Back to Journals » Infection and Drug Resistance » Volume 13
How CRISPR-Cas System Could Be Used to Combat Antimicrobial Resistance
Authors Gholizadeh P , Köse Ş, Dao S, Ganbarov K , Tanomand A , Dal T , Aghazadeh M, Ghotaslou R , Ahangarzadeh Rezaee M , Yousefi B , Samadi Kafil H
Received 25 January 2020
Accepted for publication 31 March 2020
Published 20 April 2020 Volume 2020:13 Pages 1111—1121
DOI https://doi.org/10.2147/IDR.S247271
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Suresh Antony
Pourya Gholizadeh,1,2 Şükran Köse,3 Sounkalo Dao,4 Khudaverdi Ganbarov,5 Asghar Tanomand,6 Tuba Dal,7 Mohammad Aghazadeh,8 Reza Ghotaslou,8 Mohammad Ahangarzadeh Rezaee,8 Bahman Yousefi,8 Hossein Samadi Kafil2,9
1Student Research Committee, Tabriz University of Medical Sciences, Tabriz, Iran; 2Biotechnology Research Center, Tabriz University of Medical Sciences, Tabriz, Iran; 3Department of Infectious Diseases and Clinical Microbiology, University of Health Sciences, Tepecik Training and Research Hospital, İzmir, Turkey; 4Faculté de Médecine, de Pharmacie et d’Odonto-Stomatologie (FMPOS), University of Bamako, Bamako, Mali; 5Department of Microbiology, Baku State University, Baku, Republic of Azerbaijan; 6Department of Basic Sciences, Maragheh University of Medical Sciences, Maragheh, Iran; 7Department of Clinical Microbiology, Faculty of Medicine, Ankara Yildirim Beyazit University, Ankara, Turkey; 8Immunology Research Center, Tabriz University of Medical Sciences, Tabriz, Iran; 9Drug Applied Research Center, Tabriz University of Medical Sciences, Tabriz, Iran
Correspondence: Hossein Samadi Kafil
Drug Applied Research Center, Tabriz University of Medical Sciences, Tabriz, Iran
Tel +98 9127184735
Fax +98 4133364661
Email [email protected]
Abstract: Rapid emergence of antibiotic-resistant bacteria has made it harder for us to combat infectious diseases and to develop new antibiotics. The clustered regularly interspaced short palindromic repeats – CRISPR-associated (CRISPR-Cas) system, as a bacterial adaptive immune system, is recognized as one of the new strategies for controlling antibiotic-resistant strains. The programmable Cas nuclease of this system used against bacterial genomic sequences could be lethal or could help reduce resistance of bacteria to antibiotics. Therefore, this study aims to review using the CRISPR-Cas system to promote sensitizing bacteria to antibiotics. We envision that CRISPR-Cas approaches may open novel ways for the development of smart antibiotics, which could eliminate multidrug-resistant (MDR) pathogens and differentiate between beneficial and pathogenic microorganisms. These systems can be exploited to quantitatively and selectively eliminate individual bacterial strains based on a sequence-specific manner, creating opportunities in the treatment of MDR infections, the study of microbial consortia, and the control of industrial fermentation.
Keywords: antibiotic-resistant bacteria, CRISPR-Cas system, sequence-specific manner, resensitization, genome editing
Introduction
The extensive and often injudicious use of antibiotics in agriculture and public health over the last 7 decades has exerted a significant selection pressure for antibiotic-resistant bacteria to evolve. Various strategies against antibiotic-resistant bacteria have been introduced, including production of new antibiotics, bacteriophages that target and lyse these bacteria, and discovery and production of naturally and artificial-derived peptides or enzymes that specifically target genomes or functional vital proteins of these bacteria.1,6 The widespread use and abuse of antimicrobial agents has driven bacterial populations to become resistant against antimicrobial agents through genetic additions and changes. The first global report by the World Health Organization (WHO) on antibiotic resistance in 2014 recognized that antibiotic resistance of bacteria is a serious threat to public health (https://www.who.int/mediacentre/news/releases/2014/amr-report/en/) with data collected from 114 countries. Interestingly, a small number of the pathogens are responsible for most antibiotic-resistant infections, which are highlighted by the Infectious Diseases Society of America, and they are known as ESKAPE pathogens. ESKAPE pathogens are a group of the most antibiotic-resistant bacteria, which cause difficulty in treating nosocomial infections. These pathogens include: Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter spp., which have or can acquire resistance against multiple antibiotics.7 The inherent difficulties in finding new antibiotics, along with the low economic motivation, have resulted in slow development of new antibiotics. Moreover, the pace of emerging antibiotic resistance far exceeds the pace of development of new antibiotics, which is insufficient to combat the rise in antibiotic resistance.7 Therefore, new approaches have been suggested for decreasing and limiting antibiotic resistance in pathogens, including phage therapy,2,8,9 use of peptide nucleic acid (PNA) as an ultra-narrow-spectrum antibiotic,6,10,14 zinc finger nucleases (ZFNs),15,16 and clustered regularly interspaced short palindromic repeat – CRISPR-associated (CRISPR-Cas) systems,5,17,19 which are genomic engineering tools for gene knock-out and knock-in of sequence-specific DNA antibiotic targets. Bacteriophages are bacteria-specific viruses, which can specifically infect and lyse bacteria. Phage therapy harnesses phages for treatment against bacterial pathogens and their infectious diseases. Different clinical applications have been suggested for phages: 1) use of bactericidal and virulent phages against pathogens and antibiotic-resistant bacteria,1,3,8,9,20 2) the clinical use of metabolic inhibitor properties of phage structural proteins to inhibit bacterial cell-wall synthesis,21 3) the topical use of purified transglycosides and amidase encoded by phages as bacteriolytic cell-wall hydrolyses,22,23 4) use of the coated proteins of M13 phage for the phage-display system, which fused to specific antibodies against bacterial antigens,24 5) use of bacteriophages as a carrier and vehicles for the delivery of engineered genomic materials and vaccine antigens.25,27 Owing to variable success and poor documentation of the use of phage therapy, the use of it has caused much of the controversy in the treatment of infectious diseases.9
However, most researchers focused on promising genes, which have potential targets for broad spectrum antisense growth inhibition in limited strains of one bacterial species or in different bacterial species.6,10,12 Other antisense technologies, in addition to PNAs, using against resistant bacteria, are phosphorothioate oligodeoxynucleotides (S-oligos), locked nucleic acid (LNAs), and phosphorodiamidate morpholino-oligomers (PMOs).28,30 Therefore, identification of gene targets for broad-spectrum antisense inhibition could help for the development of new antibacterial agents that could relieve the exacerbating clinical consequences caused by ESKAPE pathogens.
