Back to Journals » Drug Design, Development and Therapy » Volume 11
HMGB1 siRNA can reduce damage to retinal cells induced by high glucose in vitro and in vivo
Received 10 December 2016
Accepted for publication 22 January 2017
Published 15 March 2017 Volume 2017:11 Pages 783—795
DOI https://doi.org/10.2147/DDDT.S129913
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Anastasios Lymperopoulos
Shuang Jiang, Xiaolong Chen
Department of Ophthalmology, Shengjing Hospital of China Medical University, Shenyang, China
Background: Diabetic retinopathy (DR), one of the most common complications of late-phase diabetes, is associated with many risk factors, among which continuous low-grade inflammation is one of the principal ones. As such, lowering inflammation levels and maintain the viability of human retinal endothelial cells (HRECs) are critical for DR therapy. HMGB1 is a well-known proinflammatory cytokine. However, whether HMGB1 small interfering RNA (siRNA) can protect retina cells under a high-glucose environment from morphological changes and functional abnormalities remain undetermined. We aimed to investigate the effect of HMGB1 siRNA on retinal cells in DR.
Materials and methods: A total of 80 adult Wistar rats were randomly divided into four groups (n=20 each): normal control, diabetes mellitus (DM), scrambled (Scr) siRNA, and HMGB1 siRNA. Rats in the DM, Scr siRNA, and siRNA groups were established by intraperitoneal injection of streptozotocin. At 16 weeks after injection, rats in the siRNA and Scr-siRNA groups were intravitreally injected with 2 µL HMGB1 siRNA and 2 µL Scr-siRNA, while rats in the control and DM groups were intravitreally injected with the same dose of sterile saline. At 1 week after injections, we performed the following experiments. Immunohistochemical staining and real-time quantitative polymerase chain reaction were performed to test HMGB1 protein and messenger RNA expression in retinas. We performed TUNEL assays to detect retinal cell apoptosis and electroretinography to detect retinal function. In HRECs treated with high glucose, proliferation, morphology, apoptosis, superoxide dismutase (SOD), and reactive oxygen species production were detected. Western blot was applied to determine the expressions of HMGB1 and its related protein and apoptosis protein.
Results: Intravitreal injection of HMGB1 siRNA reduced protein and messenger RNA expression of HMGB1 (both P<0.05). Intravitreal injection of HMGB1 siRNA reduced apoptosis of retinal cells (P<0.05), protected morphological changes in the retina, and improved the function of the retina (P<0.05). In HRECs treated with high glucose, HMGB1 siRNA pretreatment increased cell viability, reduced cell apoptosis, and reduced oxidative damage to cells (all P<0.05). Western blot detection found that HMGB1 siRNA pretreatment can inhibit the expression of cleaved caspase 3 and improve the expression of BCL2 (P<0.05). HMGB1 and NFκB expression increased in a time-dependent manner in the high-glucose environment and IKKβ and NFκB protein expression decreased significantly after HMGB1 silencing.
Conclusion: As a therapeutic target, HMGB1 siRNA can reduce retinal cell damage induced by high glucose in vitro and in vivo and delay DR progress through the HMGB1–IKKβ–NFκB signaling pathway.
Keywords: diabetic retinopathy, small interfering RNA, human retinal endothelial cells, high-mobility group box 1, inhibitor of nuclear factor κB, nuclear factor κB
Background
Diabetic retinopathy (DR) is one of the main microvascular complications of diabetes mellitus (DM) and one of the leading causes of blindness worldwide.1,2 The prevalence of retinopathy is 9%–16% in patients with type 2 diabetes and 24%–27% in patients with type 1 diabetes; 0.2%–0.5% of diabetics are blind.3 It is important to understand the mechanism of underlying pathological DR to find new targets to treat it.
Strong evidence suggests that continuous low-grade inflammation is primarily involved in the pathogenesis of DR.4 HMGB1 is a kind of late inflammatory factor. HMGB1 is a nuclear DNA-binding protein released passively from necrotic cells, as well as actively from monocytes/macrophages and endothelial cells.5 As a kind of damage-associated molecular patterns, HMGB1 is through its receptors involved in physiological and pathological processes, including the release of inflammatory cytokines, cell migration, and angiogenesis.6–9
Recent research10 has shown that as one of the important inflammatory mediators, HMGB1 is associated with diabetic peripheral neuropathy and can participate in the occurrence and development of DR. All of these results indicate that HMGB1 is involved in the occurrence and development of DR. However, the effect of HMGB1 small interfering RNA (siRNA) on DR has not been explored. Therefore, our study aimed to detect whether HMGB1 siRNA has a protective effect on retinal cells in a high-glucose environment and its specific mechanism.
