Back to Journals » OncoTargets and Therapy » Volume 10
HMGB1 is negatively correlated with the development of endometrial carcinoma and prevents cancer cell invasion and metastasis by inhibiting the process of epithelial-to-mesenchymal transition
Authors Luan XR, Ma CJ, Wang P, Lou FL
Received 23 September 2016
Accepted for publication 18 November 2016
Published 3 March 2017 Volume 2017:10 Pages 1389—1402
DOI https://doi.org/10.2147/OTT.S123085
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Dr Carlos E Vigil
Xiaorong Luan,1,2 Chunjing Ma,2 Ping Wang,2 Fenglan Lou1
1Nursing College, Shandong University, 2Qilu Hospital of Shandong University, Jinan, People’s Republic of China
Abstract: High-mobility group box protein 1 (HMGB1), a nuclear protein that plays a significant role in DNA architecture and transcription, was correlated with the progression of some types of cancer. However, the role of HMGB1 in endometrial cancer cell invasion and metastasis remains unexplored. HMGB1 expression was initially assessed by immunohistochemistry and reverse transcription-quantitative polymerase chain reaction (RT-qPCR) in normal endometrial tissue and endometrial carcinoma tissue. High expressions of HMGB1 protein were detected in normal endometrial tissues; however, in endometrial cancer tissues, the expressions of HMGB1 were found to be very weak. Furthermore, HMGB1 expressions were negatively correlated with advanced stage and lymph node metastasis in endometrial cancer. Then by RT-qPCR, Western blot and immunocytochemistry, HMGB1 was also detected in primary cultured endometrial cells and four kinds of endometrial cancer cell lines (Ishikawa, HEC-1A, HEC-1B and KLE). We found that the expression of HMGB1 was much higher in normal endometrial cells than in endometrial cancer cells, and reduced expression levels of HMGB1 were observed especially in the highly metastatic cell lines. Using lentivirus transfection, HMGB1 small hairpin RNA was constructed, and this infected the lowly invasive endometrial cancer cell lines, Ishikawa and HEC-1B. HMGB1 knockdown significantly enhanced the proliferation, invasion and metastasis of endometrial cancer cells and induced the process of epithelial-to-mesenchymal transition. These results can contribute to the development of a new potential therapeutic target for endometrial cancer.
Keywords: HMGB1, endometrial cancer, invasion, metastasis, epithelial-to-mesenchymal transition
Introduction
Endometrial cancer is one of the common malignant tumors of the female reproductive system, which is commonly found in postmenopausal women. However, in recent years, the incidence of the disease has tended to be younger.1 The prognosis of endometrial cancer patients with recurrence or metastasis after surgery or radiotherapy is very poor. Gene therapy and targeted therapy have received increasing attention as the new generation strategy for cancer treatment.2,3 Presently, there is no specific marker for the treatment of endometrial cancer. The molecular mechanism for growth and metastasis of this cancer type is not well known.
High-mobility group box protein 1 (HMGB1, NP_002119), which was extracted and identified for the first time in bovine thymus in 1973, is a non-histone chromosomal binding protein present in the eukaryotic cell nucleus.4 HMGB1 is highly conserved through evolution and plays a key role in chromatin organization and transcriptional regulation.5 A series of studies have reported that HMGB1 might be associated with many physiological and pathological conditions, such as Alzheimer’s disease,6 cardiovascular disease,7 arthritis,8 ischemia,9 meningitis,10 sepsis,11 inflammation12 and cancers.13–15 HMGB1 has both pro- and anti-tumorigenic bioactivities. HMGB1 suppresses tumorigenesis by interacting with tumor suppressor genes such as p53, p73 and RB.16,17 Moreover, HMGB1 has complex effects on the hallmarks of cancer, such as limitless replicative potential, sustained angiogenesis, evasion of apoptosis, self-sufficiency in growth signals, insensitivity to growth inhibitors and tissue invasion and metastasis.18–20 However, the role of HMGB1 in the invasion and metastasis of endometrial cancer is unknown.
