwaran M, Daramola OI, et al. Phage-encoded endolysins. Antibiotics. 2021;10(2):1–31. doi:10.3390/antibiotics10020124
122. Wang I-N, Smith DL, Young R. Holins: the protein clocks of bacteriophage infections. Annu Rev Microbiol. 2000;54(1):799–825. doi:10.1146/annurev.micro.54.1.799
123. Łusiak-szelachowska M, Weber-Dąbrowska B, Górski A. Bacteriophages and lysins in biofilm control. Virol Sin. 2020;35(2):125–133. doi:10.1007/s12250-019-00192-3
124. de Miguel T, Rama JLR, Sieiro C, Sánchez S, Villa TG. Bacteriophages and lysins as possible alternatives to treat antibiotic‐resistant urinary tract infections. Antibiotics. 2020;9(8):1–11. doi:10.3390/antibiotics9080466
125. Ghose C, Euler CW. Gram-negative bacterial lysins. Antibiotics. 2020;9(2):74. doi:10.3390/antibiotics9020074
126. Huang G, Shen X, Gong Y, et al. Antibacterial properties of phage Abp1 endolysin (PlyAB1). BMC Infect Dis. 2014;14(1). doi:10.1186/s12879-014-0681-2
127. Lood R, Winer BY, Pelzek AJ, et al. Novel phage Lysin capable of killing the multidrug-resistant gram-negative bacterium Acinetobacter baumannii in a mouse bacteremia model. Antimicrob Agents Chemother. 2015;59(4):1983–1991. doi:10.1128/AAC.04641-14
128. Wang Q, Euler CW, Delaune A, Fischetti VA. Using a novel lysin to help control Clostridium difficile infections. Antimicrob Agents Chemother. 2015;59(12):7447–7457. doi:10.1128/AAC.01357-15
129. Guo Z, Huang J, Yan G, et al. Identification and characterization of Dpo42, a novel depolymerase derived from the Escherichia coli phage vB_EcoM_ECOO78. Front Microbiol. 2017;8. doi:10.3389/fmicb.2017.01460
130. Peng S-Y, You R-I, Lai M-J, Lin N-T, Chen L-K, Chang K-C. Highly potent antimicrobial modified peptides derived from the Acinetobacter baumannii phage endolysin LysAB2. Sci Rep. 2017;7(1). doi:10.1038/s41598-017-11832-7
131. Wu Y, Wang R, Xu M, et al. A Novel polysaccharide depolymerase encoded by the phage SH-KP152226 confers specific activity against multidrug-resistant Klebsiella pneumoniae via biofilm degradation. Front Microbiol. 2019;10. doi:10.3389/fmicb.2019.02768
132. Lv M, Wang S, Yan G, et al. Genome sequencing and analysis of an Escherichia coli phage vB_EcoM-ep3 with a novel lysin, Lysep3. Virus Genes. 2015;50(3):487–497. doi:10.1007/s11262-015-1195-8
133. Maciejewska B, Roszniowski B, Espaillat A, et al. Klebsiella phages representing a novel clade of viruses with an unknown DNA modification and biotechnologically interesting enzymes. Appl Microbiol Biotechnol. 2017;101(2):673–684. doi:10.1007/s00253-016-7928-3
134. Schirmeier E, Zimmermann P, Hofmann V, et al. Inhibitory and bactericidal effect of Artilysin ® Art-175 against colistin-resistant mcr-1 -positive Escherichia coli isolates. Int J Antimicrob Agents. 2018;51(3):528–529. doi:10.1016/j.ijantimicag.2017.08.027
135. Gerstmans H, Rodríguez-Rubio L, Lavigne R, Briers Y. From endolysins to Artilysin®s: novel enzyme-based approaches to kill drug-resistant bacteria. Biochem Soc Trans. 2016;44(1):123–128. doi:10.1042/BST20150192