Materials and methods
Animals
All experiments were performed in accordance with guidelines set by the animal experiment committee of Jinzhou Medical University, and the study was approved by Jinzhou Medical University’s ethics committee. Male Wistar rats (Experimental Animal Center of Jinzhou Medical University, 2016873950053732) were used in the experiment. The rats were housed in a standard laboratory environment and maintained on a 12-hour light–dark cycle at 21°C. Rats were randomly assigned to a normal control (NC) group (n=20), DM group (n=20), scrambled (Scr)-siRNA group, (n=20), and a DM siRNA (n=20) group.
DM animal model
A total of 80 male Wistar rats, specific pathogen-free grade, body weight 240–280 g were provided by the animal experiment committee of Jinzhou Medical University. Rats in the DM, Scr-siRNA, and siRNA groups were intraperitoneally injected with 1% streptozotocin (STZ; Sigma-Aldrich, St Louis, MO, USA) at 60 mg/kg, and rats in the NC group were injected with an equivalent amount of citrate-buffer solution. At 72 hours after intraperitoneal injection, blood glucose was measured via the caudal vein using a blood glucose tester (Bayer AG, Leverkusen, Germany). Random blood glucose >16.7 mmol/L indicated successful establishment of the diabetic rat model. Blood glucose was measured monthly.
Preparation of siRNA
HMGB1 and Scr-siRNA were purchased from GenePharma Co Ltd (Shanghai). The sequence of siRNA targeting of the rat HMGB1 gene is sense 5′-UCUUGACCACAGAUCUUAATT-3′, antisense 5′-UUAAGAGCUGUGGUCAAGATT-3′. The sequence of Scr-siRNA is sense 5′-UUCUCCGAACGUGUCACGUTT-3′, antisense 5′-ACGUGACACGUUCGGAGAATT-3′. The resultant siRNA was purified, quantified, and suspended in water at a concentration of 0.5 μg/μL. 0.5 μL HMGB1 siRNA was combined with 0.5 μL Lipofectamine 2000 (with the concentration of Lipofectamine at 50 nM) (Thermo Fisher Scientific, Waltham, MA, USA) for 20 minutes before injection according to the manufacturer.
HMGB1 siRNA intravitreal injection
At 16 weeks after intraperitoneal injection of STZ, we carried out HMGB1 siRNA intravitreal injections. Tropicamide phenylephrine eyedrops (Santen Pharmaceutical, Osaka, Japan) and oxybuprocaine hydrochloride eyedrops (Santen) were administered until the pupils of the rats dilated. With the aid of a surgical microscope, the needle of a microsyringe was inserted 0.5 mm posterior to the corneoscleral limbus into the vitreous cavity of right eyes in the siRNA group, avoiding the crystalline lens, and 2 μL of HMGB1 siRNA was quickly pushed in. Rats in the Scr-siRNA group were intravitreally injected with 2 μL of Scr-siRNA. Rats in the NC and DM groups were intravitreally injected with 2 μL of normal saline. Following injection, ofloxacin eye ointment (Xingqi Pharmaceutical Co Ltd, Shenyang, China) was administered for preventing infection. The animals were killed 1 week after intravitreal administration, and the retinas were carefully dissected.
Electroretinography (ERG)
Rats were dark-adapted overnight before analysis. Pupils were dilated with tropicamide phenylephrine eyedrops (Santen). Rats were anesthetized with 1% pentobarbital sodium and placed on a temperature-controlled working platform at 37°C. Contact electrodes were placed on the corneal surface, and visual responses were recorded by ERG (Ai Erxi, Chongqing, China). Flash responses were obtained.
Real-time quantitative polymerase chain reaction (qPCR)
Total RNA was extracted from the retinas using Trizol reagent (Thermo Fisher Scientific). RNA purity was determined using absorbance at 260 and 280 nm (A260/280). Complementary DNA synthesis was performed using a reverse-transcriptase kit (PrimeScript; Takara Bio, Kusatsu, Japan), according to the manufacturer’s instructions. Primer sequences used are presented in Table 1. Real-time qPCR was performed using SYBR Green PCR Master Mix (Premix Ex Taq; Takara Bio) in a total volume of 20 μL on a 7300 real-time PCR system (Thermo Fisher Scientific). All reactions involved initial denaturation at 95°C for 30 seconds, followed by 50 cycles of 95°C for 5 seconds and 60°C for 31 seconds. GAPDH was used as the reference gene. Cycle threshold (Ct) value was defined as the number of PCR cycles in which the fluorescence signal exceeded the detection threshold. First, ΔCt was calculated as follows: 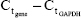 . Subsequently, the ΔΔCt was calculated as follows:
. Subsequently, the ΔΔCt was calculated as follows: 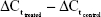 . Lastly, 2−ΔΔCt was calculated to represent the relative messenger RNA (mRNA) expression of target genes, as previously described.11
. Lastly, 2−ΔΔCt was calculated to represent the relative messenger RNA (mRNA) expression of target genes, as previously described.11
Staining
Circular shearing parallel to the direction of the corneal limbus 0.5 mm behind the corneal limbus was performed to remove the lens and vitreous gently. The cupula oculus was fixed again, dehydrated, embedded with soft and hard paraffin, and successively cut into 5.0 μm sections. The sections were conventionally stained with hematoxylin and eosin, dehydrated, mounted, observed, and imaged under light microscopy.