Materials and methods
Human primary endometrial cell isolation and culture
Using surgically resected endometrial hysterectomy specimens, primary endometrial cells were obtained by trypsin digestion,21 which utilize the enzymatic activity of trypsin to facilitate the cell separation and primary cell outgrowth. Patient-derived normal endometrial tissues were immersed in Dulbecco’s Modified Eagle’s Medium (DMEM)/F12 (Gibco BRL, Rockville, MD, USA) in a 50 mL tube at 4°C. Tissue samples were washed three times with 1× sterile phosphate-buffered saline (PBS). Using a scalpel or scissors, the tissue was cut into small pieces, immersed in 5 mL of 0.25% trypsin supplemented with 0.25 mg/mL collagenase type I (Sigma-Aldrich, St Louis, MO, USA) followed by incubation at 37°C on a rotating platform for 1 h. The sample was then centrifuged for 5 min, and the supernatant was discarded, and 10 mL of growth media (supplemented with 10% fetal bovine serum [FBS] and 1% penicillin–streptomycin) were added to the pelleted cells to deactivate the trypsin. Finally, the pelleted cells were plated into a 10 cm cell culture dish and placed in an incubator. Cells were monitored daily under a light microscope, and the media were changed every two to three days. Cells were usually visible under a light microscope after 24–48 h. When the cells reached 80%–85% confluence, 0.25% trypsin was used to passage them.
Endometrial cancer cell line culture
Endometrial cancer cell lines such as Ishikawa, KLE, HEC-1A and HEC-1B were obtained from the Shanghai Institute for Biological Sciences, Chinese Academy of Sciences. All cell lines were cultured in DMEM/F12 supplemented with 10% FBS and 1% penicillin–streptomycin and maintained at 37°C with 5% CO2.
Endometrial cancer tissue samples
Human endometrial cancer specimens (n=240) were collected from the Pathology Department of Shandong Provincial Hospital and Qilu Hospital. According to the International Federation of Gynecology and Obstetrics staging system, patients were diagnosed with the following stages: stage I, 88 cases; stage II, 75 cases; stage III, 43 cases and stage IV, 34 cases. All patients were not treated with radiotherapy or chemotherapy before surgery. By surgical hysterectomy, 60 normal endometrial tissue samples were acquired from the Pathology Department of Shandong Provincial Hospital and Qilu Hospital. Regular follow-up was performed in all patients. During the research period, 12 patients were unable to be contacted and 28 patients died. The follow-up period was between two to eight years at the end of 2015. The study was approved by the Institutional Medical Ethics Committee of Shandong University, and written informed consent was obtained from all patients.
IHC and ICC
For immunohistochemistry (IHC), paraffin-embedded sections were dewaxed in xylene and rehydrated in ethanol. After that, antigen retrieval was performed in a pressure cooker for 2 min with 0.01 M citrate buffer, pH 6.0. For immunocytochemistry (ICC), cells at log phase were collected and seeded into a 6 cm cell culture plate covered with coverslips. After 24 h, all coverslips were gathered and fixed with 4% paraformaldehyde for 30 min. Following the protocol of the Streptavidin-Peroxidase Detection Kit (ZSGB-BIO), all sections and coverslips were first incubated with 3% hydrogen peroxide for 30 min, followed by incubation with rabbit anti-human HMGB1 antibodies (ab18256; Abcam, Cambridge, MA, USA) diluted 1:200 in PBS with 0.1% Triton 100 at 4°C overnight, then incubated with the secondary antibody for 30 min at 37°C, and finally stained with the enzyme substrate 3,3-diaminobenzidine tetrahydrochloride (DAB, Sigma-Aldrich). Human ovarian cancer sections (HMGB1-positive) were used as positive control; PBS was used as a negative control instead of the primary antibody. The presence of brown granules in the nuclei was defined as positive expression of HMGB1.