136. Raz A, Serrano A, Hernandez A, Euler CW, Fischetti VA. Isolation of phage lysins that effectively kill Pseudomonas aeruginosa in mouse models of lung and skin infection. Antimicrob Agents Chemother. 2019;63(7). doi:10.1128/AAC.00024-19
137. Paradis-Bleau C, Cloutier I, Lemieux L, et al. Peptidoglycan lytic activity of the Pseudomonas aeruginosa phage φKZ gp144 lytic transglycosylase. FEMS Microbiol Lett. 2007;266(2):201–209. doi:10.1111/j.1574-6968.2006.00523.x
138. Cheng Q, Nelson D, Zhu S, Fischetti VA. Removal of group B streptococci colonizing the vagina and oropharynx of mice with a bacteriophage lytic enzyme. Antimicrob Agents Chemother. 2005;49(1):111–117. doi:10.1128/AAC.49.1.111-117.2005
139. Oliveira H, Pinto G, Oliveira A, et al. Characterization and genome sequencing of a Citrobacter freundii phage CfP1 harboring a lysin active against multidrug-resistant isolates. Appl Microbiol Biotechnol. 2016;100(24):10543–10553. doi:10.1007/s00253-016-7858-0
140. Loeffler JM, Fischetti VA. Synergistic lethal effect of a combination of phage lytic enzymes with different activities on penicillin-sensitive and -resistant Streptococcus pneumoniae strains. Antimicrob Agents Chemother. 2003;47(1):375–377. doi:10.1128/AAC.47.1.375-377.2003
141. Domenech M, Garciá E, Moscoso M. In vitro destruction of Streptococcus pneumoniae biofilms with bacterial and phage peptidoglycan hydrolases. Antimicrob Agents Chemother. 2011;55(9):4144–4148. doi:10.1128/AAC.00492-11
142. Corsini B, Díez-Martínez R, Aguinagalde L, et al. Chemotherapy with phage lysins reduces pneumococcal colonization of the respiratory tract. Antimicrob Agents Chemother. 2018;62(6). doi:10.1128/AAC.02212-17
143. Gilmer DB, Schmitz JE, Euler CW, Fischetti VA. Novel bacteriophage lysin with broad lytic activity protects against mixed infection by Streptococcus pyogenes and methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2013;57(6):2743–2750. doi:10.1128/AAC.02526-12
144. Jun SY, Jung GM, Yoon SJ, et al. Preclinical safety evaluation of intravenously administered SAL200 containing the recombinant phage endolysin SAL-1 as a pharmaceutical ingredient. Antimicrob Agents Chemother. 2014;58(4):2084–2088. doi:10.1128/AAC.02232-13
145. Xia F, Li X, Wang B, et al. Combination therapy of LysGH15 and Apigenin as a new strategy for treating pneumonia caused by Staphylococcus aureus. Appl Environ Microbiol. 2016;82(1):87–94. doi:10.1128/AEM.02581-15
146. Gil F, Grzegorzewicz AE, Catalão MJ, Vital J, McNeil MR, Pimentel M. Mycobacteriophage Ms6 LysB specifically targets the outer membrane of Mycobacterium smegmatis. Microbiology. 2010;156(5):1497–1504. doi:10.1099/mic.0.032821-0
147. Grover N, Paskaleva EE, Mehta KK, Dordick JS, Kane RS. Growth inhibition of Mycobacterium smegmatis by mycobacteriophage-derived enzymes. Enzyme Microb Technol. 2014;63:1–6. doi:10.1016/j.enzmictec.2014.04.018