Immunohistochemistry
Paraffin-embedded 5 μm eye-tissue sections were assessed by immunohistochemical analysis and treated with rabbit polyclonal antibodies against mouse HMGB1 (1:100 dilution; Abcam, Cambridge, UK) at 4°C. Retinal sections were incubated with biotinylated horse secondary antibody against mouse IgG (Zhongshan Jinqiao Biotechnology Co Ltd, Beijing, China) and reacted with the avidin-biotinylated peroxidase complex; 3,3′-diaminobenzidine was used as the chromogen. The sections were counterstained with hematoxylin, dehydrated, and mounted. Images were observed using light microscopy. Immunohistochemical images were analyzed with ImagePlus Pro 6 software. Data are presented as mean ± standard deviation of three independent experiments.
TUNEL assay
Eyeballs were enucleated and prepared for paraffin sectioning. Sections were dewaxed to water by conventional methods and repaired with sodium citrate. Then sections were treated with protein K and incubated in a wet box at 37°C for 30 minutes, 50 μL TUNEL reaction liquid added (reagent 1 [5 μL] + reagent 2 [45 μL]), and incubated for 60 minutes in a 37°C cassette. After 50 μL peroxidase had been added, sections were incubated for 30 minutes in a 37°C cassette. Instructions for the TUNEL in situ cell-death-detection kit (Hoffman-La Roche Ltd, Basel, Switzerland) were followed.
Western blot analysis
All samples were collected. For extraction of total cellular protein, tissues were lysed in radioimmunoprecipitation-assay buffer with phenylmethane sulfonyl fluoride. Protein concentration was quantified using a BCA kit (Thermo Fisher Scientific). Proteins were separated and transferred to polyvinylidene difluoride membranes. Membranes were incubated overnight at 4°C with HMGB1, cleaved caspase 3, Bcl2, MyD88, IKKβ, NFκB, and β-actin (1:1,000; Abcam). Thereafter, the membranes were incubated with horseradish peroxidase-labeled antirabbit/mouse secondary antibodies (1:1,000) for 1 hour at room temperature. Finally, membranes were visualized with an enhanced-chemiluminescence kit (Thermo Fisher Scientific). The ratio between the optical density of the protein of interest and β-actin of the same sample was calculated as the relative content of the protein detected.
Cell culture
Human retinal endothelial cells (HRECs) were purchased from the American Type Culture Collection (Manassas, VA, USA) and maintained in Dulbecco’s Modified Eagle’s Medium (DMEM)/F12 medium containing 10% fetal bovine serum (HyClone; GE Healthcare, Little Chalfont, UK) and 1% penicillin–streptomycin. Confluent cells were starved in serum-free DMEM overnight to minimize the effects of serum. Overnight-starved cells were left untreated or treated with high glucose (30 mM/L; Scharlab, Barcelona, Spain) and high glucose plus HMGB1-siRNA.
HMGB1 siRNA transfection
HMGB1 siRNA (GenePharma) was transfected into HRECs using Lipofectamine 2000 reagent (Thermo Fisher Scientific), according to the manufacturer’s instructions. The sequence of HMGB1 siRNA was sense 5′-GGCUUUCACUUAAGAACUUTT-3′, antisense 5′-AAGUUCUUAAGU GA AAGCCTT-3′.
Reactive oxygen species (ROS) assay
Intracellular ROS levels were evaluated using a cellular ROS/superoxide-detection assay kit (Beyotime, Haimen, China), according to the manufacturer’s instructions. In brief, cells were seeded in 96-well plates at a density of 105 cells/well and allowed to attach for 24 hours. After incubation with the aforementioned treatments, the culture supernatant was removed and the cells washed with 100 μL/well of assay buffer. The ROS-specific stain DCFH-DA was added to the cells and allowed to incubate in the dark for 60 minutes. After incubation, intracellular ROS levels were determined using a fluorescence microplate reader (excitation wavelength 488 nm, emission wavelength 520 nm).
Total superoxide dismutase (T-SOD) assay
Cells were seeded in six-well plates at a density of 106 cells/well and allowed to attach for 24 hours. Cells were treated by the aforementioned methods. After treatment, the cells were lysed by the freeze–thaw method three times. SOD activity was determined using a commercially available kit (Jiancheng Bioengineering Institute, Nanjing, China), according to the manufacturer’s instructions.