IHC and ICC analysis
A semiquantitative scoring system based on the percentage of stained cells and the staining intensity was used to assess HMGB1 expression in IHC and ICC experiments. The intensity of HMGB1-positive staining was assigned a score of 0–3 (negative =0, weak =1, moderate =2 or strong =3), and the percentage of positively stained cells was scored as 0 (0%), 1 (1%–25%), 2 (26%–50%), 3 (51%–75%) and 4 (76%–100%). The sum of the intensity and percentage score was used as the final staining scores (0–7), and the scores of 0, (1–3), (4–5) and (6–7) meant the sum indexes (−), (+), (++) and (+++), respectively. For statistical analysis, sum indexes (−) and (+) were defined as low HMGB1 expression, while sum indexes (++) and (+++) were defined as high HMGB1 expression. Each tissue section was analyzed independently by three pathologists.
RNA interference
The HMGB1 small hairpin RNA (shRNA) as well as a negative control was synthesized by GeneChem, Inc. (Shanghai, People’s Republic of China). The target sequence for HMGB1 was 5-GGACAAGGCCCGUUAUGAA-3; the negative control sequence was 5-TTCTCCGAACGTGTCACGT-3. According to the manufacturer’s protocol, target cells were collected, seeded into a 24-well plate and incubated at 37°C with 5% CO2 for 24 h, and then infected by lentivirus stock at a multiplicity of infection of 100. Transfected cells were cultured at 37°C in a CO2 incubator overnight. To avoid cell toxicity, new complete medium was added in place of the transfection mixture. After further incubation for 48 h, transfected cells were monitored under a fluorescence microscope. By Western blotting, real-time quantitative reverse transcription-quantitative polymerase chain reaction (RT-qPCR) and ICC techniques, transfection efficiency was assessed.
Quantitative real-time PCR (RT-qPCR)
Using TRIzol® Reagent (Ambion™) and TaqMan® Reverse Transcription Reagents (Applied Biosystems, Inc.; Thermo Fisher Scientific, Inc.), total RNA was extracted, and complementary DNA (cDNA) was reverse-transcribed. Each well (25 μL PCR reaction volume) included 12.5 μL of SYBR Green Mix (Power SYBR® Green PCR Master Mix, Applied Biosystems, Inc.), 0.2 μL of cDNA, 1 μL of primer pair mix (5 pmol/μL each primer) and 11.3 μL of DNAse/RNAse-free H2O. Each sample was prepared in three replicates. PCR was performed using ABI Prism SDS 7000 (Applied Biosystems, Inc.; Thermo Fisher Scientific, Inc.). Specific primers were synthesized by Takara Biotechnology Co., Ltd. The sequences of primers were as follows: HMGB1, forward: 5′-GCCTCCTTCGGCCTTCTT-3′ and reverse: 5′-ACAGGCCAGGATGTTCTCCTTT-3′; E-cadherin, forward: 5′-GGATTGCAAATTCCTGCCATTC-3′ and reverse: 5′-AACGTTGTCCCGGGTGTCA-3′; N-cadherin, forward: 5′-GTAGCTAATCTAACTGTGACCGATAAGG-3′ and reverse: 5′-TTGGTTTGACCACGGTGACTAA-3′; vimentin, forward: 5′-GCAGGAGGCAGAAGAATGGTA-3′ and reverse: 5′-GGGACTCATTGGTTCCTTTAAGG-3′; Snail, forward: 5′-TCGGAAGCCTAACTACAGCGA-3′ and reverse: 5′-AGATGAGCATTGGCAGCGAG-3′; Twist, forward: 5′-AGCAAGATTCAGACCCTCAAGCT-3′ and reverse: 5′-CCTGGTAGAGGAAGTCGATGTACCT-3′; β-actin, forward: 5′-CCACGAAACTACCTTCAACTCCA-3′ and reverse: 5′-GTGATCTCCTTCTGCATCCTGTC-3′.