148. Latka A, Maciejewska B, Majkowska-Skrobek G, Briers Y, Drulis-Kawa Z. Bacteriophage-encoded virion-associated enzymes to overcome the carbohydrate barriers during the infection process. Appl Microbiol Biotechnol. 2017;101(8):3103–3119. doi:10.1007/s00253-017-8224-6
149. Fernandes S, São-José C. Enzymes and mechanisms employed by tailed bacteriophages to breach the bacterial cell barriers. Viruses. 2018;10(8):396. doi:10.3390/v10080396
150. Pires DP, Melo LDR, Vilas Boas D, Sillankorva S, Azeredo J. Phage therapy as an alternative or complementary strategy to prevent and control biofilm-related infections. Curr Opin Microbiol. 2017;39:48–56. doi:10.1016/j.mib.2017.09.004
151. Yan J, Mao J, Xie J. Bacteriophage polysaccharide depolymerases and biomedical applications. BioDrugs. 2014;28(3):265–274. doi:10.1007/s40259-013-0081-y
152. Hsieh S-E, Lo -H-H, Chen S-T, Lee M-C, Tseng Y-H. Wide host range and strong lytic activity of Staphylococcus aureus Lytic Phage Stau2. Appl Environ Microbiol. 2011;77(3):756–761. doi:10.1128/AEM.01848-10
153. Oliveira A, Ribeiro HG, Silva AC, et al. Synergistic antimicrobial interaction between honey and phage against Escherichia coli biofilms. Front Microbiol. 2017;8. doi:10.3389/fmicb.2017.02407
154. Vikram A, Woolston J, Sulakvelidze A. Phage biocontrol applications in food production and processing. Curr Issues Mol Biol. 2020;40:267–302. doi:10.21775/cimb.040.267
155. Ding C, He J. Effect of antibiotics in the environment on microbial populations. Appl Microbiol Biotechnol. 2010;87(3):925–941. doi:10.1007/s00253-010-2649-5
156. Luong T, Salabarria A-C, Edwards RA, Roach DR. Standardized bacteriophage purification for personalized phage therapy. Nat Protoc. 2020;15(9):2867–2890. doi:10.1038/s41596-020-0346-0
157. Melo LDR, Oliveira H, Pires DP, Dabrowska K, Azeredo J. Phage therapy efficacy: a review of the last 10 years of preclinical studies. Crit Rev Microbiol. 2020;46(1):78–99. doi:10.1080/1040841X.2020.1729695
158. Loc-Carrillo C, Abedon ST. Pros and cons of phage therapy. Bacteriophage. 2011;1(2):111–114. doi:10.4161/bact.1.2.14590
159. Tinoco JM, Buttaro B, Zhang H, Liss N, Sassone L, Stevens R. Effect of a genetically engineered bacteriophage on Enterococcus faecalis biofilms. Arch Oral Biol. 2016;71:80–86. doi:10.1016/j.archoralbio.2016.07.001
160. Schmerer M, Molineux IJ, Bull JJ. Synergy as a rationale for phage therapy using phage cocktails. PeerJ. 2014;2014(1). doi:10.7717/peerj.590
161. Ferriol-González C, Domingo-Calap P. Phages for biofilm removal. Antibiotics. 2020;9(5):268. doi:10.3390/antibiotics9050268
162. Carrascosa C, Raheem D, Ramos F, Saraiva A, Raposo A. Microbial biofilms in the food industry—a comprehensive review. Int J Environ Res Public Health. 2021;18(4):1–31. doi:10.3390/ijerph18042014
163. Tacconelli E, Carrara E, Savoldi A, Kattula D, Burkert F. Global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics; 2017. Available from: http://www.cdc.gov/drugresistance/threat-report-2013/.