MTT assay
HRECs were cultured at 5,000 cells per well in 96-well tissue-culture plates. At 24 hours after plating, cells were washed three times with phosphate-buffered saline (PBS) and then treated with high glucose (30 mM/L) for 24 hours in DMEM/F12 medium containing 1% fetal bovine serum. At the end of the culture period, cells were washed with ice-cold PBS, MTT reagent (5 mg MTT in 10 mL PBS) was added according to the manufacturer’s instructions and absorbance measured at 490 nm using a microplate reader. Mean values were calculated from three independent experiments. After the half-maximal inhibitory concentration (IC50) of high glucose had been determined, HRECs were transfected with HMGB1 siRNA for 12 hours and then cultured as previously described.
Flow-cytometry assay
Cell apoptosis was measured by annexin V flow cytometry according to the manufacturer’s protocol (Calbiochem). Briefly after treatment, cells were washed twice with PBS and incubated in 300 μL binding buffer containing 3 μL annexin V–fluorescein isothiocyanate and 3 μL of propidium iodide in the dark for 15 minutes at room temperature. The stained samples (containing 200,000 cells/sample) were then analyzed on a FACSCalibur flow cytometer within 1 hour, following the manufacturer’s protocol (BD Biosciences, San Jose, CA, USA).
Hoechst 33342 staining
After treatment, cells were stained with Hoechst 33342 (1:1,000) and photographed using fluorescence microscopy. The incidence of DNA condensation in each preparation was analyzed by counting 300 cells and determining the percentage of DNA-condensed cells (apoptotic cells).
Statistical analysis
Each experiment was repeated a minimum of three times, the mean value of the repetitions was calculated, and this value was used in the statistical analysis. Results are presented as mean ± standard deviation. Values of ERG were analyzed by two-tailed Student’s t-test and one-way analysis of variance. Nonparametric Kruskal–Wallis tests were used for Western blotting analysis. Differences were considered significant at P<0.05.
Results
Animal model
There was no significant difference in blood glucose among the four groups at baseline (P>0.05). At 72 hours after intraperitoneal STZ injection, blood glucose was significantly increased to >16.7 mmol/L, suggesting a DM-model success rate of up to 100%. In the experimental period, blood glucose was much higher than the NC group (all P<0.05). Details of blood glucose at different time points in the four groups are shown in Table 1.
Intravitreal injection of HMGB1 siRNA significantly inhibited the expression of HMGB1 in the retina
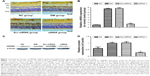
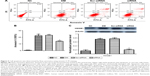
To test for HMGB1 siRNA target-specific efficacy, a negative control siRNA (Scr-siRNA) was analyzed to provide a benchmark for off-target effects on HMGB1 knockdown. In order to detect the interference effect of siRNA, we performed immunohistochemistry staining for location and real-time qPCR and Western blot for quantitative detection. The results showed that there was little HMGB1 expression in the NC group, HMGB1 expression in the DM and Scr-siRNA groups was significantly increased, mainly located in the retinal ganglion-cell layer, inner nuclear layer, and outer nuclear layer, and HMGB1 expression in the siRNA group was significantly decreased (Figure 1A and Table 2).
Real-time qPCR demonstrated that HMGB1 mRNA levels in the siRNA group were reduced compared to the Scr-siRNA group (P<0.05, Figure 1B). In addition, mRNA levels of HMGB1 in the DM and Scr-siRNA groups were upregulated significantly compared to the NC group (all P<0.05, Figure 1B), whereas there was no significant difference between the DM and Scr-siRNA groups (P>0.05, Figure 1B).
Western blot detection also supported this conclusion (Figure 1C). The protein expression of HMGB1 in the siRNA group was reduced by 77.3% compared with the Scr-siRNA group (P<0.05, Figure 1C). In addition, HMGB1 protein levels in the DM and Scr-siRNA groups were upregulated significantly compared to the NC group (all P<0.05, Figure 1D), whereas there was no significant difference between the DM and Scr-siRNA groups (P>0.05, Figure 1D).
siRNA HMGB1 significantly inhibited retinal damage and ameliorated retina-function degeneration in vivo
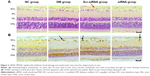
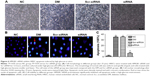
To investigate structural changes, we examined histologic changes in paraffin sections of the retina. In retinal sections, we found that in the NC the retina had a smooth surface and neatly arranged retinal ganglion cells and inner and outer nuclear layers. In the DM and Scr-siRNA groups, retinal ganglion cells were in disorder completely, and capillary endothelial cells were observed to protrude the inner limiting membranes. After pretreatment with siRNA HMGB1, compared with the DM group, the histological changes were less pronounced. The arrangement of retinal cells was relatively orderly, and few capillary endothelial cells in the inner limiting membranes were observed (Figure 2A).