ZFNs and Transcription activator-like effector nucleases (TALENs) are restriction nucleases that can be engineered and designed to cleave specific sequences of DNA, and are also gene editing tools. These proteins are fused to a nonspecific endonuclease of the type IIS FokI restriction enzyme, which confers the nuclease activity of ZFNs and TALENs.31,32 TALENs and ZFNs are similar to each other in that they can be used to knock-in or knock-out genes and generate double-strand breaks at a desired target site in the genome in the same way. However, the larger size of TALENs compared with ZFNs is a clear disadvantage, which makes it harder to deliver and express TALENs into cells.33
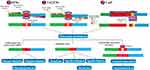
In all gene editing tools, the generated double-strand break can be repaired using either homology-directed repair (HDR) or non-homologous end joining (NHEJ) in the cells.32 NHEJ is an error-prone mechanism that can knockout the gene by a combination of nonsense-mediated decay of the mRNA transcript and pre-mature truncation of the protein mechanisms, a process that is not always particularly efficient. In addition, HDR is another mechanism to repair double-strand break in DNA, by inserting a specific mutation with the introduction of a homologous piece of DNA. These mechanisms lead to mutations that terminate the translation of the gene product and change the open reading frame (ORF)34,35 (Figure 1).
One of the most studied approaches that have been developed against antibiotic-resistant bacteria is the CRISPR-Cas systems. Several recent studies proposed CRISPR-Cas systems for controlling antibiotic-resistant strains.5,17,36,40 Therefore, in this review article, we aimed to review the role of the CRISPR-Cas system to promote sensitization of bacteria against antibiotics.
Introducing CRISPR-Cas
CRISPR-Cas systems have been identified as a bacterial adaptive immune system41 and as analogs of the mammalian immune system.42 In the recent studies, the CRISPR-Cas system has been used for specific genome editing and different applications including treating genetic diseases,43 genome engineering of various bacteria,44 plants,45 mice,46 flies,47 worms,48 and more, and reversal of antibiotic resistance by targeting resistance genes,5,17,40 as well as integration machinery of the systems, has been used to function as a molecular recording device.49 This interesting system is found in approximately 50% of bacterial genomes and 87% of archaeal genomes.50 The genetic loci of CRISPR-Cas systems contain the CRISPR array, which is comprised of short repeated sequences (repeats) and similarly sized flanking sequences (spacers). The spacers of CRISPR arrays are known as protospacers, which are acquired from DNA sequences from invading phage or plasmid. The Cas proteins are key functional elements of CRISPR systems, which are encoded upstream of the CRISPR array and determine the system activity.50,51 The mechanism of CRISPR systems is similar to RNA interference (RNAi) in eukaryotic cells, which use small RNAs (sRNA) to identify and neutralize specific sequences of invading DNA including phages, transposons, and plasmids.52,53 The mechanism of action of CRISPR-Cas systems is summarized in three stages including adaptation, expression, and interference.54 During the adaptation stage, an approximately 30bp segment of homologous invading foreign DNA integrates into the leader side of the CRISPR locus, and protospacer adjacent motifs (PAMs) have been selected from spacers sequences of the host genome. During the expression stage, RNA is transcribed from the spacers of the CRISPR locus (pre-crRNA) and processed into crRNA. Ultimately, in the interference stage, crRNA along with Cas proteins specifically detect the invading DNA, cleave it, and generate a double-strand break.54 There are six types of CRISPR-Cas system, which have been defined based on their sequence similarity, phylogenetic analysis, neighborhood analysis and comparison, distinct features of the components, and experimental data, including distinct features of the physiology, biochemistry, and molecular mechanism.55 Based on the current classification in the Makarova et al55 study, there are two classes of CRISPR-Cas system, which include six types (I–VI) and 33 subtypes. In this classification, Class 1 includes types I, III, and IV, along with 16 subtypes that contain multiple Cas proteins as effector modules, which form crRNA-binding complexes and mediate together in pre-crRNA processing and interference. Class 2 includes types II, V, and VI, along with 17 subtypes that contain a single, large, multidomain crRNA-binding protein (Cas9 in type II, Cas12 in type V, and Cas13 in type III), which is involved in all activities required for interference (in all variants), and in pre-crRNA processing (in some variants). Type I and II systems need two critical factors to effectively target DNA: 1) a PAM specific to each CRISPR-Cas system flanking the protospacer, and ii) complementarity between the target protospacer sequence and the CRISPR RNA spacer.44,56,57 Effective targeting may occur even for multiple mismatches between the protospacer and the CRISPR RNA, although mismatches are more disruptive among the “seed” region flanking the PAM.56,58 In type III systems, similar factors are required for DNA-targeting, where these systems assess base pairing between the region flanking the protospacer and the target sequence.59 Therefore, the CRISPR system can be programmed to specifically detect any DNA target provided in the CRISPR array. CRISPR-Cas has been used to target specific genes in virulence and genes that encode antibiotic resistance in bacterial populations.17,37,39,60,63 Type IV systems are highly derived variants, which require the nucleases for interference and typically have no adaptation modules.55 Other types of class 2 (ie, V and VI) contain large proteins as the effector modules, which clearly differentiate their domain structure.64 The large effector protein of type VI and subtype V-A systems also contain RNase activity of the pre-crRNA processing, while this processing activity in type II and several subtypes of type V is typically relegated to RNase III, a non-Cas enzyme.65,67 After this process, the crRNA-tracrRNA complex, as the mature guide RNA, allows for specific DNA interference through the remaining stable bound to the effectors.68,71
CRISPR-Cas Roles in the Antibiotic-Resistant Bacteria
In some studies, there was a significant reverse relation between the CRISPR-Cas system and antibiotic resistance in some species such as enterococci,72,74 but, in other studies, there was no significant relation, such as in E. coli.39 In enterococci, there are three CRISPR loci including CRISPR1-Cas, orphan CRISPR2, lack of cas genes, and CRISPR3-Cas.74 Results have shown that orphan CRISPR2 is found in all E. faecalis strains, but the presence of CRISPR1-Cas and CRISPR3-Cas is varied among strains. In addition, Palmer and Gilmore74 found that CRISPR1-Cas and CRISPR2 are functionally linked to each other. Analysis of CRISPR spacers demonstrated that pheromone-response plasmids play an important role in enterococcal genome plasticity, mobilizing chromosomally encoded virulence factors and antibiotic resistance, and are capable of promoting their own transfer, and causing displacement of CRISPR-Cas.74,75 The distribution of these spacers of pheromone-responsive plasmids suggested that certain elements have a propensity to be incorporated into CRISPR loci as spacers or are frequently encountered by E. faecalis, but no CRISPR spacers have yet been identified against Tn916, as vectors of antibiotic resistance in enterococci.76,77 The observation is that the tetM gene, which is commonly disseminated by Tn916 and other conjugative transposons,76,77 is present in E. faecalis possessing CRISPR loci,74 which may suggest that conjugate transposons evade this defense. In addition, there is no spacer for the Inc18 plasmid family, which has disseminated vancomycin-resistance genes from enterococci to MRSA.78,79 These findings may reflect the relative inefficiency of transfer of elements, which lack mechanisms for effective pair formation or the relative rarity of interspecies transfer. In addition, these roles were demonstrated in CRISPR-harboring strains of Streptococcus thermophiles, which acquired new spacers derived from the virus and became resistant to infection by phages.41