164. Khalifa L, Gelman D, Shlezinger M, et al. Defeating antibiotic- and phage-resistant Enterococcus faecalis using a phage cocktail in vitro and in a clot model. Front Microbiol. 2018;9. doi:10.3389/fmicb.2018.00326
165. Dalmasso M, De Haas E, Neve H, et al. Isolation of a novel phage with activity against Streptococcus mutans biofilms. PLoS One. 2015;10(9):e0138651. doi:10.1371/journal.pone.0138651
166. Dakheel KH, Rahim RA, Neela VK, et al. Genomic analyses of two novel biofilm-degrading methicillin-resistant Staphylococcus aureus phages. BMC Microbiol. 2019;19(1):1–23. doi:10.1186/s12866-019-1484-9
167. Bagińska N, Cieślik M, Górski A, Jończyk-Matysiak E. The role of antibiotic resistant A. baumannii in the pathogenesis of urinary tract infection and the potential of its treatment with the use of bacteriophage therapy. Antibiotics. 2021;10(3):281. doi:10.3390/antibiotics10030281
168. Montso PK, Mlambo V, Ateba CN. Efficacy of novel phages for control of multi-drug resistant Escherichia coli O177 on artificially contaminated beef and their potential to disrupt biofilm formation. Food Microbiol. 2021;94:103647. doi:10.1016/j.fm.2020.103647
169. Roach DR, Leung CY, Henry M, et al. Synergy between the host immune system and bacteriophage is essential for successful phage therapy against an acute respiratory pathogen. Cell Host Microbe. 2017;22(1):38–47.e4. doi:10.1016/j.chom.2017.06.018
170. Tabassum R, Shafique M, Amer Khawaja K, et al. Complete genome analysis of a Siphoviridae phage TSK1 showing biofilm removal potential against Klebsiella pneumoniae OPEN. Sci Rep. 2018;8(1):17904–undefined. doi:10.1038/s41598-018-36229-y
171. Li M, Shi D, Li Y, et al. Recombination of T4-like Phages and its activity against pathogenic Escherichia coli in planktonic and biofilm forms. Virol Sin. 2020;35(5):651–661. doi:10.1007/s12250-020-00233-2
172. Arumugam SN, Rudraradhya AC, Sadagopan S, Sukumaran S, Sambasivam G, Ramesh N. Analysis of susceptibility patterns of Pseudomonas aeruginosa and Isolation, Characterization of lytic bacteriophages targeting multi drug resistant pseudomonas aeruginosa. Biomed Pharmacol J. 2018;11(2):1105–1117. doi:10.13005/bpj/1471
173. Sinha A, Eniyan K, Manohar P, Ramesh N, Bajpai U. Characterization and genome analysis of B1 sub-cluster mycobacteriophage PDRPxv. Virus Res. 2020;279:197884. doi:10.1016/j.virusres.2020.197884
174. Waters EM, Neill DR, Kaman B, et al. Phage therapy is highly effective against chronic lung infections with Pseudomonas aeruginosa. Thorax. 2017;72(7):666–667. doi:10.1136/thoraxjnl-2016-209265
175. Alves DR, Perez-Esteban P, Kot W, et al. A novel bacteriophage cocktail reduces and disperses Pseudomonas aeruginosa biofilms under static and flow conditions. Microb Biotechnol. 2016;9(1):61–74. doi:10.1111/1751-7915.12316
176. Forti F, Roach DR, Cafora M, et al. Design of a broad-range bacteriophage cocktail that reduces Pseudomonas aeruginosa biofilms and treats acute infections in two animal models. Antimicrob Agents Chemother. 2018;62(6). doi:10.1128/AAC.02573-17
177. Kaur S, Harjai K, Chhibber S, Becker K. In vivo assessment of phage and Linezolid based implant coatings for treatment of Methicillin resistant S. aureus (MRSA) mediated orthopaedic device related infections. PLoS One. 2016;11(6):e0157626. doi:10.1371/journal.pone.0157626