In order to observe retinal cell damage better, we performed TUNEL detection. Positive apoptotic cells were stained with brown-yellow nuclei. The retinal ganglion-cell layer in the DM and Scr-siRNA groups had obvious brown staining, while few scattered apoptotic cells were found in the NC group (Figure 2B). In the siRNA group, the number of positive apoptotic cells was significantly decreased compared with the Scr-siRNA group.
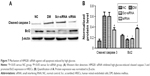
In order to verify whether siRNA HMGB1 precipitated improvement in the function of the retina, we performed flash ERG examinations at 16 weeks postinjection. Analysis of both a- and b-waves in the four groups showed that HMGB1 siRNA pretreatment dramatically upregulated the amplitude of the waves compared with the Scr-siRNA group (both P<0.05, Figure 3). HMGB1 siRNA partially improved the retinal function, implying that it can protect or rescue visual function in DR. Amplitudes of a- and b-waves in the siRNA group increased by 77.3% and 66.90% compared with the Scr-siRNA group (both P<0.05, Figure 3). In addition, a- and b-wave amplitudes in the DM and Scr-siRNA groups were downregulated significantly compared to the NC group (all P<0.05), whereas there was no significant difference between the DM and Scr-siRNA groups (P>0.05).
siRNA HMGB1 significantly inhibited high-glucose-induced HREC damage in vitro
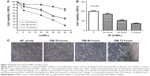
To explore which concentration of high glucose is suitable for HREC damage, MTT assays were chosen to detect the IC50 of high-glucose-induced HREC damage. HRECs were stimulated with increasing concentrations of high glucose (0, 5.5, 20, 30, and 40 mM/L) for 24, 48, and 72 hours, and their viability was measured by MTT assay. The results shown in Figure 4A demonstrated that high glucose depressed HREC viability in a dose-dependent manner. The IC50 of high glucose is about 30 mM/L at 48 hours and 22 mM/L at 72 hours. As such, 30 mM/L was chosen for the following experiments. Figure 4B shows high-glucose-induced HREC viability in a time-dependent manner. With time, HRECs viability reduced gradually with 30 mM/L glucose. This was also confirmed by morphological changes (Figure 4C). The results showed that the morphology of normal cells was uniform and spindle-shaped, and cells became smaller and density decreased after treatment with high glucose. With the extension of treatment time, changes in morphology and density were more obvious.
siRNA HMGB1 inhibited high-glucose-induced HREC apoptosis in vitro
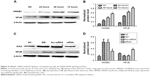
In order to detect the interference effect of HMGB1 siRNA in HRECs, we performed Western blot for quantitative detection. The results in Figure 5C show that the protein expression of HMGB1 in the siRNA group was reduced compared with the DM and Scr-siRNA groups (all P<0.05). HMGB1 protein levels in the DM and Scr-siRNA groups were upregulated significantly compared to the NC group (all P<0.05), whereas there was no significant difference between the DM and Scr-siRNA groups (P>0.05). To detect the influence of siRNA HMGB1 on HREC apoptosis, we performed flow cytometry and Hoechst 33342 staining. Results showed that HMGB1 siRNA transfection significantly inhibited the apoptosis rate of HRECs treated with Scr-siRNA (Figure 5, total apoptotic rate 23.6%±2.34% vs 47.8%±1.98%, P<0.05). These results indicated that transfection of the cells with HMGB1 siRNA exerted more prominent antiapoptotic effects on the HRECs.
Inhibitory effects of HMGB1 siRNA on HREC apoptosis were further confirmed by Hoechst 33342 staining (Figure 6B). Apoptotic nuclei were stained bright blue. Apoptotic nuclei were densely stained or showed chunky dense stains and demilune condensation. Hoechst 33342 staining revealed the apoptosis rate of HRECs. Results showed that the rate of cell apoptosis in the siRNA group decreased significantly compared with the Scr-siRNA group (6%±1.08% vs 36%±1.35%, P<0.05, Figure 6C). Taken together, these results indicated that siRNA HMGB1 not only increased cell viability in HRECs exposed to high glucose but also inhibited high-glucose-induced HREC apoptosis in vitro.
To confirm the mechanism of inhibition on high-glucose-induced HREC apoptosis, Western blot analysis was performed to detect the expressions of cleaved caspase 3 and Bcl2 in HRECs. Western blot results in Figure 7 show that HMGB1 siRNA inhibited high-glucose-induced cleaved caspase 3 expression and promoted Bcl2 expression in HRECs. This indicated that the protective effect of HMGB1 siRNA in HRECs might have been due to apoptosis inhibition.