Brouns et al53 reported that E. coli K12 strains, which carried an artificial CRISPR-Cas system with spacers that target genes of the Lambda phage, displayed decreased sensitivity to Lambda phage. In addition, Maraffini and Sontheimer80 demonstrated that S. epidermidis may have plasmid conjugation limited by the CRISPR-Cas system, which suggests a broader and more critical role for the CRISPR-Cas system in the prevention of horizontal gene transfer (HGT). There are four CRISPR loci in E. coli, including CRISPR1, CRISPR2, CRISPR3, and CRISPR4, which each have a different type of cas gene.81,82 Interestingly, all Escherichia genomes carrying CRISPR1 lack CRISPR4; consequently, no genome of Escherichia strains has more than three CRISPR.82 Touchon et al39 demonstrated that the presence of CRISPR loci of E. coli and the total number of repeats are not associated with the presence of integrons, plasmids, or antibiotic resistance. In addition, they assessed sequence similarities between available plasmid sequences and the spacers, and they found no evidence of spacers that matched elements involved in antibiotic resistance gene mobilization such as Tn3, intI, ISEcp1 or antibiotic resistance gene, ESBL production or the type of ESBL or replicon, and plasmid sequences.39 They found only relatively small numbers of spacers matching plasmid genes among the E. coli strains examined and the spacers were specific for each group of CRISPR, including one single strain with 15 spacers; 77% of the strains had one, 13.2% had two, and 7.5% had three or four spacers. In addition, Touchon et al39 found little effect of CRISPR on the epidemiology of plasmids in E. coli or on the spread of antibiotic resistant genes. These results are contrary to findings in enterococci, in which Palmer and Gilmore74 found CRISPRs are inversely associated with antibiotic resistance. Based on these data, CRISPR-Cas systems may have a different effect on the antibiotic resistance among different species, which may be due to different evolutionary histories of the CRISPR-Cas system, CRISPR locus generation by partial or total deletion in the cas genes cluster, and the presence of anti-CRISPR proteins, just like Toxin-Antitoxin, or restriction and modification systems that are used for competition between plasmids.82,85 When the cas system is complete, the number of repeats is high; when the erosion of the cas system is recent, the number of repeats is intermediate; and when only relics of the system are detectable, the number of repeats is reduced to a few copies.82 Interestingly, three very closely related strains of E.coli genomes, including BL21, BL21-DE3, and B-REL606 have a large number of repeats, which are devoid of cas genes, while a complete absence of CRISPR2 is detected in E. coli strains SMS35 due to a recent insertion of a sucrose operon.82 Generally, the number of repeats is a good indicator of the possible functionality and integrity of the system. In addition, if the CRISPR locus contains spacers matching the chromosome, the host might pose a serious danger due to acquisition of a mobile element. If the CRISPR-Cas system triggers DNA plasmid degradation, which had spacers like chromosomal DNA, the outcome could be chromosome degradation. Also, if the CRISPR-Cas system only causes gene expression interference, this might still permit host manipulation by the foreign element. Therefore, bacterial genomes might have evolved a mechanism to inhibit or escape the incoming CRISPR-Cas system. This mechanism could be the use of native CRISPR acting as anti-CRISPR, which are demonstrated in E. coli and P. aeruginosa.82,85
CRISPR-Cas System Neutralizing Antibiotic-Resistant Genes
In general, RNA-based spacers that are flanked by partial repeats direct Cas proteins to specifically target DNA and cleave it that encode matching protospacers. Therefore, the CRISPR-Cas system can be programmed to specifically target and cleave any DNA for in vivo based on the information provided in the CRISPR array, which has been exploited to target the bacterial population which carry specific genes that encode antibiotic resistance.8,60,63
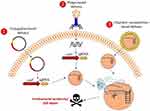
Recent work has shown that intentional or accidental targeting of the sequence of bacterial genome by the CRISPR-Cas system is cytotoxic, which can lead to cell death because of the introduction of irreversible chromosomal lesions.61,86 Because of the widely conserved CRISPR-Cas system in bacteria and archaea, the isolation, optimization, and development of delivery carriers and vectors of the CRISPR-Cas system will be needed for the creation of RNA-guided nucleases capable of targeting additional strains, including multidrug-resistant (MDR) pathogens and key members of endogenous microbiota.61 In addition, owing to the delivery system that acts in higher organisms, RNA-guided nucleases could enable us to modulate the prevalence of specific genes such as antibiotic resistance genes and virulence determinants in wild-type populations.87 The Cas9 nuclease targeting specific DNA sequences of bacterial pathogens and antibiotic resistance gene are delivered to microbial populations using polymer-derivatized CRISPR nanocomplexes,36 bacteria carrying plasmids transmissible by conjugation,61,88 and/or bacteriophages60,61 (Figure 2). Gomma et al62 used the typed I–E CRISPR-Cas system of E. coli, encoding six cas genes in two operons including casABCDE and cas3,53 to specific and essential sequences in the genomes of different sequences. They concluded that potent elimination can be achieved by targeting of multiple and divers locations including ftsA, nusB, msbA, and asd throughout the genome, and extents of elimination exhibited by simultaneous targeting of multiple locations similar to those with targeting of only one of the locations.62 In this study, they used transformation in spite of using a potent procedure for delivering a type I CRISPR-cas system into bacteria, as well as targeting chromosomal genes, which are unique to part of the mixed population, in spite of targeting resistance genes on extrachromosomal elements. Bikard et al60 exploited the Cas9 phagemid, which is a plasmid that is designed to be packaged in phage capsids,89 and to eradicate MRSA strains from a mixed population of bacteria. In addition, they designed a CRISPR-Cas9 plasmid that targets tetracycline-resistant plasmids including pUSA01 and pUSA02. They are constructed cas9 and crRNA of methicillin resistance gene mecA90 in the phagemid (pDB121::mecA), to treat the clinical isolate of S. aureus USA300Φ. In all cases of Cas9 plasmid that target tetracycline-resistant plasmids, cell death was not observed, but tetracycline sensitivity was found in more than 99.99% of the cells, and a decrease in proportion of S. aureus USA300Φ was observed from 50% before treatment to 0.4% after treatment of pDB121:mecA phagemid. In addition, they constructed enterotoxin sek gene91 in the phagemid and they observed the phagemid able to kill all S. aureus with comparable efficiencies. In another study, Citorik et al61 used two approaches including M13-based phagemid and conjugative plasmid to deliver the Cas9 nuclease into bacteria for targeting blaNDM-1 and blaSHV-18, which encode pan-resistant to beta-lactams and extended-spectrum resistant to antibiotics, respectively.92,93 Treatment of the resistant strains with the phages resulted in 2–3-log reductions in viable cells. In addition, they demonstrated that the CRISPR-Cas system could discriminate between resistant and susceptible strains, which found that programmed gyrA phagemid was specifically cytotoxic only for chromosomal gyrA mutations that are responsible to quinolone-resistant E. coli94 and not cytotoxic for isogenic strains with the wild type gyrA gene.61
In another study, Kang et al36 introduced a nonviral delivery method based on a Cas9-nanocomplex, which used Cas9, sgRNA targeting mecA, and a cationic polymer, known as branched polyethyleneimine (bPEI). The bPEI is one of the most commonly used carriers for gene delivery95,96 and was used as the carrier for packaging sgRNA, which can enhance Cas9 delivery into MRSA strains.36 Kang et al36 demonstrated that Cas9 conjugated with bPEI could uptake into the bacteria greater than native Cas9 simply mixed with bPEI and native Cas9 mixed with lipofectamine (a carrier for gene delivery in mammalian cells97,98), which did not show any sign of uptake. They suggested that this phenomenon could be due to the resultant enhancement in polarity of the protein or highly cationic characteristic of the bPEI polymer.36 In addition, the cultured MRSA strains treated with Cas9-bPEI could not be able to grow in agar media, including 6 µg/mL oxacillin, while the strains that were not treated could grow in the media. Also, treatment with the Cas9-bPEI could significantly decrease the growth (a 32% decrease) compared to treating with the Cas9-bPEI complex without sgRNA as the control, while native Cas9/sgRNA complexed with a lipofectamine carrier showed no decrease in growth.36 This finding could be a forward step to design CRISPR-based antimicrobials drugs, because vector-free delivery of CRISPR can avoid off-target effects and immunogenicity problems, as well as could simply induce phenotypic changes and edit the bacterial genome.