178. Ooi ML, Drilling AJ, Morales S, et al. Safety and tolerability of bacteriophage therapy for chronic rhinosinusitis due to Staphylococcus aureus. JAMA Otolaryngol - Head Neck Surg. 2019;145(8):723–729. doi:10.1001/jamaoto.2019.1191
179. Yilmaz C, Colak M, Yilmaz BC, Ersoz G, Kutateladze M, Gozlugol M. Bacteriophage therapy in implant-related infections: an experimental study. J Bone Jt Surg - Ser A. 2013;95(2):117–125. doi:10.2106/JBJS.K.01135
180. Kishor C, Mishra R, Saraf S, Kumar M, Srivastav A, Nath G. Phage therapy of staphylococcal chronic osteomyelitis in experimental animal model. Indian J Med Res. 2016;143(1). doi:10.4103/0971-5916.178615
181. Gibb BP, Hadjiargyrou M. Bacteriophage therapy for bone and joint infections. Bone Joint J. 2021;103-B(2):234–244. doi:10.1302/0301-620X.103B2.BJJ-2020-0452.R2
182. Maszewska A, Zygmunt M, Grzejdziak I, Różalski A. Use of polyvalent bacteriophages to combat biofilm of Proteus mirabilis causing catheter-associated urinary tract infections. J Appl Microbiol. 2018;125(5):1253–1265. doi:10.1111/jam.14026
183. Fu W, Forster T, Mayer O, Curtin JJ, Lehman SM, Donlan RM. Bacteriophage cocktail for the prevention of biofilm formation by Pseudomonas aeruginosa on catheters in an in vitro model system. Antimicrob Agents Chemother. 2010;54(1):397–404. doi:10.1128/AAC.00669-09
184. Wasfi R, Hamed SM, Amer MA, Fahmy LI. Proteus mirabilis biofilm: development and therapeutic strategies. Front Cell Infect Microbiol. 2020;10. doi:10.3389/fcimb.2020.00414
185. Melo LDR, Veiga P, Cerca N, et al. Development of a phage cocktail to control Proteus mirabilis catheter-associated urinary tract infections. Front Microbiol. 2016;7:1024. doi:10.3389/fmicb.2016.01024
186. Gomaa S, Serry F, Abdellatif H, Abbas H. Elimination of multidrug-resistant Proteus mirabilis biofilms using bacteriophages. Arch Virol. 2019;164(9):2265–2275. doi:10.1007/s00705-019-04305-x
187. Lehman SM, Donlan RM. Bacteriophage-mediated control of a two-species biofilm formed by microorganisms causing catheter-associated urinary tract infections in an in vitro urinary catheter model. Antimicrob Agents Chemother. 2015;59(2). doi:10.1128/AAC.03786-14
188. Malik S, Sidhu PK, Rana JS, Nehra K. Managing urinary tract infections through phage therapy: a novel approach. Folia Microbiol (Praha). 2020;65(2):217–231. doi:10.1007/s12223-019-00750-y
189. Nishikawa H, Yasuda M, Uchiyama J, et al. T-even-related bacteriophages as candidates for treatment of Escherichia coli urinary tract infections. Arch Virol. 2008;153(3). doi:10.1007/s00705-007-0031-4
190. Chibeu A, Lingohr EJ, Masson L, et al. Bacteriophages with the ability to degrade uropathogenic Escherichia coli biofilms. Viruses. 2012;4(4). doi:10.3390/v4040471
191. Chadha P, Katare OP, Chhibber S. In vivo efficacy of single phage versus phage cocktail in resolving burn wound infection in BALB/c mice. Microb Pathog. 2016;99:68–77. doi:10.1016/j.micpath.2016.08.001
192. Motlagh AM, Bhattacharjee AS, Goel R. Biofilm control with natural and genetically-modified phages. World J Microbiol Biotechnol. 2016;32(4). doi:10.1007/s11274-016-2009-4
193. Manohar P, Loh B, Athira S, et al. Phage antibiotic therapy. Front Microbiol. 2020;11:1134–1149. doi:10.3389/fmicb.2020.01434
194. Morrisette T, Kebriaei R, Lev KL, Morales S, Rybak MJ. Bacteriophage therapeutics: a primer for clinicians on phage-antibiotic combinations. Pharmacotherapy. 2020;40(2):153–168. doi:10.1002/phar.2358