HMGB1 siRNA ameliorated high-glucose-induced oxidative stress in vitro
To detect oxidative stress levels, we examined ROS and T-SOD expression. ROS production in the DM and Scr-siRNA groups increased by about 50% compared to the NC group, and the difference between the two was not statistically significant (P>0.05). HMGB1 siRNA inhibited the increasing trend of ROS production (P<0.05, Figure 8A). The SOD assay also showed the same trend (Figure 8B). As such, HMGB1 siRNA inhibited high-glucose-induced HREC apoptosis and oxidative stress in vitro.
HMGB1 siRNA inhibited DR through the IKKβ–NFκB signaling pathway
In order to verify the mechanism of HMGB1 protection, we initially used Western blot assays to detect the expression of HMGB1. The results showed that under a high-glucose environment, the expression of HMGB1 and NFκB protein increased gradually in a time-dependent manner (P<0.05, Figure 9A and B). After HMGB1 siRNA transfection, IKKβ and NFκB protein expressions were inhibited significantly (P<0.05, Figure 9C and D). These results suggested that HMGB1 may play a role through the IKKβ–NFκB signaling pathway.
Discussion
DR is a multifactor disease that affects retinal vessels, and ultimately leads to blindness in millions of people worldwide. Diabetes-induced microvascular disease is characterized by progressive alterations in the retinal microvasculature, including endothelial cell dysfunction, breakdown of the blood–retina barrier, ischemia, and retinal neovascularization.12 Current clinical data show that DR has affected nearly 3 billion people. Half the world’s population will suffer from DR by 2020. Therefore, researchers urgently need to find effective therapeutic methods to control DR progression.
Strong evidence indicates that persistent low-grade inflammation is implicated in the pathogenesis of diabetes.13 As a recognized late inflammatory factor, HMGB1 has been repeatedly proved to be related to the occurrence and development of DR.14,15 Recent research has shown that HMGB1 regulates the expression of VEGF, ICAM1, and integrity of the blood–retina barrier in the diabetic retina.16,17 Mohammad et al investigated the expression of HMGB1-signaling pathway components in the retinas of diabetic rats, and proved that HMGB1 activated ERK1/2 and NFκB to generate inflammatory responses and disrupt the retinal vascular barrier.16 Zhang et al found that HMGB1 regulated AGE-induced proinflammatory cytokines in HRECs.18 Zhao et al19 found that in retinal ganglion cells treated with high glucose, protein and mRNA expression of HMGB1 increased, while cell viability decreased. After HMGB1 siRNA transfection, these changes could be reversed. These results indicated that HMGB1 promoted the development of DR. However, HMGB1 silencing to treat or prevent DR has not been reported. Therefore, this study selected siRNA to silence HMGB1 to observe whether it can reduce or even reverse the development of DR.
RNA interference is a powerful tool for posttranscriptional gene silencing, and is a new method to study gene function.20 A previous study has shown that transfer of the siRNA gene to the retina was accomplished at 1 day after intravitreal injection.21 We first observed the effect of HMGB1 siRNA on the retina in vivo. First, we set up a diabetes model of rats. At 16 weeks after the model was established, we performed HMGB1 siRNA intravitreal injection. By immunohistochemistry, Western blot, and real-time qPCR, we found that the expression of HMGB1 in retina was significantly decreased after injection. The rat model is suitable for further studies. Intravitreal injection is one of the important methods for the treatment of ocular fundus diseases. Due to local administration, intravitreal injection reduces systemic side effects.22 Therefore, this study also chose this method of intravitreal injection.
Figure 2 shows that high glucose can lead to tremendous retinal morphological changes, mainly retinal ganglion-cell damage and inner and outer nuclear layer change. This is the same as some previous studies.23,24 After administration of HMGB1 siRNA, morphological changes in the retina significantly improved. These results suggested that HMGB1 siRNA can effectively improve the retinal morphology damage induced by high glucose. In addition, intravitreal injection of HMGB1 siRNA can significantly improve the function of the retina based on ERG.
Based on the protective effect of HMGB1 siRNA in vivo, we tried to explore the specific protection mechanism of HMGB1 siRNA in vitro. We first observed the effect of high glucose on HRECs in vitro. We found that high glucose can lead to apoptosis of HRECs in a significant time- and dose-dependent manner, similar to other research.25 The apoptosis rate of HRECs pretreated with HMGB1 siRNA declined. Most interestingly, HMGB1 siRNA seems only to inhibit the early stage of apoptosis, but necrosis was not much affected. These findings should be further studied in the future experiments. By Western blot assay, we found that HMGB1 siRNA reduced the expression of cleaved caspase 3 and increase the expression of Bcl2. This indicated that the protective effect of HMGB1 siRNA is due to the inhibition of cleaved caspase 3 and promotion of Bcl2.