Western blotting
Using radioimmunoprecipitation assay buffer supplemented with 1 mM phenylmethylsulfonyl fluoride, cells were lysed on ice. Protein samples (40 μg/lane) were subjected to sodium dodecyl sulfate polyacrylamide gel electrophoresis, transferred to polyvinyl difluoride membranes and blocked with 5% bovine serum albumin. These membranes were then cultured with primary antibodies (E-cadherin sc-8426, N-cadherin sc-7939, vimentin sc-6260, Santa Cruz; HMGB1 ab18256, Snail ab167609, Twist ab50887, Abcam) with working dilutions 1:1,000 at 4°C overnight. The next day, these membranes were incubated with secondary antibody with working dilutions 1:2,000 at room temperature for 1 h, and blots were developed using the enhanced chemiluminescence method (Pierce™ ECL Western Blotting Substrate; Thermo Fisher Scientific, Inc.).
Growth curves
Cells in the log phase were collected, seeded into 24-well plates (1×104 cells/well) and cultured at 37°C with 5% CO2. Three wells were singled out every day, the average cell numbers for seven consecutive days were calculated and the growth curves were drawn accordingly.
Soft agar colony formation assay
Cells were suspended in complete culture medium. For the bottom layer, 1.5 mL of DMEM supplemented with 20% FBS was mixed with 1.5 mL of 1.2% agar, and the mixture was allowed to solidify in 3.5 cm dishes. For the upper layer, 1.5 mL of 0.7% agar was mixed with 1.5 mL of DMEM (20% FBS) and 200 μL of cell suspension (including 600 cells), and then the mixture was immediately poured into the 3.5 cm dishes with solidified bottom layer. The assay was repeated for three replicates, and all dishes were cultured at 37°C with 5% CO2 for two weeks. A cluster of ten or more cells was defined as a colony and assessed under an inverted microscope. Using horizontal and vertical lines, the 3.5 cm dishes were divided into four quadrants, colony numbers were counted in each quadrant and the mean was calculated, and the results were expressed as mean ± standard error (SE).
Cell invasion assay and migration assay
Cell invasion assay and migration assay were performed as described by Albini and Noonan.22 For the cell invasion assay, polyvinylpyrrolidone-free polycarbonate (PVPF) membranes of 8.0 μm pore size were covered with 50 μL of Matrigel diluted with serum-free culture medium at 1:3. The cells (2×105) suspended in the serum-free culture medium (200 μL) were added to the upper Boyden chambers. As a chemotactic factor, serum-free NIH3T3-conditioned culture medium (600 μL) was added to the bottom Boyden chamber. The Boyden chambers were then cultured at 37°C with 5% CO2 for 24 h. Non-invading cells on the upper surface were removed, and the cells on the lower surface were fixed with 4% paraformaldehyde for 30 min, stained with hematoxylin and eosin (H&E) and counted in four quadrants divided by horizontal and vertical lines under an inverted microscope. The assay was repeated thrice. Meanwhile, cell migration assays were performed as described above, except without the Matrigel covering on the PVPF membranes. All data are expressed as mean ± SE.
Tumor xenografts in nude mice
BALB/c-nu/nu nude mice were obtained from the National Resource Center for Rodent Laboratory Animal of China. In each group, five mice were injected subcutaneously with 5.0×106 cells, and five mice were injected through the tail vein with the same number of cells. All mice were kept under observation in a sterile environment, and the growth of the tumor was measured by two people at different time periods. After eight weeks, the mice were sacrificed and dissected, and lung tissues were collected and processed into paraffin-embedded sections and stained with H&E. These animal experiments were conducted in compliance with National Institutes of Health (NIH) guidelines (NIH Pub. No 85-23, revised 2010) and approved by the Institutional Animal Care and Use Committee of Shandong University.
Statistical analyses
We analyzed IHC data using the χ2 test. We used a two-tailed t-test to compare the mean values between the two groups and a one-way analysis of variance to compare the mean values among three groups. Statistical Package for the Social Sciences (SPSS) software version 13.0 (SPSS, Inc., Chicago, IL, USA) was used to analyze all data. P<0.05 (two-sided) was considered statistically significant.