195. Comeau AM, Té F, Trojet SN. Phage-antibiotic synergy (PAS): β-Lactam and quinolone antibiotics stimulate virulent phage growth. PLoS One. 2007;2(8):e799–undefined. doi:10.1371/journal.pone.0000799
196. Tkhilaishvili T, Lombardi L, Klatt AB, Trampuz A, Di Luca M. Bacteriophage Sb-1 enhances antibiotic activity against biofilm, degrades exopolysaccharide matrix and targets persisters of Staphylococcus aureus. Int J Antimicrob Agents. 2018;52(6). doi:10.1016/j.ijantimicag.2018.09.006
197. Coulter LB, Rohde RE, Aron GM. Effect of bacteriophage infection in combination with tobramycin on the emergence of resistance in Escherichia coli and Pseudomonas aeruginosa biofilms. Viruses. 2014;6(10):3778–3786. doi:10.3390/v6103778
198. Oechslin F, Piccardi P, Mancini S, et al. Synergistic interaction between phage therapy and antibiotics clears Pseudomonas aeruginosa infection in endocarditis and reduces virulence. J Infect Dis. 2017;215(5):703–712. doi:10.1093/infdis/jiw632
199. Chaudhry WN, Concepció N-Acevedo J, Park T, Andleeb S, Bull JJ, Levin BR. Synergy and order effects of antibiotics and phages in killing Pseudomonas aeruginosa biofilms. PLoS One. 2017;12(1):e0168615. doi:10.1371/journal.pone.0168615
200. Sass P, Bierbaum G. Lytic activity of recombinant bacteriophage φ11 and φ12 endolysins on whole cells and biofilms of Staphylococcus aureus. Appl Environ Microbiol. 2007;73(1):347–352. doi:10.1128/AEM.01616-06
201. Gondil VS, Harjai K, Chhibber S. Endolysins as emerging alternative therapeutic agents to counter drug-resistant infections. Int J Antimicrob Agents. 2020;55(2). doi:10.1016/j.ijantimicag.2019.11.001
202. Schuch R, Khan BK, Raz A, Rotolo JA, Wittekind M. Bacteriophage lysin CF-301, a potent antistaphylococcal biofilm agent. Antimicrob Agents Chemother. 2017;61(7). doi:10.1128/AAC.02666-16
203. Vander EN, Linden SB, Lavigne R, Meyer E, Briers Y, Nelson DC. Characterization of the bacteriophage-derived endolysins plyss2 and plyss9 with in vitro lytic activity against bovine mastitis Streptococcus uberis. Antibiotics. 2020;9(9). doi:10.3390/antibiotics9090621
204. Wang S, Gu J, Lv M, et al. The antibacterial activity of E. coli bacteriophage lysin lysep3 is enhanced by fusing the Bacillus amyloliquefaciens bacteriophage endolysin binding domain D8 to the C-terminal region. J Microbiol. 2017;55(5):403–408. doi:10.1007/s12275-017-6431-6
205. Gutiérrez D, Ruas-Madiedo P, Martínez B, Rodríguez A, García P. Effective removal of staphylococcal biofilms by the endolysin LysH5. PLoS One. 2014;9(9):e107307. doi:10.1371/journal.pone.0107307
206. Hernandez-Morales AC, Lessor LL, Wood TL, et al. Genomic and biochemical characterization of Acinetobacter podophage petty reveals a novel lysis mechanism and tail-associated depolymerase activity. J Virol. 2018;92(6). doi:10.1128/jvi.01064-17
207. Mi L, Liu Y, Wang C, et al. Identification of a lytic Pseudomonas aeruginosa phage depolymerase and its anti-biofilm effect and bactericidal contribution to serum. Virus Genes. 2019. doi:10.1007/s11262-019-01660-4
208. Olsen NMC, Thiran E, Hasler T, et al. Synergistic removal of static and dynamic Staphylococcus aureus biofilms by combined treatment with a bacteriophage endolysin and a polysaccharide depolymerase. Viruses. 2018;10(8). doi:10.3390/v10080438