One of the limiting factors in CRISPR-mediated killing by plasmids is the low frequency of conjugation, which can be developed phagemid- or bacteriophage-mediated delivery that are much more efficient. Nonetheless, delivery of CRISPR nucleases by the conjugative plasmid delivery remains an interested option due to conjugative plasmids which do not require a cellular receptor,99 are easy to engineer with large coding capacities,100 are resistant to restriction-modification systems,101 and have broad host ranges.102 Conjugative plasmids that encode and promote biofilm formation could increase rates of conjugative plasmid transfer due to the enhanced cell-to-cell contact,103 which may be suited for delivery of molecular tools such as CRISPR nucleases for modulating composition of microbial communities104,106 that many of them exist as biofilms. In a study by Hamilton et al,107 they developed a cis-conjugative system that the plasmid encodes both CRISPR nuclease and conjugative machinery. They designed 65 total sgRNA, which target 38, 23, and 4 essential genes, non-essential genes and genes with unresolved phenotypes. They found that plasmids containing conjugative machinery and CRISPR nuclease under conditions that enhance cell-to-cell contact have a higher frequency of conjugative transfer from E. coli to Salmonella enterica. They concluded that single or multiplexed sgRNAs targeting non-essential genes such as katG (catalase reductase), yghJ (putative lipoprotein), aegA (putative oxidoreductase), and gltJ (glutamate/aspartate transporter) result in high killing efficacy of S. enterica compared to essential genes. In addition, they demonstrated that when the bacteria receive the cis-conjugative plasmids, recipients become potential donors for subsequent rounds of conjugation and could be potentially leading to exponentially enhancing numbers of conjugative donor bacteria in the population. They suggested that this delivery system combined by CRISPR nucleases could be an effective tool for modification of microbiome.
Citorik et al61 demonstrated that treatment of enterohemorrhagic E. coli (EHEC) with phagemid eae targeting intimin, which is a chromosomally encoded virulence factor of E. coli O157:H7 and is necessary for intestinal pathology and colonization,108 resulted in a 20-fold reduction in viable cell counts. These data demonstrated that the CRISPR-Cas system activity can selectivity remove bacteria with specific genome, which could reduce the prevalence of unwanted genes, such as virulence loci, antibiotic resistance, or metabolic pathways from the bacterial population without affecting bystanders. In addition, the CRISPR-Cas system can be used to modulate the composition of complex bacterial communities, which current therapies that use a drug to modify the human microbiota, prebiotic or probiotic have potential for reducing suffering of various diseases, but such therapies remain poorly characterized in terms of the specific mechanisms by which they act.109
The efficiency of the Cas9 phagemid against pathogens were tested in an in-vivo mouse model.60 Topical treatment of the back of CD1 mice, which were infected by RNKΦ cells, with the CRISPR-Cas9 antimicrobial pDB121::aph phagemid (against kanamycin resistance gene aph110), demonstrated a decrease in the proportion of RNKΦ cells that was significantly different from 2% mupirocin or streptomycin (200 mg/mouse) treatments (P<0.0001), of which streptomycin treatment eliminated the infection, but mupirocin did not.60 In another study, Citorik et al61 demonstrated that treatment of EHEC with cas9-eae phagemid in Galleria mellonella larva was significantly improved survival over no treatment control and was significantly more efficient than chloramphenicol treatment, to which the strain was resistant, as well as was inferior to carbenicillin, to which the strain was susceptible. G. mellonella is an infection model to evaluate the efficiency of phage therapy and antimicrobials against various Gram-positive and -negative and fungal pathogens.111 Other studies, such as Kiga et al,112 designed Cas13a-based phage to target carbapenem-resistant E. coli and methicillin-resistant S. aureus. These data support the CRISPR-Cas system and use of phagemid as viable alternatives for cases where bacterial strains are highly resistant to existing antimicrobial agents.
However, the use of phagemid has two key weaknesses. First, the phagemid does not produce more phages after infections, which means that the amount of phagemid needed to treatments are much larger than the size of the target population. Second, the narrow-host range and large scale population of phagemids could preclude their extensive use.
The advantage of programmed Cas9-mediated killing is the possibility of a nuclease with two or more crRNA guides to trigger different plasmid and/or chromosomal sequences, which could decrease resistant clones that desert phagemid treatment through the generation of target mutations, as well as extend the range of targeted cells. In addition, delivery of the sequence-specific Cas9 nuclease and easy reprogramming to target different sequences reduces the plasmid content in a bacterial population without killing the cells, which can be immunized non-pathogenic strains against the transfer of antibiotic-resistant and/or virulence plasmids.
As future directions, depending on the nature of the dysbiosis and microbiome, most of the researchers focused on the phage-, conjugative-, and polymeric nanoparticles-based CRISPR delivery systems and the best killing efficiency target genes. Most of the studies suggest that a combination of the phage-, conjugative-, and polymeric nanoparticles-based delivery systems may be appropriate for planktonic and biofilm conditions. In addition, a central problem in microbiome manipulation, microbiology, and infectious disease control is complete elimination of the target organism(s) and is the lack of special tools to control pathogenic species or to alter the composition of microbial communities.113 CRISPR-based nucleases as sequence-specific antimicrobial agents could reduce the relative abundance of the target and infectious bacteria, yet the development of a broadly applicable and robust delivery system remains a key milestone. Therefore, the challenges ahead against CRISPR-Cas-based antibacterials could be CRISPR-Cas delivery vectors or vehicles and architecture of them, complex bacterial communities, different mechanisms of resistance to one antibacterial agents in the organism(s), possibility of mutations in the target genes, legislation, and social responsibilities of CRISPR-Cas-based antibacterials.
Conclusion
Owing to the rational design, sequence-informed and additional of facile to a field that has been dominated by cost- and time-intensive screening for broad spectrum, the small-molecules and CRISPR-Cas-based antibiotics have the potential to reinvigorate novel horizons of development for new antimicrobials.