Results
HMGB1 expressions in human endometrial tissues
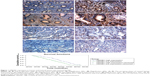
As shown in Figure 1, high expression of HMGB1 protein was detected in normal endometrial stroma and the endometrial glandular epithelial cell nucleus (Figure 1A and B). However, in most endometrial cancer tissues, the immunoreactivity of HMGB1 was very weak. Compared to the highly differentiated endometrial cancer tissues (Figure 1C and D), the expression of HMGB1 was detected at very low levels in the poorly differentiated endometrial cancer tissues (Figure 1E and F) and was mainly found in the endometrial cancer cell nuclei, not the stroma. Furthermore, HMGB1 expression was negatively correlated with advanced stage and lymph node metastasis in endometrial cancer (Table 1). To assess the prognostic value of HMGB1 in human endometrial cancer, survival analysis was performed by the Kaplan–Meier method. The result showed that patients with low HMGB1 expression had a much worse prognosis than those with high HMGB1 expression (log rank, P<0.05; Figure 1G). The expression of HMGB1 was negatively correlated with the prognosis of patients with endometrial cancer.
  | Table 1 HMGB1 protein expressions in human endometrial tissues |
Acquisition of endometrial epithelial cells and mesenchymal cells
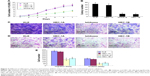
The endometrial glandular epithelial cells were observed to grow into spiral-shaped, monolayer cell colonies (Figure 2A). In the cytoplasm of glandular epithelial cells, cytokeratin (epithelial marker) protein staining was positive by ICC (Figure 2B). However, endometrial mesenchymal cells were mostly spindle-shaped (Figure 2C), and vimentin (mesenchymal marker) staining was positive in the cytoplasm of mesenchymal cells (Figure 2D). After passage, endometrial glandular epithelial cells died, but endometrial mesenchymal cells survived and could be passaged for about five generations.
Different proliferation and invasion abilities of endometrial cancer cell lines
Endometrial cancer cell lines such as Ishikawa and HEC-1B had significantly lower proliferation abilities compared with KLE and HEC-1A cells (Figure 3A and B). Furthermore, in the cell migration and invasion assays (Figure 3C and D), Ishikawa and HEC-1B cells were detected to have weaker migration and invasion abilities. The average migrating and invading cell counts of Ishikawa and HEC-1B cells were much lower than those of KLE and HEC-1A cells (Figure 3E).
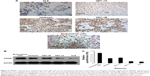
Different expressions of HMGB1 in human endometrial cell lines
As shown in Figure 4, in normal endometrial cells, the expression of HMGB1 was very strong. However, endometrial cancer cell lines such as KLE and HEC-1A with higher proliferation and invasion abilities had lower expression of HMGB1. Conversely, endometrial cancer cell lines such as Ishikawa and HEC-1B, which had lower proliferation and invasion abilities, had higher expression of HMGB1. These results indicated that the expression of HMGB1 was inversely related to the proliferation and invasion abilities of endometrial cancer cells.
Downregulation of HMGB1 expression by RNA interference
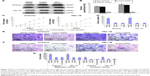
In order to study the role of HMGB1 in endometrial cancer cell proliferation and invasion, the expression of HMGB1 was disrupted in the less invasive endometrial cancer cell lines such as Ishikawa and HEC-1B by RNA interference. After lentivirus transfection, downregulation of HMGB1 expression was confirmed by real-time qRT-PCR and Western blotting for both mRNA and protein levels, which indicated high efficiency in the transfection experiments (Figure 5A and B).
Effects of HMGB1 knockdown on endometrial cancer cell proliferation and invasion abilities
The cell proliferation abilities of the less invasive endometrial cancer cell lines such as Ishikawa and HEC-1B were significantly increased by HMGB1 knockdown (Figure 5C). Accordingly, in the soft agar colony formation assay, the colony-forming ability of HMGB1 shRNA-infected cells was increased by HMGB1 knockdown (Figure 5D). In the cell migration and invasion assays, HMGB1 knockdown promoted the invasion and migration abilities of the less invasive endometrial cancer cell lines, such as Ishikawa and HEC-1B (Figure 5E and F). The average migrating and invading cell counts of HMGB1 shRNA-infected cells were much higher than those of non-infected and negative control cells (Figure 5G). No significant differences were found between non-infected group and negative control group.