209. Geredew Kifelew L, Mitchell JG, Speck P. Mini-review: efficacy of lytic bacteriophages on multispecies biofilms. Biofouling. 2019;35(4). doi:10.1080/08927014.2019.1613525
210. Chan B, Abedon S. Bacteriophages and their enzymes in biofilm control. Curr Pharm Des. 2014;21(1). doi:10.2174/1381612820666140905112311
211. Lampp JW, Griswold KE. Alginate lyase exhibits catalysis-independent biofilm dispersion and antibiotic synergy. Antimicrob Agents Chemother. 2013;57(1). doi:10.1128/AAC.01789-12
212. Lu TK, Collins JJ. Dispersing biofilms with engineered enzymatic bacteriophage. Proc Natl Acad Sci U S A. 2007;104(27):11197–11202. doi:10.1073/pnas.0704624104
213. Møller-Olsen C, Ho SFS, Shukla RD, Feher T, Sagona AP. Engineered K1F bacteriophages kill intracellular Escherichia coli K1 in human epithelial cells. Sci Rep. 2018;8(1). doi:10.1038/s41598-018-35859-6
214. Lemon DJ, Kay MK, Titus JK, et al. Construction of a genetically modified T7Select phage system to express the antimicrobial peptide 1018. J Microbiol. 2019;57(6). doi:10.1007/s12275-019-8686-6
215. Yosef I, Manor M, Kiro R, Qimron U. Temperate and lytic bacteriophages programmed to sensitize and kill antibiotic-resistant bacteria. Proc Natl Acad Sci U S A. 2015;112(23). doi:10.1073/pnas.1500107112
216. Vázquez R, García E, García P. Phage lysins for fighting bacterial respiratory infections: a new generation of antimicrobials. Front Immunol. 2018;9. doi:10.3389/fimmu.2018.02252
217. Poonacha N, Nair S, Desai S, et al. Efficient killing of planktonic and biofilm-embedded coagulase-negative staphylococci by bactericidal protein P128. Antimicrob Agents Chemother. 2017;61(8). doi:10.1128/AAC.00457-17
218. Sharma U, Vipra A, Channabasappa S. Phage-derived lysins as potential agents for eradicating biofilms and persisters. Drug Discov Today. 2018;23(4). doi:10.1016/j.drudis.2018.01.026
219. Channabasappa S, Chikkamadaiah R, Durgaiah M, et al. Efficacy of chimeric ectolysin P128 in drug-resistant Staphylococcus aureus bacteraemia in mice. J Antimicrob Chemother. 2018;73(12). doi:10.1093/jac/dky365
220. Nair S, Desai S, Poonacha N, Vipra A, Sharma U. Antibiofilm activity and synergistic inhibition of Staphylococcus aureus biofilms by bactericidal protein P128 in combination with antibiotics. Antimicrob Agents Chemother. 2016;60(12). doi:10.1128/AAC.01118-16
221. Singh PK, Donovan DM, Kumar A. Intravitreal injection of the chimeric phage endolysin Ply187 protects mice from Staphylococcus aureus endophthalmitis. Antimicrob Agents Chemother. 2014;58(8). doi:10.1128/AAC.00126-14
222. Huang L, Luo D, Gondil VS, et al. Construction and characterization of a chimeric lysin ClyV with improved bactericidal activity against Streptococcus agalactiae in vitro and in vivo. Appl Microbiol Biotechnol. 2020;104(4):1609–1619. doi:10.1007/s00253-019-10325-z
223. Fowler VG, Das AF, Lipka-Diamond J, et al. Exebacase for patients with Staphylococcus aureus bloodstream infection and endocarditis. J Clin Invest. 2020;130(7). doi:10.1172/JCI136577
224. Watson A, Sauve K, Cassino C, Schuch R. Exebacase demonstrates in vitro synergy with a broad range of antibiotics against both methicillin-resistant and methicillin-susceptible staphylococcus aureus. Antimicrob Agents Chemother. 2020;64(2). doi:10.1128/AAC.01885-19