Modern antibiotic therapy has shifted toward the straightforward acquisition of antibiotic resistance genes, among other things, increasing plasticity or decreasing genome stability, and enabling the colonization of new habitats such as the antibiotic-laden hospital environment.
We envision that CRISPR-Cas approaches may open novel ways for the development of smart antibiotics, which can eliminate MDR pathogens and make differentiation between beneficial and pathogenic microorganisms. These systems can be exploited to quantitatively and selectively eliminate individual bacterial strains based on the sequence-specific manner, creating opportunities in the treatment of MDR infections, the study of microbial consortia, and the control of industrial fermentations.
Ethics and Consent Statement
All figures in this study are original and not reproduced from any other study.
Acknowledgment
We thank all staff of the DARC research center for all their support and collaboration. We also thank Imam Reza Hospital staff, Rohollah Safari, Laleh Jamali, and Ms Kalani, who helped us to get our goals for infection control.
Funding
This study was supported by Iran National Science Foundation (INSF) with the grant number 97015174 and Faculty of Medicine, Tabriz University of Medical Sciences.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Matsuzaki S, Yasuda M, Nishikawa H, et al. Experimental protection of mice against lethal Staphylococcus aureus infection by novel bacteriophage ϕMR11. J Infect Dis. 2003;187(4):613–624. doi:10.1086/374001
2. Heo Y-J, Lee Y-R, Jung -H-H, Lee J, Ko G, Cho Y-H. Antibacterial efficacy of phages against pseudomonas aeruginosa infections in mice and drosophila melanogaster. Antimicrob Agents Chemother. 2009;53(6):2469–2474. doi:10.1128/AAC.01646-08
3. Watanabe R, Matsumoto T, Sano G, et al. Efficacy of bacteriophage therapy against gut-derived sepsis caused by Pseudomonas aeruginosa in mice. Antimicrob Agents Chemother. 2007;51(2):446–452. doi:10.1128/AAC.00635-06
4. Verhoef TI, Morris S. Cost‐effectiveness and pricing of antibacterial drugs. Chem Biol Drug Des. 2015;85(1):4–13. doi:10.1111/cbdd.12417
5. Goren M, Yosef I, Qimron U. Sensitizing pathogens to antibiotics using the CRISPR-Cas system. Drug Resist Updates. 2017;30:1–6. doi:10.1016/j.drup.2016.11.001
6. Narenji H, Gholizadeh P, Aghazadeh M, Rezaee MA, Asgharzadeh M, Kafil HS. Peptide nucleic acids (PNAs): currently potential bactericidal agents. Biomed Pharmacother. 2017;93:580–588. doi:10.1016/j.biopha.2017.06.092
7. Boucher HW, Scheld M, Bartlett J, et al. Bad bugs, no drugs: no ESKAPE! An update from the infectious diseases society of America. Clin Infect Dis. 2009;48(1):1–12. doi:10.1086/595011
8. Yosef I, Manor M, Kiro R, Qimron U. Temperate and lytic bacteriophages programmed to sensitize and kill antibiotic-resistant bacteria. Proc Natl Acad Sci. 2015;112(23):7267–7272. doi:10.1073/pnas.1500107112
9. Lin DM, Koskella B, Lin HC. Phage therapy: an alternative to antibiotics in the age of multi-drug resistance. World J Gastrointest Pharmacol Ther. 2017;8(3):162–173. doi:10.4292/wjgpt.v8.i3.162
10. Bai H, You Y, Yan H, et al. Antisense inhibition of gene expression and growth in gram-negative bacteria by cell-penetrating peptide conjugates of peptide nucleic acids targeted to rpoD gene. Biomaterials. 2012;33(2):659–667. doi:10.1016/j.biomaterials.2011.09.075
11. Wright GD. Making sense of antisense in antibiotic drug discovery. Cell Host Microbe. 2009;6(3):197–198. doi:10.1016/j.chom.2009.08.009
12. Singh SB, Phillips JW, Wang J. Highly sensitive target-based whole-cell antibacterial discovery strategy by antisense RNA silencing. Curr Opin Drug Discov Devel. 2007;10(2):160.
13. Abdi SN, Ghotaslou R, Asgharzadeh M, et al. AdeB efflux pump gene knockdown by mRNA mediated peptide nucleic acid in multidrug resistance Acinetobacter baumannii. Microb Pathog. 2020;139:103825. doi:10.1016/j.micpath.2019.103825
14. Narenji H, Teymournejad O, Rezaee MA, et al. Antisense peptide nucleic acids againstftsZ andefaA genes inhibit growth and biofilm formation of Enterococcus faecalis. Microb Pathog. 2020;139:103907. doi:10.1016/j.micpath.2019.103907
15. Dastjerdeh MS, Kouhpayeh S, Sabzehei F, et al. Zinc finger nuclease: a new approach to overcome beta-lactam antibiotic resistance. Jundishapur J Microbiol. 2016;9(1).
16. Gupta RM, Musunuru K. Expanding the genetic editing tool kit: ZFNs, TALENs, and CRISPR-Cas9. J Clin Invest. 2014;124(10):4154–4161. doi:10.1172/JCI72992
17. Gholizadeh P, Aghazadeh M, Asgharzadeh M, Kafil H. Suppressing the CRISPR/Cas adaptive immune system in bacterial infections. Eur J Clin Microbiol Infect Dis. 2017;36(11):2043–2051. doi:10.1007/s10096-017-3036-2
18. van Belkum A, Soriaga LB, LaFave MC, et al. Phylogenetic distribution of CRISPR-Cas systems in antibiotic-resistant Pseudomonas aeruginosa. MBio. 2015;6(6):e01796–15. doi:10.1128/mBio.01796-15
19. Yao R, Liu D, Jia X, Zheng Y, Liu W, Xiao Y. CRISPR-Cas9/Cas12a biotechnology and application in bacteria. Synth Syst Biotechnol. 2018;3(3):135–149. doi:10.1016/j.synbio.2018.09.004
20. Biswas B, Adhya S, Washart P, et al. Bacteriophage therapy rescues mice bacteremic from a clinical isolate of vancomycin-resistant Enterococcus faecium. Infect Immun. 2002;70(1):204–210. doi:10.1128/IAI.70.1.204-210.2002
21. Bernhardt TG, Wang N, Struck DK, Young R. A protein antibiotic in the phage Qβ virion: diversity in lysis targets. Science. 2001;292(5525):2326–2329. doi:10.1126/science.1058289
22. Schuch R, Nelson D, Fischetti VA. A bacteriolytic agent that detects and kills Bacillus anthracis. Nature. 2002;418(6900):884. doi:10.1038/nature01026
23. Loeffler JM, Nelson D, Fischetti VA. Rapid killing of Streptococcus pneumoniae with a bacteriophage cell wall hydrolase. Science. 2001;294(5549):2170–2172. doi:10.1126/science.1066869
24. Cao J, Sun Y-Q, Berglindh T, et al. Helicobacter pylori-antigen-binding fragments expressed on the filamentous M13 phage prevent bacterial growth. Biochim Biophys Acta Bioenerg. 2000;1474(1):107–113. doi:10.1016/S0304-4165(00)00005-2
25. Clark JR, March JB. Bacterial viruses as human vaccines? Expert Rev Vaccines. 2004;3(4):463–476. doi:10.1586/14760584.3.4.463
26. March JB, Clark JR, Jepson CD. Genetic immunisation against hepatitis B using whole bacteriophage lambda particles. Vaccine. 2004;22(13–14):1666–1671. doi:10.1016/j.vaccine.2003.10.047
27. Jepson CD, March JB. Bacteriophage lambda is a highly stable DNA vaccine delivery vehicle. Vaccine. 2004;22(19):2413–2419. doi:10.1016/j.vaccine.2003.11.065
28. Sully EK, Geller BL. Antisense antimicrobial therapeutics. Curr Opin Microbiol. 2016;33:47–55. doi:10.1016/j.mib.2016.05.017
29. Singh P, Panda D. FtsZ inhibition: a promising approach for antistaphylococcal therapy. Drug News Perspect. 2010;23(5):295–304. doi:10.1358/dnp.2010.23.5.1429489
30. Lopez C, Arivett BA, Actis LA, Tolmasky ME. Inhibition of AAC (6′)-Ib-mediated resistance to amikacin in Acinetobacter baumannii by an antisense peptide-conjugated 2′, 4′-bridged nucleic acid-NC-DNA hybrid oligomer. Antimicrob Agents Chemother. 2015;59(9):5798–5803. doi:10.1128/AAC.01304-15