Effect of HMGB1 knockdown on the tumor growth in nude mice
HMGB1 shRNA-infected Ishikawa cells and negative control shRNA-infected Ishikawa cells were injected subcutaneously or through the tail veins into five nude mice. The tumor formation rate of negative control cells was only 60%, with very slow tumor growth. However, 100% tumor formation rate was observed in HMGB1 shRNA-infected cells, with relatively rapid tumor growth. Moreover, the average tumor volume of the negative control was much smaller than that of HMGB1 shRNA-infected cells (44.75±8.75 mm3 vs 505.49±34.64 mm3, P<0.01) (Figure 6A and B). The negative control group had no lung metastasis, while the lung metastasis rate of HMGB1 shRNA-infected cells was 100% (Figure 6C). In conclusion, HMGB1 knockdown could promote tumor formation, growth and metastasis in vivo.
Effects of HMGB1 knockdown on EMT genes
It is well known that epithelial-to-mesenchymal transition (EMT) is closely related to cancer cell invasion and metastasis. We examined the effects of HMGB1 knockdown on several key EMT genes, including E-cadherin, N-cadherin, vimentin, Snail and Twist. The results showed that HMGB1 knockdown significantly increased the expression of N-cadherin, vimentin, Snail and Twist but decreased the expression of E-cadherin. Collectively, HMGB1 knockdown could play a role in endometrial cancer cell invasion and metastasis by promoting the process of EMT (Figure 7).
Discussion
In our study, we initially detected that HMGB1 expression was negatively correlated with the prognosis of patients with endometrial cancer, and HMGB1 knockdown could promote endometrial cancer cell proliferation, invasion and metastasis by inducing the process of EMT.
We found that the expression of HMGB1 in endometrial cancer tissues and cells was much lower than that in normal endometrial tissues and cells. Moreover, HMGB1 was negatively related to the clinical and pathological characteristics of endometrial cancer. In the current study, the elevated expression levels of HMGB1 in both tissues and sera of patients are more commonly found in solid tumors, including hepatocellular,23 ovarian,24 colorectal25 and non-small cell lung cancers.26 The overexpression of HMGB1 was positively correlated with the prognosis of patients with various types of cancer.27 In melanoma tumor cells,19 knockdown of HMGB1 expression increased apoptosis and reduced both invasion and levels of p-NF-κB. Conversely, exogenous HMGB1 significantly decreased apoptosis and increased both invasion and levels of p-NF-κB. Knockdown of HMGB1 in human lung cancer A549 cells significantly increased cell β-actin polymerization, cell skeleton formation, cancer cell migration and invasion in vitro, as well as metastasis in vivo.20 In human cutaneous squamous cell carcinoma, administration of HMGB1 to SCC13 cells caused an increase in cell migration in a time- and dose-dependent manner.28 However, in different types of breast cancers, HMGB1 exhibits a dual role. HMGB1 silencing inhibited invasion and migration and promoted apoptosis in the human breast cancer cell line MCF-7.29 However, some research found that HMGB1 could interact with tumor suppressor genes such as p53, p73 and RB and suppressed tumorigenesis.30,31 In endometrial cancer, our results revealed that HMGB1 was a negative regulator in the tumor development.
EMT promotes the malignant epithelial cells to migrate from the primary site and invade the vascular system.32–34 We therefore investigated whether HMGB1 could prevent the progression of endometrial cancer by inhibiting EMT. Our related results indicate that this may be the case. Knockdown of HMGB1 decreased the expression of E-cadherin (epithelial marker) and increased the expression of N-cadherin and vimentin (mesenchymal markers). The transcription factors such as Snail and Twist, which could inhibit the expression of E-cadherin and induce EMT, were both upregulated when HMGB1 was knocked down. These data indicated that HMGB1 knockdown could promote the process of EMT. However, in intrahepatic cholangiocarcinoma, HMGB1 knockdown inhibited EMT by increasing the expression of E-cadherin and decreasing the expression of N-cadherin, vimentin and Snail in RBE cells.35 Similar results were also found in colorectal carcinoma, where HMGB1 was found to act as a promoter for EMT through the RAGE/Snail/NF-κB-signaling pathways.36 Collectively, HMGB1 could play a role in the process of EMT, but the mechanism of action, promotion or inhibition may be dependent on the type of cancer.