225. Briers Y, Walmagh M, Grymonprez B, et al. Art-175 is a highly efficient antibacterial against multidrug-resistant strains and persisters of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2014;58(7). doi:10.1128/AAC.02668-14
226. Heselpoth RD, Euler CW, Schuch R, Fischetti VA. Lysocins: bioengineered antimicrobials that deliver lysins across the outer membrane of Gram-negative bacteria. Antimicrob Agents Chemother. 2019;63(6). doi:10.1128/AAC.00342-19
227. Harcombe WR, Bull JJ. Impact of phages on two-species bacterial communities. Appl Environ Microbiol. 2005;71(9). doi:10.1128/AEM.71.9.5254-5259.2005
228. Akanda ZZ, Taha M, Abdelbary H. Current review—The rise of bacteriophage as a unique therapeutic platform in treating peri-prosthetic joint infections. J Orthop Res. 2018;36(4):1051–1060. doi:10.1002/jor.23755
229. Brovko LY, Anany H, Griffiths MW. Bacteriophages for Detection and control of bacterial pathogens in food and food-processing environment. In: Advances in Food and Nutrition Research. Vol. 67. Academic Press Inc.; 2012:241–288. doi:10.1016/B978-0-12-394598-3.00006-X
230. Malik DJ. Bacteriophage encapsulation using spray drying for phage therapy. Curr Issues Mol Biol. 2021;40:303–316. doi:10.21775/cimb.040.303
231. Pinto AM, Cerqueira MA, Bañobre-Lópes M, Pastrana LM, Sillankorva S. Bacteriophages for chronic wound treatment: from traditional to novel delivery systems. Viruses. 2020;12(2):235. doi:10.3390/v12020235
232. Morozova VV, Vlassov VV, Tikunova NV. Applications of bacteriophages in the treatment of localized infections in humans. Front Microbiol. 2018;9:1696. doi:10.3389/fmicb.2018.01696
233. Górski A, Borysowski J, Międzybrodzki R. Phage therapy: towards a successful clinical trial. Antibiotics. 2020;9(11):1–7. doi:10.3390/antibiotics9110827
234. Yan W, Banerjee P, Liu Y, et al. Development of thermosensitive hydrogel wound dressing containing Acinetobacter baumannii phage against wound infections. Int J Pharm. 2021;602:120508. doi:10.1016/j.ijpharm.2021.120508
235. Markoishvili K, Tsitlanadze G, Katsarava R, Morris JG, Sulakvelidze A. A novel sustained-release matrix based on biodegradable poly (ester amide) s and impregnated with bacteriophages and an antibiotic shows promise in management of infected venous stasis ulcers and other poorly healing wounds. Int J Dermatol. 2002;41:453–458. doi:10.1046/j.1365-4362.2002.01451.x
236. Sarhan WA, Azzazy HM. Apitherapeutics and phage-loaded nanofibers as wound dressings with enhanced wound healing and antibacterial activity. Nanomedicine. 2017;12(17). doi:10.2217/nnm-2017-0151
237. Tkhilaishvili T, Wang L, Tavanti A, Trampuz A, Di Luca M. Antibacterial efficacy of two commercially available bacteriophage formulations, Staphylococcal bacteriophage and PYO bacteriophage, against Methicillin-resistant Staphylococcus aureus: prevention and eradication of biofilm formation and control of a systemic infection of Galleria mellonella larvae. Front Microbiol. 2020;11. doi:10.3389/fmicb.2020.00110
238. Principi N, Silvestri E, Esposito S. Advantages and limitations of bacteriophages for the treatment of bacterial infections. Front Pharmacol. 2019;10:1–9. doi:10.3389/fphar.2019.00513
239. Dennehy JJ, Abedon ST. Bacteriophage Ecology. In: Bacteriophages. Springer International Publishing; 2020:1–42. doi:10.1007/978-3-319-40598-8_8-1
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