31. Liu Y, Zhao H, Cheng CH. Mutagenesis in Xenopus and Zebrafish Using TALENs. Springer; 2016:207–227.
32. Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Genome editing with engineered zinc finger nucleases. Nat Rev Genet. 2010;11(9):636. doi:10.1038/nrg2842
33. Holkers M, Maggio I, Liu J, et al. Differential integrity of TALE nuclease genes following adenoviral and lentiviral vector gene transfer into human cells. Nucleic Acids Res. 2012;41(5):e63–e. doi:10.1093/nar/gks1446
34. Rémy S, Tesson L, Ménoret S, Usal C, Scharenberg AM, Anegon I. Zinc-finger nucleases: a powerful tool for genetic engineering of animals. Transgenic Res. 2010;19(3):363–371. doi:10.1007/s11248-009-9323-7
35. Rouet P, Smih F, Jasin M. Expression of a site-specific endonuclease stimulates homologous recombination in mammalian cells. Proc Natl Acad Sci. 1994;91(13):6064–6068. doi:10.1073/pnas.91.13.6064
36. Kang YK, Kwon K, Ryu JS, Lee HN, Park C, Chung HJ. Nonviral genome editing based on a polymer-derivatized CRISPR nanocomplex for targeting bacterial pathogens and antibiotic resistance. Bioconjug Chem. 2017;28(4):957–967. doi:10.1021/acs.bioconjchem.6b00676
37. Müller V, Rajer F, Frykholm K, et al. Direct identification of antibiotic resistance genes on single plasmid molecules using CRISPR/Cas9 in combination with optical DNA mapping. Sci Rep. 2016;6:37938. doi:10.1038/srep37938
38. Price VJ, McBride SW, Duerkop B, Palmer KL. CRISPR-Cas blocks antibiotic resistance plasmid transfer between Enterococcus faecalis strains in the gastrointestinal tract. bioRxiv. 2018;312751.
39. Touchon M, Charpentier S, Pognard D, et al. Antibiotic resistance plasmids spread among natural isolates of Escherichia coli in spite of CRISPR elements. Microbiology. 2012;158(12):2997–3004. doi:10.1099/mic.0.060814-0
40. Zhang H, Cheng Q-X, Liu A-M, Zhao G-P WJ. A novel and efficient method for bacteria genome editing employing both CRISPR/Cas9 and an antibiotic resistance cassette. Front Microbiol. 2017;8:812. doi:10.3389/fmicb.2017.00812
41. Barrangou R, Fremaux C, Deveau H, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315(5819):1709–1712. doi:10.1126/science.1138140
42. Goren M, Yosef I, Edgar R, Qimron U. The bacterial CRISPR/Cas system as analog of the mammalian adaptive immune system. RNA Biol. 2012;9(5):549–554. doi:10.4161/rna.20177
43. Hussain W, Mahmood T, Hussain J, et al. CRISPR/Cas system: a game changing genome editing technology, to treat human genetic diseases. Gene. 2019;685:70–75. doi:10.1016/j.gene.2018.10.072
44. Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol. 2013;31(3):233. doi:10.1038/nbt.2508
45. Feng Z, Zhang B, Ding W, et al. Efficient genome editing in plants using a CRISPR/Cas system. Cell Res. 2013;23(10):1229–1232. doi:10.1038/cr.2013.114
46. Li D, Qiu Z, Shao Y, et al. Heritable gene targeting in the mouse and rat using a CRISPR-Cas system. Nat Biotechnol. 2013;31(8):681–683. doi:10.1038/nbt.2661
47. Gratz SJ, Cummings AM, Nguyen JN, et al. Genome engineering of Drosophila with the CRISPR RNA-Guided Cas9 nuclease. Genetics. 2013;194(4):1029–1035. doi:10.1534/genetics.113.152710
48. Friedland AE, Tzur YB, Esvelt KM, Colaiácovo MP, Church GM, Calarco JA. Heritable genome editing in C. elegans via a CRISPR-Cas9 system. Nat Methods. 2013;10(8):741–743. doi:10.1038/nmeth.2532
49. Shipman SL, Nivala J, Macklis JD, Church GM. Molecular recordings by directed CRISPR spacer acquisition. Science. 2016;353(6298):aaf1175. doi:10.1126/science.aaf1175
50. Makarova KS, Wolf YI, Alkhnbashi OS, et al. An updated evolutionary classification of CRISPR–Cas systems. Nat Rev Microbiol. 2015;13:722. doi:10.1038/nrmicro3569
51. Horvath P, Barrangou R. CRISPR/Cas, the immune system of bacteria and archaea. Science. 2010;327(5962):167–170. doi:10.1126/science.1179555
52. Makarova KS, Grishin NV, Shabalina SA, Wolf YI, Koonin EV. A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol Direct. 2006;1(1):7. doi:10.1186/1745-6150-1-7
53. Brouns SJ, Jore MM, Lundgren M, et al. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321(5891):960–964. doi:10.1126/science.1159689
54. Makarova KS, Haft DH, Barrangou R, et al. Evolution and classification of the CRISPR–Cas systems. Nat Rev Microbiol. 2011;9(6):467. doi:10.1038/nrmicro2577
55. Makarova KS, Wolf YI, Iranzo J, et al. Evolutionary classification of CRISPR–Cas systems: a burst of class 2 and derived variants. Nat Rev Microbiol. 2020;18(2):67–83. doi:10.1038/s41579-019-0299-x
56. Semenova E, Jore MM, Datsenko KA, et al. Interference by clustered regularly interspaced short palindromic repeat (CRISPR) RNA is governed by a seed sequence. Proc Natl Acad Sci. 2011;108(25):10098–10103. doi:10.1073/pnas.1104144108
57. Mojica FJ, Díez-Villaseñor C, García-Martínez J, Almendros C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology. 2009;155(3):733–740. doi:10.1099/mic.0.023960-0
58. Wiedenheft B, van Duijn E, Bultema JB, et al. RNA-guided complex from a bacterial immune system enhances target recognition through seed sequence interactions. Proc Natl Acad Sci. 2011;108(25):10092–10097. doi:10.1073/pnas.1102716108
59. Marraffini LA, Sontheimer EJ. Self versus non-self discrimination during CRISPR RNA-directed immunity. Nature. 2010;463(7280):568. doi:10.1038/nature08703
60. Bikard D, Euler CW, Jiang W, et al. Exploiting CRISPR-Cas nucleases to produce sequence-specific antimicrobials. Nat Biotechnol. 2014;32:1146. doi:10.1038/nbt.3043
61. Citorik RJ, Mimee M, Lu TK. Sequence-specific antimicrobials using efficiently delivered RNA-guided nucleases. Nat Biotechnol. 2014;32:1141. doi:10.1038/nbt.3011