In conclusion, HMGB1 was negatively correlated with the progression of endometrial cancer. HMGB1 knockdown could promote endometrial cancer cell invasion and metastasis by inducing EMT. We believe that this study on HMGB1 may contribute to early diagnosis and treatment of endometrial cancer.
Acknowledgments
The study was supported by funding from Natural Science Foundation of Shandong Province (2014ZRE27067). The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Disclosure
The authors report no conflicts of interest in this work.
References
Jin C, Liang R. miR-205 promotes epithelial-mesenchymal transition by targeting AKT signaling in endometrial cancer cells. J Obstet Gynaecol Res. 2015;41(10):1653–1660. | ||
Kito M, Motoyama S, Fujita K, et al. CRP 1846C>T genetic polymorphism is associated with lymph node metastasis and/or severe lymphatic invasion in endometrial cancer. Tohoku J Exp Med. 2015;237(1):25–30. | ||
Bakkum-Gamez JN, Mariani A, Dowdy SC, et al. Efficacy of contemporary chemotherapy in stage IIIC endometrial cancer: a histologic dichotomy. Gynecol Oncol. 2014;132(3):578–584. | ||
Goodwin GH, Johns EW. Isolation and characterization of two calf-thymus chromatin non-histone proteins with high contents of acidic and basic amino acids. Eur J Biochem. 1973;40:215–219. | ||
Bianchi ME. HMGB1 loves company. J Leukoc Biol. 2009;86(3):573–576. | ||
Leclerc E, Sturchler E, Vetter SW, Heizmann CW. Crosstalk between calcium, amyloid beta and the receptor for advanced glycation endproducts in Alzheimer’s disease. Rev Neurosci. 2009;20(2):95–110. | ||
Park S, Yoon SJ, Tae HJ, Shim CY. RAGE and cardiovascular disease. Front Biosci (Landmark Ed). 2011;16:486–497. | ||
Ostberg T, Kawane K, Nagata S, et al. Protective targeting of high mobility group box chromosomal protein 1 in a spontaneous arthritis model. Arthritis Rheum. 2010;62(10):2963–2972. | ||
Evankovich J, Cho SW, Zhang R, et al. High mobility group box 1 release from hepatocytes during ischemia and reperfusion injury is mediated by decreased histone deacetylase activity. J Biol Chem. 2010;285(51):39888–39897. | ||
Tang D, Kang R, Cao L, et al. A pilot study to detect high mobility group box 1 and heat shock protein 72 in cerebrospinal fluid of pediatric patients with meningitis. Crit Care Med. 2008;36(1):291–295. | ||
Huang W, Tang Y, Li L. HMGB1, a potent proinflammatory cytokine in sepsis. Cytokine. 2010;51(2):119–126. | ||
Yang H, Tracey KJ. Targeting HMGB1 in inflammation. Biochim Biophys Acta. 2010;1799:149–156. | ||
Ellerman JE, Brown CK, de Vera M, et al. Masquerader: high mobility group box-1 and cancer. Clin Cancer Res. 2007;13(10):2836–2848. | ||
Tang D, Kang R, Zeh HJ 3rd, Lotze MT. High-mobility group box 1 and cancer. Biochim Biophys Acta. 2010;1799(1–2):131–140. | ||
Campana L, Bosurgi L, Rovere-Querini P. HMGB1: a two-headed signal regulating tumor progression and immunity. Curr Opin Immunol. 2008;20(5):518–523. | ||
McKinney K, Prives C. Efficient specific DNA binding by p53 requires both its central and C-terminal domains as revealed by studies with high-mobility group 1 protein. Mol Cell Biol. 2002;22(19):6797–6808. | ||
Jiao Y, Wang HC, Fan SJ. Growth suppression and radiosensitivity increase by HMGB1 in breast cancer. Acta Pharmacol Sin. 2007;28(12):1957–1967. | ||