Further, we detected ROS in HRECs treated with high glucose. Although the impact of hyperglycemia on increasing the oxidative stress of endothelial cells has been confirmed,26–28 few studies have examined oxidative stress in HRECs treated with HMGB1 siRNA. Oxidative stress refers to the production of ROS, overwhelming their clearance and leading to their accumulation.29 The level of ROS production is significantly increased after treatment with high glucose, indicating oxidative stress is a result of high glucose. After pretreatment with HMGB1 siRNA, we observed that HMGB1 siRNA can retard this trend to a certain extent. The same trend was also found in SOD detection. In other studies, ROS production was also found increased in diabetic rats, and oxidative stress levels were decreased by drug treatment.30–32
Reduction in oxidative stress levels may be a new strategy to reduce DR
In order to detect the mechanism of HMGB1 in diabetic rats, we examined the expression of HMGB1 in HRECs treated with high glucose by Western blot detection. The results showed that the protein expression of HMGB1 increased in a time-dependent manner. At the same time, NFκB expression also gradually increased. Recent findings suggest that the cytoplasmic translocation of HMGB1 may be caused by diabetes and high glucose in retinal pericytes, and that the pathogenic role of HMGB1 may be dependent on the expression of receptor for advanced glycation end product and activation of NFκB.33 In our study, IKKβ and NFκB protein expression was also decreased after inhibiting the expression of HMGB1. This indicated that the increase in HMGB1 induced by high glucose promoted the occurrence and development of DR by activating the IKKβ–NFκB signaling pathway. We did not study the translocation of HMGB1 in the cells, and further study is needed.
Conclusion
Our results demonstrated that HMGB1 plays an important role in vivo and in vitro under hyperglycemia conditions via the IKKβ–NFκB signaling pathway. As such, we suggest that HMGB1 is a potential target for the prevention and treatment of DR. This study also provides strong support for the view that siRNA therapy may play an important role in future therapeutic strategies.
Acknowledgment
This work was partly supported by the National Natural Science Foundation of China (81570866).
Disclosure
The authors report no conflicts of interest in this work.
References
Agarwal A, Soliman MK, Sepah YJ, Do DV, Nguyen QD. Diabetic retinopathy: variations in patient therapeutic outcomes and pharmacogenomics. Pharmgenomics Pers Med. 2014;7:399–409. | ||
Wang SY, Andrews CA, Herman WH, Gardner TW, Stein JD. Incidence and risk factors for developing diabetic retinopathy among youths with type 1 or type 2 diabetes throughout the United States. Ophthalmology. Epub 2016 Nov 30. | ||
Schorr SG, Hammes HP, Müller UA, Abholz HH, Landgraf R, Bertram B. The prevention and treatment of retinal complications in diabetes. Dtsch Arztebl Int. 2016;113(48):816–823. | ||
Cheung N, Mitchell P, Wong TY. Diabetic retinopathy. Lancet. 2010;376(9735):124–136. | ||
Lohani N, Rajeswari MR. Dichotomous life of DNA binding high mobility group box1 protein in human health and disease. Curr Protein Pept Sci. 2016;17(8):762–775. | ||
Tsung A, Tohme S, Billiar TR. High-mobility group box-1 in sterile inflammation. J Intern Med. 2014;276(5):425–443. | ||
Kang R, Tang D, Schapiro NE, et al. The HMGB1/RAGE inflammatory pathway promotes pancreatic tumor growth by regulating mitochondrial bioenergetics. Oncogene. 2014;33(5):567–577. | ||
Huebener P, Hernandez C, Schwabe RF. HMGB1 and injury amplification. Oncotarget. 2015;6(27):23048–23049. | ||
Pandolfi F, Altamura S, Frosali S, Conti P. Key role of DAMP in inflammation, cancer, and tissue repair. Clin Ther. 2016;38(5):1017–1028. | ||
Mudaliar H, Pollock C, Ma J, Wu H, Chadban S, Panchapakesan U. The role of TLR2 and 4-mediated inflammatory pathways in endothelial cells exposed to high glucose. PLoS One. 2014;9(10):e108844. | ||
Di Y, Zhang Y, Yang H, Wang A, Chen X. The mechanism of CCN1-enhanced retinal neovascularization in oxygen-induced retinopathy through PI3K/Akt-VEGF signaling pathway. Drug Des Devel Ther. 2015;9:2463–2473. | ||