62. Gomaa AA, Klumpe HE, Luo ML, Selle K, Barrangou R, Beisel CL. Programmable removal of bacterial strains by use of genome-targeting CRISPR-Cas systems. mBio. 2014;5(1):e00928–13. doi:10.1128/mBio.00928-13
63. Hale Caryn R, Majumdar S, Elmore J, et al. Essential features and rational design of CRISPR RNAs that function with the Cas RAMP module complex to cleave RNAs. Mol Cell. 2012;45(3):292–302. doi:10.1016/j.molcel.2011.10.023
64. Shmakov S, Smargon A, Scott D, et al. Diversity and evolution of class 2 CRISPR–Cas systems. Nat Rev Microbiol. 2017;15(3):169. doi:10.1038/nrmicro.2016.184
65. Fonfara I, Richter H, Bratovič M, Le Rhun A, Charpentier E. The CRISPR-associated DNA-cleaving enzyme Cpf1 also processes precursor CRISPR RNA. Nature. 2016;532(7600):517–521. doi:10.1038/nature17945
66. East-Seletsky A, O’Connell MR, Knight SC, et al. Two distinct RNase activities of CRISPR-C2c2 enable guide-RNA processing and RNA detection. Nature. 2016;538(7624):270–273. doi:10.1038/nature19802
67. Liu L, Li X, Wang J, et al. Two distant catalytic sites are responsible for C2c2 RNase activities. Cell. 2017;168(1–2):121–34. e12. doi:10.1016/j.cell.2016.12.031
68. Chylinski K, Makarova KS, Charpentier E, Koonin EV. Classification and evolution of type II CRISPR-Cas systems. Nucleic Acids Res. 2014;42(10):6091–6105. doi:10.1093/nar/gku241
69. Briner AE, Barrangou R. Guide RNAs: a glimpse at the sequences that drive CRISPR–Cas systems. Cold Spring Harb Protoc. 2016;2016(7):
70. Faure G, Shmakov SA, Makarova KS, et al. Comparative genomics and evolution of trans-activating RNAs in Class 2 CRISPR-Cas systems. RNA Biol. 2019;16(4):435–448. doi:10.1080/15476286.2018.1493331
71. Chyou T-Y, Brown CM. Prediction and diversity of tracrRNAs from type II CRISPR-Cas systems. RNA Biol. 2019;16(4):423–434. doi:10.1080/15476286.2018.1498281
72. Burley KM, Sedgley CM. CRISPR-Cas, a prokaryotic adaptive immune system, in endodontic, oral, and multidrug-resistant hospital-acquired enterococcus faecalis. J Endod. 2012;38(11):1511–1515. doi:10.1016/j.joen.2012.07.004
73. Lindenstrauß AG, Pavlovic M, Bringmann A, Behr J, Ehrmann MA, Vogel RF. Comparison of genotypic and phenotypic cluster analyses of virulence determinants and possible role of CRISPR elements towards their incidence in Enterococcus faecalis and Enterococcus faecium. Syst Appl Microbiol. 2011;34(8):553–560. doi:10.1016/j.syapm.2011.05.002
74. Palmer KL, Gilmore MS. Multidrug-resistant enterococci lack CRISPR-cas. mBio. 2010;1(4):e00227–10. doi:10.1128/mBio.00227-10
75. Manson JM, Hancock LE, Gilmore MS. Mechanism of chromosomal transfer of Enterococcus faecalis pathogenicity island, capsule, antimicrobial resistance, and other traits. Proc Natl Acad Sci. 2010;107(27):12269–12274. doi:10.1073/pnas.1000139107
76. Gilmore MS, Clewell DB. The Enterococci: Pathogenesis, Molecular Biology, and Antibiotic Resistance. Zondervan; 2002.
77. Roberts AP, Mullany P. A modular master on the move: the Tn916 family of mobile genetic elements. Trends Microbiol. 2009;17(6):251–258. doi:10.1016/j.tim.2009.03.002
78. Flannagan SE, Chow JW, Donabedian SM, et al. Plasmid content of a vancomycin-resistant Enterococcus faecalis isolate from a patient also colonized by Staphylococcus aureus with a VanA phenotype. Antimicrob Agents Chemother. 2003;47(12):3954–3959. doi:10.1128/AAC.47.12.3954-3959.2003
79. Zhu W, Clark NC, McDougal LK, Hageman J, McDonald LC, Patel JB. Vancomycin-resistant Staphylococcus aureus isolates associated with Inc18-like vanA plasmids in Michigan. Antimicrob Agents Chemother. 2008;52(2):452–457. doi:10.1128/AAC.00908-07
80. Marraffini LA, Sontheimer EJ. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science. 2008;322(5909):1843–1845. doi:10.1126/science.1165771
81. Díez-Villaseñor C, Almendros C, García-Martínez J, Mojica FJ. Diversity of CRISPR loci in Escherichia coli. Microbiology. 2010;156(5):1351–1361. doi:10.1099/mic.0.036046-0
82. Touchon M, Rocha EP. The small, slow and specialized CRISPR and anti-CRISPR of Escherichia and Salmonella. PLoS One. 2010;5(6):e11126. doi:10.1371/journal.pone.0011126
83. Bondy-Denomy J, Garcia B, Strum S, et al. Multiple mechanisms for CRISPR–Cas inhibition by anti-CRISPR proteins. Nature. 2015;526(7571):136. doi:10.1038/nature15254
84. Pawluk A, Bondy-Denomy J, Cheung VH, Maxwell KL, Davidson AR. A new group of phage anti-CRISPR genes inhibits the type IE CRISPR-Cas system of Pseudomonas aeruginosa. MBio. 2014;5(2):e00896–14. doi:10.1128/mBio.00896-14
85. Pawluk A, Staals RH, Taylor C, et al. Inactivation of CRISPR-Cas systems by anti-CRISPR proteins in diverse bacterial species. Nat Microbiol. 2016;1(8):16085. doi:10.1038/nmicrobiol.2016.85
86. Vercoe RB, Chang JT, Dy RL, et al. Cytotoxic chromosomal targeting by CRISPR/Cas systems can reshape bacterial genomes and expel or remodel pathogenicity islands. PLoS Genet. 2013;9(4):e1003454. doi:10.1371/journal.pgen.1003454
87. Esvelt KM, Smidler AL, Catteruccia F, Church GM. Concerning RNA-guided gene drives for the alteration of wild populations. eLife. 2014;3:e03401. doi:10.7554/eLife.03401
88. Kim J-S, Cho D-H, Park M, et al. CRISPR/Cas9-mediated re-sensitization of antibiotic-resistant Escherichia coli harboring extended-spectrum beta-lactamases. J Microbiol Biotechnol. 2016;26(2):394–401. doi:10.4014/jmb.1508.08080
89. Melnikov AA, Tchernov AP, Fodor I, Bayev AA. Lambda phagemids and their transducing properties. Gene. 1984;28(1):29–35. doi:10.1016/0378-1119(84)90084-2
90. Ubukata K, Nonoguchi R, Matsuhashi M, Konno M. Expression and inducibility in Staphylococcus aureus of the mecA gene, which encodes a methicillin-resistant S. aureus-specific penicillin-binding protein. J Bacteriol. 1989;171(5):2882–2885.