Yang S, Xu L, Yang T, Wang F. High-mobility group box-1 and its role in angiogenesis. J Leukoc Biol. 2014;95(4):563–574. | ||
Xie B, Cao K, Li J, et al. Hmgb1 inhibits Klotho expression and malignant phenotype in melanoma cells by activating NF-κB. Oncotarget. Epub 2016 Oct 13. | ||
Zuo Z, Che X, Wang Y, et al. High mobility group Box-1 inhibits cancer cell motility and metastasis by suppressing activation of transcription factor CREB and nWASP expression. Oncotarget. 2014;5(17):7458–7470. | ||
Jividen K, Movassagh MJ, Jazaeri A, Li H. Two methods for establishing primary human endometrial stromal cells from hysterectomy specimens. J Vis Exp. 2014;(87):e51513. | ||
Albini A, Noonan DM. The ‘chemoinvasion’ assay, 25 years and still going strong: the use of reconstituted basement membranes to study cell invasion and angiogenesis. Curr Opin Cell Biol. 2010;22(5):677–689. | ||
Zhang L, Han J, Wu H, et al. The association of HMGB1 expression with clinicopathological significance and prognosis in hepatocellular carcinoma: a meta-analysis and literature review. PLoS One. 2014;9(10):e110626. | ||
Wang H, Li Z, Sun Y, et al. Relationship between high-mobility group box 1 overexpression in ovarian cancer tissue and serum: a meta-analysis. Onco Targets Ther. 2015;8:3523–3531. | ||
Zhang X, Yu J, Li M, Zhu H, Sun X, Kong L. The association of HMGB1 expression with clinicopathological significance and prognosis in Asian patients with colorectal carcinoma: a meta-analysis and literature review. Onco Targets Ther. 2016;9:4901–4911. | ||
Feng A, Tu Z, Yin B. The effect of HMGB1 on the clinicopathological and prognostic features of non-small cell lung cancer. Oncotarget. 2016;7(15):20507–20519. | ||
Wu T, Zhang W, Yang G, et al. HMGB1 overexpression as a prognostic factor for survival in cancer: a meta-analysis and systematic review. Oncotarget. Epub 2016 Jul 6. | ||
Sun Y, Tu Y, He LI, Ji C, Cheng BO. High mobility group box 1 regulates tumor metastasis in cutaneous squamous cell carcinoma via the PI3K/AKT and MAPK signaling pathways. Oncol Lett. 2016;11(1):59–62. | ||
Ni P, Zhang Y, Liu Y, et al. HMGB1 silence could promote MCF-7 cell apoptosis and inhibit invasion and metastasis. Int J Clin Exp Pathol. 2015;8(12):15940–15946. | ||
Exner R, Sachet M, Arnold T, et al. Prognostic value of HMGB1 in early breast cancer patients under neoadjuvant chemotherapy. Cancer Med. Epub 2016 Jul 25. | ||
Sohun M, Shen H. The implication and potential applications of high-mobility group box 1 protein in breast cancer. Ann Transl Med. 2016;4(11):217. | ||
Steinestel K, Eder S, Schrader AJ, Steinestel J. Clinical significance of epithelial–mesenchymal transition. Clin Transl Med. 2014;3:17. | ||
Wendt MK, Balanis N, Carlin CR, Schiemann WP. STAT3 and epithelial–mesenchymal transitions in carcinomas. JAKSTAT. 2014;3(1):e28975. | ||
Singh A, Settleman J. EMT, cancer stem cells and drug resistance: an emerging axis of evil in the war on cancer. Oncogene. 2010;29(34):4741–4751. | ||
Xu YF, Ge FJ, Han B, et al. High-mobility group box 1 expression and lymph node metastasis in intrahepatic cholangiocarcinoma. World J Gastroenterol. 2015;21(11):3256–3265. | ||
Zhu L, Li X, Chen Y, Fang J, Ge Z. High-mobility group box1: a novel inducer of the epithelial–mesenchymal transition in colorectal carcinoma. Cancer Lett. 2015;357(2):527–534. |
 © 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.