Mohammad G, Jomar D, Siddiquei MM, Alam K, El-Asrar AM. High-mobility group box-1 protein mediates the regulation of signal transducer and activator of transcription-3 in the diabetic retina and in human retinal Müller cells. Ophthalmic Res. Epub 2016 Aug 25. | ||
Yang S, Xu L, Yang T, Wang F. High-mobility group box-1 and its role in angiogenesis. J Leukoc Biol. 2014;95(4):563–574. | ||
Yu Y, Yang L, Lv J, et al. The role of high mobility group box 1 (HMGB-1) in the diabetic retinopathy inflammation and apoptosis. Int J Clin Exp Pathol. 2015;8(6):6807–6813. | ||
El-Asrar AM, Nawaz MI, Kangave D, Abouammoh M, Mohammad G. High-mobility group box-1 and endothelial cell angiogenic markers in the vitreous from patients with proliferative diabetic retinopathy. Mediators Inflamm. 2012;2012:697489. | ||
Mohammad G, Siddiquei MM, Othman A, Al-Shabrawey M, El-Asrar AM. High-mobility group box-1 protein activates inflammatory signaling pathway components and disrupts retinal vascular-barrier in the diabetic retina. Exp Eye Res. 2013;107:101–109. | ||
El-Asrar AM, Mohammad G, Nawaz MI, Siddiquei MM. High-mobility group box-1 modulates the expression of inflammatory and angiogenic signaling pathways in diabetic retina. Curr Eye Res. 2015;40(11):1141–1152. | ||
Zhang YF, Wei W, Li L, et al. Sirt1 and HMGB1 regulate the AGE-induced pro-inflammatory cytokines in human retinal cells. Clin Lab. 2015;61(8):999–1008. | ||
Zhao H, Zhang J, Yu J. HMGB-1 as a potential target for the treatment of diabetic retinopathy. Med Sci Monit. 2015;21:3062–3067. | ||
Hanrahan F, Humphries P, Campbell M. RNAi-mediated barrier modulation: synergies of the brain and eye. Ther Deliv. 2010;1(4): 587–594. | ||
Masuda I, Matsuo T, Yasuda T, Matsuo N. Gene transfer with liposomes to the intraocular tissues by different routes of administration. Invest Ophthalmol Vis Sci. 1996;37(9):1914–1920. | ||
Li B, Zhang HQ, Shi Y, et al. Overexpression of nuclear transport factor 2 may protect against diabetic retinopathy. Mol Vis. 2009;15:861–869. | ||
Jiang N, Chen XL, Yang HW, Ma YR. Effects of nuclear factor κB expression on retinal neovascularization and apoptosis in a diabetic retinopathy rat model. Int J Ophthalmol. 2015;8(3):448–452. | ||
Gong Y, Jin X, Wang QS, et al. The involvement of high mobility group 1 cytokine and phospholipases A2 in diabetic retinopathy. Lipids Health Dis. 2014;13:156. | ||
Chen XL, Zhang XD, Li YY, Chen XM, Tang DR, Ran RJ. Involvement of HMGB1 mediated signalling pathway in diabetic retinopathy: evidence from type 2 diabetic rats and ARPE-19 cells under diabetic condition. Br J Ophthalmol. 2013;97(12):1598–1603. | ||
Monnier L, Mas E, Ginet C, et al. Activation of oxidative stress by acute glucose fluctuations compared with sustained chronic hyperglycemia in patients with type 2 diabetes. JAMA. 2006;295(14):1681–1687. | ||
Suh SW, Gum ET, Hamby AM, Chan PH, Swanson RA. Hypoglycemic neuronal death is triggered by glucose reperfusion and activation of neuronal NADPH oxidase. J Clin Invest. 2007;117(4):910–918. | ||
Weidig P, McMaster D, Bayraktutan U. High glucose mediates pro-oxidant and antioxidant enzyme activities in coronary endothelial cells. Diabetes Obes Metab. 2004;6(6):432–441. | ||
Wu N, Shen H, Liu H, Wang Y, Bai Y, Han P. Acute blood glucose fluctuation enhances rat aorta endothelial cell apoptosis, oxidative stress and pro-inflammatory cytokine expression in vivo. Cardiovasc Diabetol. 2016;15(1):109. | ||
Kumar B, Gupta SK, Nag TC, et al. Retinal neuroprotective effects of quercetin in streptozotocin-induced diabetic rats. Exp Eye Res. 2014;125:193–202. | ||
Wang H, Zheng Z, Gong Y, Zhu B, Xu X. U83836E inhibits retinal neurodegeneration in early-stage streptozotocin-induced diabetic rats. Ophthalmic Res. 2011;46(1):19–24. | ||
Fan Y, Qiao Y, Huang J, Tang M. Protective effects of Panax notoginseng saponins against high glucose-induced oxidative injury in rat retinal capillary endothelial cells. Evid Based Complement Alternat Med. 2016;2016:5326382. | ||
Kim J, Kim CS, Sohn E, Kim JS. Cytoplasmic translocation of high-mobility group box-1 protein is induced by diabetes and high glucose in retinal pericytes. Mol Med Rep. 2016;14(4):3655–3661. |
 © 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.