91. Isnard C, Malbruny B, Leclercq R, Cattoir V. Genetic basis for in vitro and in vivo resistance to lincosamides, streptogramins A, and pleuromutilins (LSAP phenotype) in Enterococcus faecium. Antimicrob Agents Chemother. 2013;57(9):4463–4469. doi:10.1128/AAC.01030-13
92. Rasheed JK, Anderson GJ, Yigit H, et al. Characterization of the extended-spectrum β-lactamase reference strain, Klebsiella pneumoniae K6 (ATCC 700603), which produces the novel enzyme SHV-18. Antimicrob Agents Chemother. 2000;44(9):2382–2388. doi:10.1128/AAC.44.9.2382-2388.2000
93. Rasheed JK, Kitchel B, Zhu W, et al. New Delhi Metallo-β-Lactamase–producing Enterobacteriaceae, United States. Emerg Infect Dis. 2013;19(6):870. doi:10.3201/eid1906.121515
94. Jacoby GA. Mechanisms of resistance to quinolones. Clin Infect Dis. 2005;41(Supplement_2):S120–S6. doi:10.1086/428052
95. Patnaik S, Gupta KC. Novel polyethylenimine-derived nanoparticles for in vivo gene delivery. Expert Opin Drug Deliv. 2013;10(2):215–228. doi:10.1517/17425247.2013.744964
96. Chung HJ, Hong CA, Lee SH, Jo SD, Park TG. Reducible siRNA dimeric conjugates for efficient cellular uptake and gene silencing. Bioconjug Chem. 2011;22(2):299–306. doi:10.1021/bc100438m
97. Zuris JA, Thompson DB, Shu Y, et al. Cationic lipid-mediated delivery of proteins enables efficient protein-based genome editing in vitro and in vivo. Nat Biotechnol. 2015;33(1):73. doi:10.1038/nbt.3081
98. Wang M, Zuris JA, Meng F, et al. Efficient delivery of genome-editing proteins using bioreducible lipid nanoparticles. Proc Natl Acad Sci. 2016;113(11):2868–2873. doi:10.1073/pnas.1520244113
99. Pérez-Mendoza D. de la Cruz F. Escherichia coli genes affecting recipient ability in plasmid conjugation: are there any? BMC Genom. 2009;10(1):71. doi:10.1186/1471-2164-10-71
100. Smillie C, Garcillán-Barcia MP, Francia MV, Rocha EP, de la Cruz F. Mobility of plasmids. Microbiol Mol Biol Rev. 2010;74(3):434–452. doi:10.1128/MMBR.00020-10
101. Oliveira PH, Touchon M, Rocha EP. The interplay of restriction-modification systems with mobile genetic elements and their prokaryotic hosts. Nucleic Acids Res. 2014;42(16):10618–10631. doi:10.1093/nar/gku734
102. Jain A, Srivastava P. Broad host range plasmids. FEMS Microbiol Lett. 2013;348(2):87–96. doi:10.1111/1574-6968.12241
103. Hausner M, Wuertz S. High rates of conjugation in bacterial biofilms as determined by quantitative in situ analysis. Appl Environ Microbiol. 1999;65(8):3710–3713. doi:10.1128/AEM.65.8.3710-3713.1999
104. Peters JM, Koo B-M, Patino R, et al. Enabling genetic analysis of diverse bacteria with Mobile-CRISPRi. Nat Microbiol. 2019;4(2):244–250. doi:10.1038/s41564-018-0327-z
105. Brophy JA, Triassi AJ, Adams BL, et al. Engineered integrative and conjugative elements for efficient and inducible DNA transfer to undomesticated bacteria. Nat Microbiol. 2018;3(9):1043–1053. doi:10.1038/s41564-018-0216-5
106. López-Igual R, Bernal-Bayard J, Rodríguez-Patón A, Ghigo J-M MD. Engineered toxin–intein antimicrobials can selectively target and kill antibiotic-resistant bacteria in mixed populations. Nat Biotechnol. 2019;37(7):755–760. doi:10.1038/s41587-019-0105-3
107. Hamilton TA, Pellegrino GM, Therrien JA, et al. Efficient inter-species conjugative transfer of a CRISPR nuclease for targeted bacterial killing. Nat Commun. 2019;10(1):4544. doi:10.1038/s41467-019-12448-3
108. Kaper JB, Nataro JP, Mobley HL. Pathogenic escherichia coli. Nat Rev Microbiol. 2004;2(2):123. doi:10.1038/nrmicro818
109. Sonnenburg JL, Fischbach MA. Community health care: therapeutic opportunities in the human microbiome. Sci Transl Med. 2011;3(78):78ps12. doi:10.1126/scitranslmed.3001626
110. Gray GS, Fitch WM. Evolution of antibiotic resistance genes: the DNA sequence of a kanamycin resistance gene from Staphylococcus aureus. Mol Biol Evol. 1983;1(1):57–66. doi:10.1093/oxfordjournals.molbev.a040298
111. Desbois AP, Coote PJ. Utility of greater wax moth larva (Galleria mellonella) for evaluating the toxicity and efficacy of new antimicrobial agents. Adv Appl Microbiol. 2012;78:25–53. Elsevier
112. Kiga K, Tan X-E, Ibarra-Chávez R, et al. Development of CRISPR-Cas13a-based antimicrobials capable of sequence-specific killing of target bacteria. bioRxiv. 2019;808741.
113. Roach DR, Leung CY, Henry M, et al. Synergy between the host immune system and bacteriophage is essential for successful phage therapy against an acute respiratory pathogen. Cell Host Microbe. 2017;22(1):38–47. e4. doi:10.1016/j.chom.2017.06.018
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.